Abstract
There are two major pathways that mammalian cells use to supply themselves with cholesterol, one involving the synthesis of sterols from acetyl-CoA and the other the metabolism of cholesterol-rich lipoprotein particles via receptor-mediated endocytosis. There also are several pathways that mammalian cells use to break down cholesterol, and these disposal pathways are equal in physiological importance to the supply pathways. A major catabolic route involves conversion of cholesterol into conjugated bile salts, a transformation mediated by 16 or more liver enzymes. This review highlights findings in cholesterol catabolism from the last five decades with special emphasis on advances in bile acid synthesis, transport, and regulation.
Keywords: cholesterol, catabolism, liver, transporters, nuclear receptors
The first paper on bile acids published in the Journal of Lipid Research appeared in volume 1, issue 5. The year was 1960, a time when the “lipid hypothesis” of atherosclerosis was anathema to most and searches for cholesterol lowering drugs almost nonexistent (1). Frank J. Wolf, a scientist at Merck who would gain subsequent fame as the discoverer of streptavidin, was the senior author of this study, which documented the ability of oral bile acid binding resins to increase fecal excretion of bile acids and in so doing to lower plasma cholesterol levels. These findings led the way to the development of bile acid binding resins such as cholestyramine and colesevelam, which are still in use today for the treatment of hypercholesterolemia. The Wolf paper was followed by many more on bile acids in the Journal, which continues to publish findings from high caliber research in the field.
Prior to Wolf's contribution, studies dating back to the 1920s by Adolf Windaus, Heinrich Wieland, Rudolf Schoenheimer, Konrad Bloch, Sune Bergstrom, Bengt Samuelsson, and other giants of lipid research defined the chemistry and physiology of bile acids. These polar derivatives of cholesterol were shown to be synthesized in the liver; secreted into the bile; and in smaller amounts, to be excreted from the body in the stool. Bile acids facilitated the solubilization of essential dietary nutrients such as fat-soluble vitamins in the gut, and in the liver, they were required for the solubilization and excretion of lipophilic metabolites such as bilirubins. With this large body of work as a background, biochemists and molecular biologists entered the fields of bile acid synthesis and metabolism in the 1980s. What have we learned from their efforts?
MULTIPLE CHOLESTEROL CATABOLISM PATHWAYS
We now know that cells utilize several enzymic mechanisms to clear excess cholesterol. The major pathway, accounting for approximately 90% of cholesterol breakdown, is the conversion of cholesterol into bile acids by enzymes in the liver. More minor routes of disposal include the conversion of cholesterol into steroid hormones by endocrine tissues (2); the synthesis of oxysterols by the lung (3) and brain (4); and the chemical and enzymatic conversion of an intermediate in the cholesterol biosynthetic pathway (7-dehydrocholesterol) into activated vitamin D3 by the skin, liver, and kidney (5). The latter three pathways account for less than 10% of whole-body cholesterol catabolism, but the end products that arise from them have biological roles that greatly exceed their masses.
ENZYMES OF BILE ACID SYNTHESIS
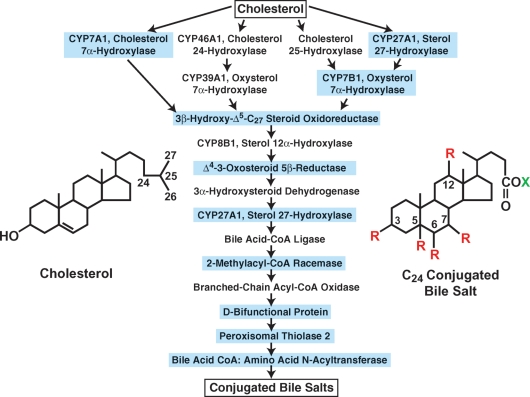
In the mid 1980s, textbooks outlined a single pathway by which cholesterol was converted into bile acids (6). Conversion was known to involve multiple hydroxylations of the ring structures and the oxidation and shortening of the side chain of cholesterol to produce a C24 bile acid (Fig. 1); but the precise number of steps, the identities of the enzymes involved, and the mechanisms by which the pathway was regulated were largely unknown at the time. This situation began to change when Kjell Wikvall reported the purification of two enzymes in the pathway, sterol 27-hydroxylase and 3β-hydroxy-Δ5-C27-steroid oxidoreductase (7, 8). His pioneering biochemical efforts led directly to the first cDNA cloning of a bile acid biosynthetic enzyme (the sterol 27-hydroxylase) (9), and the start of the “molecular era” of bile acid metabolism in which many of the early mysteries concerning the pathway and its regulation were solved.
Fig. 1.
Enzymes of bile acid synthesis. The 16 enzymes and the 17 reactions they catalyze in converting cholesterol into a conjugated bile salt are shown. Mutations causing human disease have been identified in the nine enzymes that are boxed in blue (adapted from Ref. 33). The structures of cholesterol and a generic conjugated bile salt are shown. R = OH or H; X = glycine or taurine.
Wikvall's purification successes inspired the subsequent isolation of the enzyme catalyzing the initiating and rate-limiting step in bile acid synthesis, cholesterol 7α-hydroxylase (CYP7A1). If there is a near-universal chemical signature of the bile acid, it is the presence of a hydroxyl group on carbon 7 of the B-ring of the molecule in either the α-stereochemical configuration (rodents, primates, other species) or the β-configuration (bears) (Fig. 1). The rodent enzyme that 7α-hydroxylates cholesterol to initiate bile acid synthesis was known to be unstable, membrane-bound, and present in only trace amounts in the liver, but by 1989/1990, advances in membrane protein purification and scale-up allowed three laboratories to exploit earlier observations of Wikvall et al. (10) to isolate the protein in near homogeneous form and to clone the encoding cDNA (as reviewed in Ref. 11). With the cDNA and shortly thereafter, the cholesterol 7α-hydroxylase gene in hand, bile acids were shown to repress and cholesterol was shown to induce the enzyme at the transcriptional level in rats. These findings laid the foundation for future work that elucidated the molecular mechanisms by which bile acid synthesis is controlled (see later discussion).
Biochemical studies and experiments with cholesterol 7α-hydroxylase knockout mice revealed that there must be alternate pathways by which bile acids are synthesized, and subsequently two oxysterol 7α-hydroxylases were isolated that utilized side-chain oxysterols rather than cholesterol as substrates for bile acid synthesis (as reviewed in Ref. 12). The oxysterol substrates of these enzymes differed from cholesterol by the presence of an additional hydroxyl group on carbons 24, 25, or 27 of the side chain of the molecule (Fig. 1). The identification of the oxysterol 7α-hydroxylases showed that cholesterol catabolism was initiated by two mechanisms: 1) direct 7α-hydroxylation of cholesterol (the classic pathway), and 2) an initial hydroxylation on the side chain of the molecule followed by 7α-hydroxylation (the alternate pathway).
The enzymes that synthesize side-chain oxysterols and those that act on 7α-hydroxylated cholesterol and 7α-hydroxylated oxysterols were isolated in due time and their roles in cholesterol breakdown defined (12). This international effort led to the annotation of the bile acid synthesis pathways as we know them today, which comprise at least 16 enzymes that catalyze as many as 17 reactions to produce a conjugated bile salt (Fig. 1). The physiochemical transformation brought about by these enzymes is impressive; their actions convert an almost insoluble cholesterol molecule (Ksp ∼4 μg/l) into an exceedingly soluble conjugated bile salt (Ksp ∼670 g/l) that is readily excreted from the liver into the bile. As of this writing, inherited mutations in genes encoding nine of these enzymes have been identified in subjects presenting with symptoms ranging from hypercholesterolemia to liver failure to progressive neuropathy (13), which underscores the physiological importance of cholesterol catabolism and of bile acids themselves.
ONE ENZYME, MULTIPLE FUNCTIONS
With the elucidation of the bile acid pathways has come the realization that 12 of the 16 participating enzymes play multiple roles in intermediary metabolism (Table 1). For example, the three enzymes that initiate the alternate pathway of bile acid synthesis produce oxysterols, which also are ligands for nuclear hormone receptors (14), facilitators of sterol excretion from the lung and brain (3, 4), and potent regulators of cholesterol synthesis (15). Another trio of enzymes act on both intermediates in the bile acid pathway and steroid hormones, serving for the most part to inactivate the latter and thus to terminate signaling through classic steroid hormone receptors. Five bile acid enzymes metabolize very long chain fatty acids (16), such as dietary pristanic acid, and gene mutations that impair these enzymes cause defects in hepatic bile acid synthesis and the accumulation of very long chain fatty acids leading to neurological dysfunction (as reviewed in Ref. 12).
TABLE 1.
Multiple metabolic functions of bile acid biosynthetic enzymesa
| Function | Enzyme |
|---|---|
| Oxysterol biosynthesis | Cholesterol 24-hydroxylase |
| Cholesterol 25-hydroxylase | |
| Sterol 27-hydroxylase |
|
| Steroid metabolism | Oxysterol 7α-hydroxylase (CYP7B1) |
| Steroid 5β-reductase | |
| 3α-Hydroxysteroid dehydrogenase |
|
| Very long chain fatty acid metabolism | Bile acid-CoA ligase |
| 2-Methylacyl-CoA racemase | |
| Branched-chain acyl-CoA oxidase | |
| D-Bifunctional protein | |
| Peroxisomal thiolase 2 |
|
| Vitamin D biosynthesis | Sterol 27-hydroxylase |
Adapted from Ref. 33.
One of the 16 bile acid enzymes, the sterol 27-hydroxylase, may be trifunctional. This mitochondrial cytochrome P450 is involved in the synthesis of oxysterols, bile acids, and vitamin D (Table 1). The initial step in the synthesis of vitamin D takes place in the skin and involves the UV light-mediated cleavage of 7-dehydrocholesterol to produce previtamin D, followed by the spontaneous rearrangement of this compound into the secosteroid vitamin D3. The newly generated vitamin D3 is transported to the liver where it is subject to 25-hydroxylation in a reaction that is catalyzed by sterol 27-hydroxylase or by a microsomal vitamin D 25-hydroxylase (17). The final step to generate a high affinity ligand for the vitamin D receptor involves 1α-hydroxylation in the kidney and is accomplished by a second mitochondrial P450, the CYP27B1 enzyme. Several lines of evidence support a role for sterol 27-hydroxylase in the synthesis of bile acids and oxysterols, but the enzyme's proposed role in vitamin D synthesis is less well established and arises largely from in vitro biochemical studies. Whereas inherited mutations in microsomal vitamin D 25-hydroxylase cause vitamin D deficiency in humans (18), mutations in sterol 27-hydroxylase disrupt bile acid and oxysterol synthesis, but do not appear to affect vitamin D action (13).
BILE ACID TRANSPORTERS
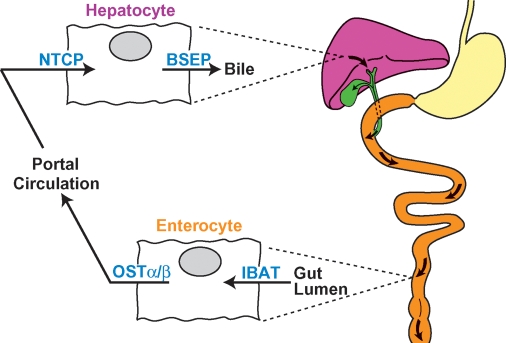
Following synthesis in the liver and secretion into the bile, conjugated primary bile salts travel an enterohepatic itinerary in which they are stored in the gallbladder, released into the duodenum of the small intestine in response to gut hormones, and thereafter are progressively moved along the small intestine via peristalsis (Fig. 2). Upon reaching the ileum, bile acids are taken up by specific transport proteins, moved across the enterocyte, and transferred into the portal vein for return to the liver. The cycle is completed by cell surface proteins on hepatocytes that transport bile acids from the portal circulation into the liver from which they are once more secreted into the bile to travel again through the enterohepatic circulation. A conjugated bile salt may travel this route 4 to 12 times a day in the human (19).
Fig. 2.
The enterohepatic circulation and bile acid transporters. Conjugated bile salts are moved through the enterohepatic circulation by transporters located on the apical and basolateral surfaces of hepatocytes and enterocytes. Redrawn from Ref. 19.
The last two decades have witnessed the isolation of the major transport proteins that shuttle bile acids between the liver and gut (as reviewed in Refs. 20, 21). Secretion from the liver into bile is accomplished by the bile salt export protein (BSEP), a member of the large ATP-binding cassette transporter family (ABCB11). Mutations in the encoding human and mouse genes cause progressive and nonprogressive forms of liver disease, respectively. A majority of bile acids are imported into the liver from the portal circulation by the sodium taurocholate cotransporting protein (NTCP, SLC10A1).
The ileal bile acid transporter (IBAT, SLC10A2) moves bile acids from the gut lumen into the enterocyte, and loss of this protein in humans and mice has profound consequences for bile acid metabolism. The organic solute transporter (OST)α/β (22), a heterodimeric protein, exports bile acids into the portal circulation, and mutation of the gene specifying OSTα in mice causes accumulation of bile acids in the enterocyte (23). The BSEP, NTCP, IBAT, and OSTα/β transporters account for a majority of bile acid movement within the enterohepatic circulation; however, when these proteins or their resident tissues are impaired, compensatory mechanisms involving other transporters are induced in an effort to maintain bile acid transport (20).
REGULATION OF BILE ACID SYNTHESIS
Work in the late 1960s showed that cholesterol 7α-hydroxylase catalyzed the rate-limiting step in bile acid synthesis and that the activity of this enzyme was suppressed by dietary bile acids (as reviewed in Ref. 11). Additional findings showed that cholesterol feeding induced the activity of the enzyme in rats. As previously noted, experiments with the cloned cholesterol 7α-hydroxylase gene indicated that bile acids and cholesterol acted at the transcriptional level, which suggested DNA sequences in the gene were mediating these feed-back and feed-forward responses. Multiple transcription factors that recognized these regulatory elements were subsequently isolated in the 1990s and shown to be members of the nuclear receptor gene family (as reviewed in Ref. 12).
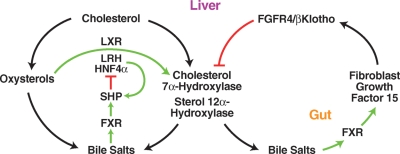
Induction of the gene by cholesterol is mediated by the liver X receptor (LXR), the liver receptor homolog (LRH), and hepatocyte nuclear factor 4α (HNF4α) (Fig. 3). In response to the binding of oxysterols, LXR interacts with a DNA sequence that is present in the 5′-flanking regions of the rat and mouse cholesterol 7α-hydroxylase genes and in turn recruits LRH or HNF4α to the promoter to activate transcription. The LXR-responsive DNA sequence is not present in the cholesterol 7α-hydroxylase genes of other species, including humans. Repression of cholesterol 7α-hydroxylase by bile acids occurs through two mechanisms. In one, the binding of excess bile acids by the nuclear receptor farnesoid X receptor (FXR) leads to the expression of the short heterodimer partner (SHP), an atypical nuclear receptor lacking a DNA binding domain, and SHP subsequently binds to and antagonizes the positive actions of LRH and HNF4α on the cholesterol 7α-hydroxylase promoter. The roles of LXR, LRH, HNF4α, FXR, and SHP in this regulatory scheme were confirmed in studies of knockout mice lacking these receptors, and several of these transcription factors were shown to similarly regulate the sterol 12α-hydroxylase gene in the pathway (12).
Fig. 3.
Regulation of bile acid synthesis. Intermediates and endproducts of the bile acid pathway regulate the expression of two genes in the liver (cholesterol 7α-hydroxylase and sterol 12α-hydroxylase) that synthesize conjugated bile salts through intricate regulatory schemes involving nuclear receptors [farnesoid X receptor (FXR), short heterodimer partner (SHP), liver X receptor (LXR), liver receptor homolog (LRH), hepatocyte nuclear factor 4α (HNF4α)], polypeptide hormones (FGF15) produced in the gut, and hepatic cell surface receptors [fibroblast growth factor 4 receptor (FGFR4)/βKlotho]. See text for details.
Unexpectedly, removing components of the negative feedback pathway such as SHP or FXR decreased but did not eliminate the ability of bile acids to repress their synthesis in the liver, suggesting that a second negative regulatory pathway existed. The first clue to the components of this system came from the finding of unregulated bile acid synthesis in mice lacking the fibroblast growth factor 4 receptor (FGFR4) (24). The second clue came when a similar phenotype of unregulated bile acid synthesis was reported in a line of mice lacking a protein termed βKlotho (25). The final two clues came with the identification of FGF15 as an FXR-responsive endocrine hormone secreted by enterocytes of the small intestine in response to excess bile acids (26), and the ability of this protein to repress additional synthesis through binding to a heterodimeric receptor composed of FGFR4 and βKlotho expressed on hepatocytes (27, 28). Subsequently, FGF15 together with FGFR4/βKlotho were shown to regulate gallbladder filling (29). The second regulatory pathway thus provides a mechanism by which the gut can sense the bile acid pool and signal appropriately to the liver to regulate bile acid synthesis and the flow of bile through the enterohepatic circulation.
BILE ACIDS AS G PROTEIN-COUPLED RECEPTOR LIGANDS
As of this writing, the newest finding in bile acid synthesis and metabolism concerns the ability of bile acids to act as ligands for a G protein-coupled receptor termed M-BAR/TG5, and in so doing, regulate diverse physiological processes ranging from thyroid hormone action to energy homeostasis. This story began with the identification of M-BAR/TGR5 as a cell surface receptor for bile acids (30, 31), and continued with the findings that the in vivo interaction of a bile acid with this receptor on brown fat cells led to increased production of thyroid hormone and a subsequent enhancement of metabolic rate (32). These results suggest endocrine roles for bile acids in metabolism, and they expand the purview of what were once thought to be little more than detergents and cholesterol breakdown products.
PERSPECTIVES
The answer to the question, what does the future hold for bile acids? is “a lot!” We can look forward to gazing at the three-dimensional structures of the enzymes, transporters, and receptors of bile acid synthesis and metabolism. Additional insights into the roles of minor cholesterol catabolism pathways in the lung and brain will be forthcoming, as will knowledge regarding new cholesterol turnover pathways. And who knows, perhaps a bile acid will be found to regulate food intake via actions in the central nervous system. The prospects for discovery are bright.
Acknowledgments
We thank colleagues who have inspired, stimulated, and facilitated our work on bile acids over the last 20 years and apologize to those whose papers were not cited due to space limitations. Jay Horton and Jan Sjovall provided critical comments on this manuscript.
Research in the author's laboratory is supported by National Institutes of Health Grants P01 HL-20948 and PL1 DK-81182, and by grants from the Robert A. Welch Foundation (I-0971) and the Perot Family Fund.
Published, JLR Papers in Press, September 24, 2008.
References
- 1.Steinberg, D. 2007. The Cholesterol Wars. Elsevier, Amsterdam.
- 2.Orth, D. N., and W. J. Kovacs. 1998. The adrenal cortex. In Williams Textbook of Endocrinology. J. D. Wilson, D. F. Foster, H. M. Kronenberg, and P. R. Larsen, editors. W.B. Saunders, Philadelphia. 517–664.
- 3.Babiker A., O. Andersson, D. Lindblom, J. van der Linden, B. Wiklund, D. Lutjohann, U. Diczfalusy, and I. Bjorkhem. 1999. Elimination of cholesterol as cholestenoic acid in human lung by sterol 27-hydroxylase: evidence that most of this steroid in the circulation is of pulmonary origin. J. Lipid Res. 40 1417–1425. [PubMed] [Google Scholar]
- 4.Bjorkhem I., D. Lutjohann, U. Diczfalusy, L. Stahle, G. Ahlborg, and J. Wahren. 1998. Cholesterol homeostasis in human brain: turnover of 24S-hydroxycholesterol and evidence for a cerebral origin of most of this oxysterol in the circulation. J. Lipid Res. 39 1594–1600. [PubMed] [Google Scholar]
- 5.Prosser D. E., and G. Jones. 2004. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem. Sci. 29 664–673. [DOI] [PubMed] [Google Scholar]
- 6.Bjorkhem, I. 1985. Mechanism of bile acid biosynthesis in mammalian liver. In Sterols and Bile Acids. H. Danielsson and J. Sjovall, editors. Elsevier, Amsterdam-New York-Oxford. 231–278.
- 7.Wikvall K. 1984. Hydroxylations in biosynthesis of bile acids: isolation of a cytochrome P-450 from rabbit liver mitochondria catalyzing 26-hydroxylation of C27-steroids. J. Biol. Chem. 259 3800–3804. [PubMed] [Google Scholar]
- 8.Wikvall K. 1981. Purification and properties of a 3β-hydroxy-Δ5-C27-steroid oxidoreductase from rabbit liver microsomes. J. Biol. Chem. 256 3376–3380. [PubMed] [Google Scholar]
- 9.Andersson S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264 8222–8229. [PubMed] [Google Scholar]
- 10.Andersson S., H. Bostrom, H. Danielsson, and K. Wikvall. 1985. Purification from rabbit and rat liver of cytochromes P-450 involved in bile acid biosynthesis. Methods Enzymol. 111 364–377. [DOI] [PubMed] [Google Scholar]
- 11.Russell D. W., and K. D. R. Setchell. 1992. Bile acid biosynthesis. Biochemistry. 31 4737–4749. [DOI] [PubMed] [Google Scholar]
- 12.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72 137–174. [DOI] [PubMed] [Google Scholar]
- 13.Bjorkhem, I., K. M. Boberg, and E. Leitersdorf. 2001. Inborn errors in bile acid biosynthesis and storage of sterols other than cholesterol. In The Metabolic and Molecular Bases of Inherited Disease. C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler, and B. Vogelstein, editors. McGraw-Hill, Inc., New York. 2961–2988.
- 14.Tontonoz P., and D. J. Mangelsdorf. 2003. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 17 985–993. [DOI] [PubMed] [Google Scholar]
- 15.Radhakrishnan A., Y. Ikeda, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanders, R. J. A., P. G. Barth, and H. S. A. Heymans. 2001. Single peroxisomal enzyme deficiencies. In The Metabolic & Molecular Bases of Inherited Disease. C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler, and B. Vogelstein, editors. McGraw-Hill, New York. 3219–3256.
- 17.Cheng J. B., D. L. Motola, D. J. Mangelsdorf, and D. W. Russell. 2003. De-orphanization of cytochrome P450 2R1, a microsomal vitamin D 25-hydroxylase. J. Biol. Chem. 278 38084–38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng J. B., M. A. Levine, N. H. Bell, D. J. Mangelsdorf, and D. W. Russell. 2004. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl. Acad. Sci. USA. 101 7711–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey, M. C. 1982. The enterohepatic circulation. In The Liver, Biology and Pathobiology. Raven Press, New York. 429–465.
- 20.Magnus-Pauli C., and P. J. Meier. 2006. Hepatobiliary transporters and drug-induced cholestasis. Hepatology. 44 778–787. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann A. F. 2007. Biliary secretion and excretion in health and disease: current concepts. Ann. Hepatol. 6 15–27. [PubMed] [Google Scholar]
- 22.Dawson P. A., M. Hubbert, J. Haywood, A. L. Craddock, N. Zerangue, W. V. Christian, and N. Ballatori. 2005. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J. Biol. Chem. 280 6960–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao A., J. Haywood, A. L. Craddock, M. G. Belinsky, G. D. Kruh, and P. A. Dawson. 2008. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc. Natl. Acad. Sci. USA. 105 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C., F. Wang, M. Kan, C. Jin, R. B. Jones, M. Weinstein, C. X. Deng, and W. L. McKeehan. 2000. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275 15482–15489. [DOI] [PubMed] [Google Scholar]
- 25.Ito S., T. Fujimori, A. Furuya, J. Satoh, Y. Nabeshima, and Y-I. Nabeshima. 2005. Impaired negative feedback suppression of bile acid synthesis in mice lacking βKlotho. J. Clin. Invest. 115 2202–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inagaki T., M. Choi, A. Moschetta, L. Peng, C. L. Cummins, J. G. McDonald, G. Luo, S. A. Jones, B. Goodwin, J. A. Richardson, et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2 217–225. [DOI] [PubMed] [Google Scholar]
- 27.Kurosu H., M. Choi, Y. Ogawa, A. S. Dickson, R. Goetz, A. V. Eliseenkova, M. Mohammadi, K. P. Rosenblatt, S. A. Kliewer, and M. Kuro-o. 2007. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282 26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X., H. Ge, J. Gupte, J. Weiszmann, G. Shimamoto, J. Stevens, N. Hawkins, B. Lemon, W. Shen, J. Xu, et al. 2007. Co-receptor requirements for fibroblast growth factor-19 signaling. J. Biol. Chem. 282 29069–29072. [DOI] [PubMed] [Google Scholar]
- 29.Choi M., A. Moschetta, A. L. Bookout, L. Peng, M. Umetani, S. R. Holmstrom, K. Suino-Powell, H. E. Xu, J. A. Richardson, R. D. Gerard, et al. 2006. Identification of a hormonal basis for gallbladder filling. Nat. Med. 12 1253–1255. [DOI] [PubMed] [Google Scholar]
- 30.Maruyama T., Y. Miyamoto, T. Nakamura, Y. Tamai, H. Okada, E. Sugiyama, T. Nakamura, H. Itadani, and K. Tanaka. 2002. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298 714–719. [DOI] [PubMed] [Google Scholar]
- 31.Kawamata Y., R. Fujii, M. Hosoya, M. Harada, H. Yoshida, M. Miwa, S. Fukusumi, Y. Habata, T. Itoh, Y. Shintani, et al. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278 9435–9440. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe M., S. M. Houten, C. Mataki, M. A. Christoffolete, B. W. Kim, H. Sato, N. Messaddeq, J. W. Harney, O. Ezaki, T. Kodama, et al. 2006. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 439 484–489. [DOI] [PubMed] [Google Scholar]
- 33.Russell, D. W., and J. B. Cheng. 2005. Multifunctional roles of bile acid biosynthetic enzymes and the isolation of a new vitamin D 25-hydroxylase. In Bile Acid Biology and its Therapeutic Implications. G. Paumgartner, D. Keppler, U. Leuschner, and A. Stiehl, editors. Springer, Dordrecht. 19–26.