Abstract
The human genome expresses nine patatin-like phospholipase domain containing proteins (PNPLA1–9). Members of this family share a protein domain discovered initially in patatin, the most abundant protein of the potato tuber. Patatin is a lipid hydrolase with an unusual folding topology that differs from classical lipases. Mammalian PNPLAs include lipid hydrolases with specificities for diverse substrates such as triacylglycerols, phospholipids, and retinol esters. Analysis of induced mutant mouse models and the clinical phenotype of patients with mutations revealed important insights into the physiological role of several members of the PNPLA family. This review aims to summarize current knowledge of PNPLA proteins and to document their emerging importance in lipid and energy homeostasis.
Keywords: PNPLA, ATGL, adiponutrin, GS2, GS2-like, NTE, NRE, iPLA2γ, PLA2G6
The large group of lipid hydrolases, diverse enzymes involved in the hydrolysis of ester and amide bonds of fatty acids (FA) in lipid molecules, include triacylglycerol (TG) hydrolases (lipases), phospholipases, ceraminidases as well as cholesterol ester, and retinol ester (RE) hydrolases. These enzymes are generally important to a plethora of intra- and extracellular processes, including maintenance of membrane integrity, lipid signaling, and regulation of energy homeostasis. In recent years, a mammalian family of lipid hydrolases, designated patatin-like phospholipase domain containing (PNPLA), has attracted attention, as its members were found to serve critical roles in diverse aspects of lipid metabolism and signaling. Several gain- and loss-of-function mouse models revealed important implications for PNPLA proteins in physiological processes. Moreover, the phenotypic consequences of mutations and gene polymorphisms in humans identified enzyme family members as key players in disease states linked to energy homeostasis, neuronal integrity, and age-related bone morphology.
As the name implies, PNPLA proteins are members of the patatin family (Pfam01734). The eponym of this protein family, potato patatin, harbors the evolutionarily conserved consensus serine lipase motif Gly-X-Ser-X-Gly and exhibits nonspecific acyl-hydrolase activity (1). The 3-D structure of Pat17, an isoenzyme of potato patatin, localized the active site of the enzyme to the so-called patatin-like phospholipase domain (2). The name of this domain as well as the nomenclature of the PNPLA family is, unfortunately, somewhat misleading, because several of the subsequently discovered mammalian proteins exhibit hydrolase but not phospholipase activity. The patatin-like phospholipase domain is characterized by a three-layer α/β/α architecture employing a catalytic Ser-Asp catalytic dyad instead of the classical catalytic triad (2). The observed folding topology and the 3-D arrangement of the catalytic site of patatin (oxyanion hole, catalytic Ser within a nucleophilic elbow, spatial position of the catalytic Asp) differ from those found in canonical lipase α/β hydrolase folds.
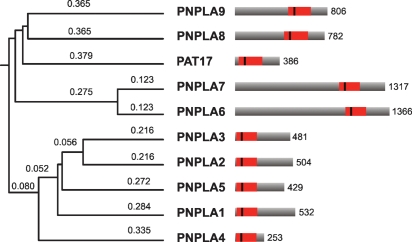
The phylogenetic relationship among mammalian PNPLA protein family members (PNPLA1–9) is displayed in Fig. 1. This review aims to summarize advances in the understanding of the physiological function of the PNPLA family on the basis of biochemical studies, the characterization of induced mutant mouse models, and the disease pattern of patients with mutations in the respective genes (summarized in Table 1).
Fig. 1.
Phylogenetic relationship and structural comparison of proteins within the PNPLA family. The patatin domain in the full-length PNPLA proteins is shown as red box, the vertical line indicates the predicted active site serine. Numbers on the right denote protein lengths in amino acids.
TABLE 1.
Phenotypes of genetic variants in murine and human PNPLAs
| Protein | Mouse Models | Human Mutations | Human Polymorphisms |
|---|---|---|---|
| PNPLA2 | Global deletion: decreased plasma FA levels, TG accumulation in multiple tissues, obesity, premature death from heart failure, defective cold adaptation, enhanced glucose and insulin tolerance (13). | Neutral lipid storage disease with myopathy, further various symptoms: cardiomyopathy, short stature, hepatomegaly, splenomegalie, diabetes, chronic pancreatitis (18, 20). | Association with decreased plasma FA and TG as well as increased fasting plasma glucose and risk for type 2 diabetes (22). |
| ATGL (7) | |||
| Desnutrin (5) | |||
| iPLA2ζ (6) | |||
| PEDF-R (9) | |||
| PLA2GVIE (47) | |||
| PNPLA3 | Association with hepatosteatosis and nonalcoholic fatty liver disease (25), liver transaminases (26), obesity, insulin secretion (27). | ||
| Adiponutrin (24) | |||
| iPLA2ɛ (6) | |||
| PLA2GVID (47) | |||
| PNPLA6 | Global deletion: embryonic lethality (30). | NTE-related motor neuron disease: distal muscle wasting, progressive spastic paraplegia (34). | |
| NTE (31) | Brain-specific deletion: neurodegeneration, vacuolation of nerve cell bodies, disruption of ER (33). | ||
| iPLA2δ (47) | |||
| PLA2GVIC (47) | |||
| PNPLA8 | Global deletion: mitochondrial dysfunction, decreased myocardial cardiolipin content, altered cardiolipin molecular species composition, multiple bioenergetic defects (39). | ||
| iPLA2γ (36) | Cardiac overexpression: decreased phospholipid mass, impaired mitochondrial function, fasting-induced TG accumulation and hemodynamic dysfunction (37). | ||
| PLA2GVIB (41) | |||
| PNPLA9 | Global deletion: reduced male fertility, decreased glucose tolerance, impaired insulin secretion (41, 44), age-related loss of bone mass (43), and neurodegeneration (45). | INAD and idiopathic NBIA (46). | |
| iPLA2β (41) | β-Cell-specific overexpression: increased glucose tolerance and insulin secretion (44). | ||
| PLA2G6 | |||
| PLA2GVIA (47) |
PNPLA1: AN ENIGMATIC MEMBER OF THE PNPLA FAMILY
Essentially, no published data exist on the structure, enzymatic activity, or physiological relevance of PNPLA1. Low mRNA levels in various human tissues have been observed (3) but not consistently (4).
PNPLA2: A KEY ENZYME IN THE HYDROLYSIS OF STORED TG
Three groups independently discovered PNPLA2 (5–7), and recent findings on this enzyme have been reviewed (8). In accordance with its predominant function, we named the enzyme adipose triglyceride lipase (ATGL) (7). ATGL localizes to cellular lipid droplets and catalyzes the initial step in TG hydrolysis, yielding diacylglycerols and FA (6, 7). Notably, ATGL also exhibits transacylase and phospholipase A (PLA)2 activity in vitro (6, 9). The significance of these activities in vivo is unclear.
ATGL mRNA is expressed in multiple tissues. The highest expression levels are found in white and brown adipose tissue (WAT and BAT, respectively) (5, 7, 10, 11). Upregulation of ATGL transcript levels is observed during adipocyte differentiation as well as upon dexamethasone treatment or fasting in mouse WAT (4, 5, 10, 12). Consistent with this nutritional regulation, ATGL mRNA levels are downregulated by insulin and, conversely, increased in WAT of mouse models with abrogated insulin action (10, 11).
The phenotype of ATGL-knockout (-ko) mice (13) confirmed in vitro indications of its critical role in TG hydrolysis in multiple tissues. Impaired TG hydrolase activity resulted in enhanced lipid deposition in WAT, BAT, and numerous nonadipose tissues of ATGL-ko mice, thereby demonstrating that ATGL loss is not compensated by hormone-sensitive lipase (HSL) or other lipases. TG accumulation was most pronounced in the heart (>20-fold) and caused severe cardiomyopathy, congestive heart failure, and the premature death of ATGL-ko animals. Impaired lipolysis in BAT of ATGL-ko mice resulted in defective cold adaptation. Despite massive lipid accumulation in multiple insulin target tissues, ATGL-ko mice shifted toward increased glucose utilization and displayed enhanced whole-body insulin sensitivity and glucose tolerance.
An activator of ATGL was discovered soon after the identification of ATGL. Comparative gene identification-58 (CGI-58), also named α/β hydrolase domain containing-5 (ABHD5), drastically enhances ATGL-mediated TG hydrolysis without affecting HSL activity (12). The molecular mechanism underlying ATGL activation is currently unknown. However, it was shown that in hormonally unstimulated cells, CGI-58/ABHD5 binds to perilipin, a major lipid droplet protein (14). Upon hormonal stimulation, protein kinase-A phosphorylates perilipin, resulting in CGI-58/ABHD5 dissociation followed by its colocalization with ATGL (15).
The recent discovery of mutations in the human genes for ATGL and CGI-58/ABHD5 brought significant insight to their function. Mutations in both genes cause neutral lipid storage disease characterized by the accumulation of TG in multiple tissues, including white blood cells (Jordans' anomaly) (16). Additional clinical phenotypes differ significantly. In contrast to patients with ATGL mutations, individuals that lack functional CGI-58/ABHD5 are affected with severe ichthyosis (17). Accordingly, the disorder was named neutral lipid storage disease with ichthyosis. On the other hand, mutations in the ATGL gene are commonly associated with severer forms of skeletal- and cardiomyopathy than mutations in CGI-58/ABHD5, and the disorder was named neutral lipid storage disease with myopathy (18). The molecular basis for these differences is currently not understood. Possibly, CGI-58/ABHD5 may exercise additional functions besides activating ATGL-mediated TG hydrolysis (19). Reported mutations in ATGL include several C-terminal truncations of the enzyme and a point mutation. Only one mutation reported so far affected the patatin domain (20). Interestingly, the truncated forms of ATGL are enzymatically highly active when artificial lipid substrates are used but fail to hydrolyze TG in lipid droplets within cells, implying that cellular lipid droplet association requires the C-terminal region of ATGL (21).
To date, a single study has associated polymorphisms in PNPLA2 with decreased plasma FA and TG levels, increased fasting glucose concentrations, and an increased risk for type 2 diabetes (22). Collectively, these results imply that ATGL is a key factor in the hydrolytic catabolism of fat stores in multiple tissues. The fact that ATGL and HSL together account for more than 95% of TG hydrolysis in murine WAT (23) suggests that these lipases are the principal enzymes for fat cell lipolysis.
PNPLA3: AN INDICATOR OF THE NUTRITIONAL STATE
PNPLA3, also named adiponutrin, was originally identified as a highly adipose-specific transcript that rapidly responds to nutritional status (24). Human adiponutrin localizes to cellular membranes of adipocytes (24). Abundance of mRNA levels increases during adipocyte differentiation or by insulin and drastically decreases during fasting (3–6, 10, 24). The biochemical function of the protein remains unclear. In vitro, human adiponutrin exhibited TG hydrolase, transacylase, and modest PLA2 activity (4, 6). However, in a cellular context, these activities appear minor. Overexpression or knockdown of adiponutrin did not affect lipolysis or the cellular TG level (4, 10). Although these data are insufficient to assign a conclusive biochemical or physiological function to adiponutrin, two recent genome-wide screens in humans found an association between PNPLA3 gene variants and hepatic fat content as well as liver function. The first study reported a robust association between allelic variation in the adiponutrin gene and the ancestry-related predisposition toward nonalcoholic fatty liver disease (25). In this context, it should be noted that adiponutrin mRNA levels are severely upregulated in the (steatotic) liver of ob/ob mice (4). The second study associated a single nucleotide polymorphism within PNPLA3 with increased plasma levels of liver transaminases indicative of liver dysfunction (26). Moreover an association between polymorphisms in PNPLA3 and insulin secretion as well as obesity was reported (27).
PNPLA4: A KERATINOCYTE RE HYDROLASE
Human PNPLA4 was originally described as gene sequence-2 (GS2) (28). A GS2 gene was identified in several mammalian genomes but is absent in the mouse (4, 29). Human GS2 mRNA is expressed in multiple tissues, including adipose, muscle, heart, liver, and skin (keratinocytes) (3, 4, 28, 29). Importantly, human GS2, but not rat GS2, is a potent RE hydrolase (29) and by regulating cellular levels of all-trans retinoic acid, human GS2 may contribute to epidermal morphogenesis. GS2 was also shown to act as acylglycerol and retinol transacylase (6, 29), TG hydrolase (4, 29), and PLA2 (6). The physiological relevance of these activities remains to be determined, particularly in light of the fact that rat GS2 fails to exhibit RE hydrolase and transacylase activities (29) and that the mouse genome lacks a GS2 gene. Skepticism is also appropriate, since Lee et al. (28) found that X-linked ichthyosis patients lacking the STS gene and the nearby GS2 gene present equivalent phenotypes as those lacking STS alone.
PNPLA5: REGULATED BY THE NUTRITIONAL STATUS AND OBESITY
The mRNA of PNPLA5, also named GS2-like, is expressed at low levels in both mouse and human tissues, including lung, WAT, BAT, brain, and pituitary gland (3, 4). Murine GS2-like expression is regulated similarly to adiponutrin mRNA: strong inhibition by fasting, upregulation during adipocyte differentiation as well as low WAT levels, plus enormous induction in the liver of ob/ob mice (4). In vitro, GS2-like exhibited TG hydrolase activity, and overexpression of GS2-like resulted in a decrease of the cellular TG content (4). However, a second study found no evidence for TG hydrolase, RE hydrolase, or transacylase activities (29). Thus, the biochemical function and physiological roles of GS2-like remain elusive.
PNPLA6: AN ESSENTIAL LYSOPHOSPHOLIPASE IN THE BRAIN
PNPLA6 is commonly termed neuropathy target esterase (NTE). Murine NTE is primarily expressed in the nervous system (30), and the protein localizes to the endoplasmic reticulum (ER) and Golgi apparatus (31). NTE exhibits in vitro esterase and hydrolase activity against membrane lipids, preferentially lysophospholipids (31). Importantly, the capacity of NTE to hydrolyze lysophosphatidylcholine (LPC) in mouse brain hints at its conceivable physiological substrate (32). NTE was originally identified as a target enzyme for the poisonous effect of organophosphates. These compounds cause a severe neurological disorder in vertebrates known as organophosphate induced delayed neuropathy, characterized by degeneration of long axons in the spinal cord and peripheral nerves, leading to paralysis of the lower limbs. In mice, global NTE deficiency is embryonically lethal (30). Brain-specific deletion of NTE in mice resulted in severe neuropathologic symptoms concomitant with disruption of the ER, vacuolation of nerve cell bodies, and abnormal reticular aggregates (33). In humans, NTE mutations near the catalytic site were found to cause the severe NTE-related motor neuron disease, which is characterized by progressive spastic paraplegia and distal muscle wasting (34). Thus, the lysophospholipase activity of NTE has a critical function in maintaining axonal integrity across species.
PNPLA7: AN INSULIN-REGULATED LYSOPHOSPHOLIPASE IN MUSCLE AND FAT
PNPLA7 is closely related to PNPLA6 and was accordingly designated NTE-related esterase (NRE). Human NRE is primarily expressed in prostate, pancreas, and WAT (3). Mouse NRE mRNA was predominant in testis and insulin target tissues such as skeletal muscle, heart, BAT, and WAT (35). NRE transcript levels are strongly induced by fasting, suggesting nutritional regulation of the enzyme (35). Enhanced NRE mRNA expression is detected during adipocyte differentiation, while insulin downregulates NRE mRNA levels. NRE associates with the ER as well as with small lipid droplets (35). Similar to its paralog NTE, NRE exhibits prominent in vitro hydrolase activity against lysophospholipid substrates, but no hydrolase activity was detected against phosphatidylcholine (PC), TG, monoacylglycerols, RE, or cholesterol esters (35). Collectively, these findings suggest an involvement of NRE in energy metabolism, but clarification requires the characterization of mutant mouse models.
PNPLA8: A MYOCARDIAL PHOSPHOLIPASE MAINTAINING MITOCHONDRIAL INTEGRITY
PNPLA8 is synonymous with calcium-independent PLA2 (iPLA2)γ and was originally cloned from human heart cDNA (36). iPLA2γ mRNA expression is most prominent in human heart; however, the transcript is also detected at lower levels in other tissues. Immunohistochemical analysis in mouse myocardium demonstrated that iPLA2γ associates with mitochondria and peroxisomes, consistent with the protein's dual localization motifs (37). Interestingly, the enzyme exhibits both PLA1 and PLA2 activity for saturated or monounsaturated FA in phospholipids (38). However, incubation of purified iPLA2γ with 1-palmitoyl-2-arachidonoyl-sn-glycero-3-PC substrates leads to accumulation of 2-arachidonoyl-LPC. This product is a naturally occurring lysophospholipid in human myocardium and a lipid intermediate that may serve either as an endocannabinoid precursor or a source of arachidonic acid for eicosanoid production. Purified iPLA2γ was also shown to release arachidonic acid from the sn-2 position of plasmalogen (38).
The importance of iPLA2γ in cardiac phospholipid homeostasis and mitochondrial function was revealed by gain- and loss-of-function experiments in mice (37, 39). Cardiac-specific overexpression of human iPLA2γ resulted in a substantial loss of myocardial phospholipid mass, disorganized mitochondrial cristae, and compromised mitochondrial function as evident from decreased mitochondrial respiration (37). Increased levels of 2-arachidonoyl- and 2-docosahexaenoyl-LPC in the myocardium of iPLA2γ transgenic mice confirmed the enzyme's PLA1 activity for sn-2 polyunsaturated aliphatic chains in vivo. Interestingly, fasting induced excessive deposition of TG in the heart of transgenic mice overexpressing human iPLA2γ, which coincided with hemodynamic dysfunction (37). Impaired mitochondrial function was also observed in iPLA2γ-deficient mice as manifested by decreased complex IV-mediated oxygen consumption (39). This was associated with decreased myocardial cardiolipin content and altered composition of molecular species of cardiolipin, indicating that iPLA2γ is involved in cardiolipin remodeling. As a result, iPLA2γ-deficient mice exhibit overt bioenergetic defects, which are manifested in growth retardation, cold intolerance, decreased exercise endurance, and increased mortality due to cardiac stress (39).
PNPLA9: A PHOSPHOLIPASE ASSOCIATED WITH NEUROLOGIC DISEASE
PNPLA9 is also named PLA2 group VI (PLA2G6). The structure and function of the protein was recently reviewed by Turk and Ramanadham (40). PLA2G6 was cloned from various species and tissues. PLA2G6 mRNA is ubiquitously expressed in tissues of rodents and humans (3, 41–43). In vitro, PLA2G6 preferentially hydrolyzes the FA at the sn-2 position of glycerophospholipids (38, 42). Notably, PLA2G6 also possesses acetyl hydrolase activity against platelet-activating factor at the sn-2 position, suggesting that its PLA2 activity is not restricted to long-chain FA (42). PLA2G6 mainly localizes to the cytoplasm of resting cells; however, upon stimulation, the enzyme translocates to cellular membrane compartments in the perinuclear area (40). Consistent with its ubiquitous expression and stimulus-specific localization, PLA2G6 is involved in a number of cellular processes, including phospholipid remodeling, signal transduction, cell proliferation, and apoptotic cell death (40).
Despite multiple functions of PLA2G6 in in vitro assays and cell culture systems, mice lacking the enzyme were viable and appeared relatively normal (41). Specifically, male PLA2G6-ko mice displayed compromised fertility due to impaired mobility of spermatozoa. Unexpectedly, PLA2G6 deletion did not result in altered levels of arachidonate-containing glycerophosphocholine lipids in testes, suggesting that PLA2G6 is dispensable in the acylation/reacylation cycle leading to the formation of these lipid species (41). Male PLA2G6-ko mice also exhibit reduced glucose tolerance, which is associated with decreased insulin secretion from islets of these mice (44). Consistent with this observation, mice with β-cell-specific overexpression of PLA2G6 display increased glucose tolerance, and their islets show enhanced amplification of glucose-induced insulin secretion by forskolin (44). In addition to altered glucose metabolism and insulin secretion, age-dependent phenotypes were observed, including accelerated decrease in bone mass due to an increase in bone marrow fat content in PLA2G6-ko mice (43). Undifferentiated bone marrow stromal cells (BMSC) from PLA2G6-ko mice display enhanced adipogenesis and decreased osteogenesis, suggesting a role of PLA2G6 in BMSC differentiation (43). Furthermore, aging PLA2G6-ko mice develop neurological impairment due to disrupted axonal membrane homeostasis and accumulation of ubiquitinated proteins (45).
Recently, mutations in the human gene for PLA2G6 were shown to cause two childhood neurodegenerative disorders: infantile neuroaxonal dystrophy (INAD) and idiopathic neurodegeneration with brain iron accumulation (NBIA) (46). Axonal degeneration, ataxia, gait instability, optic and cerebellar atrophy, as well as brain iron accumulation are common characteristics of these diseases. PLA2G6 mutations are responsible for the majority (80%) of INAD cases and a considerable number (20%) of patients with NBIA (46).
OUTLOOK
Essential lipid hydrolases belong to a family of structurally unique PNPLA proteins. These enzymes play a fundamental role in lipid remodeling and catabolism and include TG hydrolases, (lyso)phospholipases, and a RE hydrolase. However, the biochemical and physiological function of the majority of PNPLA proteins remains unknown. An emerging interest in this protein family and the increasing availability of various mutant mouse models will help to elucidate the (patho)physiological role of all PNPLAs.
Supplementary Material
Acknowledgments
We would like to thank Dr. Ellen Zechner for critically reviewing this manuscript.
Abbreviations
ABHD5, α/β hydrolase domain containing-5
ATGL, adipose triglyceride lipase
BAT, brown adipose tissue
BMSC, bone marrow stromal cell
CGI-58, comparative gene identification-58
ER, endoplasmic reticulum
FA, fatty acid
GS2, gene sequence-2
HSL, hormone-sensitive lipase
INAD, infantile neuroaxonal dystrophy
iPLA2, calcium-independent phospholipase A2
ko, knockout
LPC, lysophosphatidylcholine
NBIA, neurodegeneration with brain iron accumulation
NRE, neuropathy target esterase-related esterase
NTE, neuropathy target esterase
PC, phosphatidylcholine
PLA, phospholipase A
PLA2G6, phospholipase A2 group VI
PNPLA, patatin-like phospholipase domain containing
RE, retinol ester
TG, triacylglycerol
WAT, white adipose tissue
This research was supported by the grant GOLD - Genomics of Lipid-Associated Disorders, which is part of the Austrian Genome Project GEN-AU Genome research in Austria funded by the Austrian Ministry of Science and Research and by the Austrian Science Foundation (FWF) grant W901-B05DK (Doktoratskolleg Molecular Enzymology) and F30-B05 (SFB Lipotox).
Published, JLR Papers in Press, November 23, 2008.
References
- 1.Andrews D. L., B. Beames, M. D. Summers, and W. D. Park. 1988. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J. 252 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rydel T. J., J. M. Williams, E. Krieger, F. Moshiri, W. C. Stallings, S. M. Brown, J. C. Pershing, J. P. Purcell, and M. F. Alibhai. 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 42 6696–6708. [DOI] [PubMed] [Google Scholar]
- 3.Wilson P. A., S. D. Gardner, N. M. Lambie, S. A. Commans, and D. J. Crowther. 2006. Characterization of the human patatin-like phospholipase family. J. Lipid Res. 47 1940–1949. [DOI] [PubMed] [Google Scholar]
- 4.Lake A. C., Y. Sun, J. L. Li, J. E. Kim, J. W. Johnson, D. Li, T. Revett, H. H. Shih, W. Liu, J. E. Paulsen, et al. 2005. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J. Lipid Res. 46 2477–2487. [DOI] [PubMed] [Google Scholar]
- 5.Villena J. A., S. Roy, E. Sarkadi-Nagy, K. H. Kim, and H. S. Sul. 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279 47066–47075. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins C. M., D. J. Mancuso, W. Yan, H. F. Sims, B. Gibson, and R. W. Gross. 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279 48968–48975. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann R., J. G. Strauss, G. Haemmerle, G. Schoiswohl, R. Birner-Gruenberger, M. Riederer, A. Lass, G. Neuberger, F. Eisenhaber, A. Hermetter, et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306 1383–1386. [DOI] [PubMed] [Google Scholar]
- 8.Zechner R., P. C. Kienesberger, G. Haemmerle, R. Zimmermann, and A. Lass. 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 50 3–21. [DOI] [PubMed] [Google Scholar]
- 9.Notari L., V. Baladron, J. D. Aroca-Aguilar, N. Balko, R. Heredia, C. Meyer, P. M. Notario, S. Saravanamuthu, M. L. Nueda, F. Sanchez-Sanchez, et al. 2006. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J. Biol. Chem. 281 38022–38037. [DOI] [PubMed] [Google Scholar]
- 10.Kershaw E. E., J. K. Hamm, L. A. Verhagen, O. Peroni, M. Katic, and J. S. Flier. 2006. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 55 148–157. [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J. Y., K. Tillison, J. H. Lee, D. A. Rearick, and C. M. Smas. 2006. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3–L1 adipocytes and is a target for transactivation by PPARgamma. Am. J. Physiol. Endocrinol. Metab. 291 E115–127. [DOI] [PubMed] [Google Scholar]
- 12.Lass A., R. Zimmermann, G. Haemmerle, M. Riederer, G. Schoiswohl, M. Schweiger, P. Kienesberger, J. G. Strauss, G. Gorkiewicz, and R. Zechner. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3 309–319. [DOI] [PubMed] [Google Scholar]
- 13.Haemmerle G., A. Lass, R. Zimmermann, G. Gorkiewicz, C. Meyer, J. Rozman, G. Heldmaier, R. Maier, C. Theussl, S. Eder, et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312 734–737. [DOI] [PubMed] [Google Scholar]
- 14.Brasaemle D. L., G. Dolios, L. Shapiro, and R. Wang. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279 46835–46842. [DOI] [PubMed] [Google Scholar]
- 15.Granneman J. G., H. P. Moore, R. L. Granneman, A. S. Greenberg, M. S. Obin, and Z. Zhu. 2007. Analysis of lipolytic protein trafficking and interactions in adipocytes. J. Biol. Chem. 282 5726–5735. [DOI] [PubMed] [Google Scholar]
- 16.Igal R. A., J. M. Rhoads, and R. A. Coleman. 1997. Neutral lipid storage disease with fatty liver and cholestasis. J. Pediatr. Gastroenterol. Nutr. 25 541–547. [DOI] [PubMed] [Google Scholar]
- 17.Lefevre C., F. Jobard, F. Caux, B. Bouadjar, A. Karaduman, R. Heilig, H. Lakhdar, A. Wollenberg, J. L. Verret, J. Weissenbach, et al. 2001. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am. J. Hum. Genet. 69 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer J., C. Lefevre, E. Morava, J. M. Mussini, P. Laforet, A. Negre-Salvayre, M. Lathrop, and R. Salvayre. 2007. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 39 28–30. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh A. K., G. Ramakrishnan, C. Chandramohan, and R. Rajasekharan. 2008. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J. Biol. Chem. 283 24525–24533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama M., K. Sakai, M. Ogawa, J. R. McMillan, D. Sawamura, and H. Shimizu. 2007. Novel duplication mutation in the patatin domain of adipose triglyceride lipase (PNPLA2) in neutral lipid storage disease with severe myopathy. Muscle Nerve. 36 856–859. [DOI] [PubMed] [Google Scholar]
- 21.Schweiger M., G. Schoiswohl, A. Lass, F. P. Radner, G. Haemmerle, R. Malli, W. Graier, I. Cornaciu, M. Oberer, R. Salvayre, et al. 2008. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J. Biol. Chem. 283 17211–17220. [DOI] [PubMed] [Google Scholar]
- 22.Schoenborn V., I. M. Heid, C. Vollmert, A. Lingenhel, T. D. Adams, P. N. Hopkins, T. Illig, R. Zimmermann, R. Zechner, S. C. Hunt, et al. 2006. The ATGL gene is associated with free fatty acids, triglycerides, and type 2 diabetes. Diabetes. 55 1270–1275. [DOI] [PubMed] [Google Scholar]
- 23.Schweiger M., R. Schreiber, G. Haemmerle, A. Lass, C. Fledelius, P. Jacobsen, H. Tornqvist, R. Zechner, and R. Zimmermann. 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281 40236–40241. [DOI] [PubMed] [Google Scholar]
- 24.Baulande S., F. Lasnier, M. Lucas, and J. Pairault. 2001. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J. Biol. Chem. 276 33336–33344. [DOI] [PubMed] [Google Scholar]
- 25.Romeo S., J. Kozlitina, C. Xing, A. Pertsemlidis, D. Cox, L. A. Pennacchio, E. Boerwinkle, J. C. Cohen, and H. H. Hobbs. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan X., D. Waterworth, J. R. Perry, N. Lim, K. Song, J. C. Chambers, W. Zhang, P. Vollenweider, H. Stirnadel, T. Johnson, et al. 2008. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 83 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson L. E., U. Lindblad, C. A. Larsson, L. Rastam, and M. Ridderstrale. 2008. Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. Eur. J. Endocrinol. 159 577–583. [DOI] [PubMed] [Google Scholar]
- 28.Lee W. C., E. Salido, and P. H. Yen. 1994. Isolation of a new gene GS2 (DXS1283E) from a CpG island between STS and KAL1 on Xp22.3. Genomics. 22 372–376. [DOI] [PubMed] [Google Scholar]
- 29.Gao J. G., and M. Simon. 2007. A comparative study of human GS2, its paralogues, and its rat orthologue. Biochem. Biophys. Res. Commun. 360 501–506. [DOI] [PubMed] [Google Scholar]
- 30.Winrow C. J., M. L. Hemming, D. M. Allen, G. B. Quistad, J. E. Casida, and C. Barlow. 2003. Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat. Genet. 33 477–485. [DOI] [PubMed] [Google Scholar]
- 31.van Tienhoven M., J. Atkins, Y. Li, and P. Glynn. 2002. Human neuropathy target esterase catalyzes hydrolysis of membrane lipids. J. Biol. Chem. 277 20942–20948. [DOI] [PubMed] [Google Scholar]
- 32.Quistad G. B., C. Barlow, C. J. Winrow, S. E. Sparks, and J. E. Casida. 2003. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc. Natl. Acad. Sci. USA. 100 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akassoglou K., B. Malester, J. Xu, L. Tessarollo, J. Rosenbluth, and M. V. Chao. 2004. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc. Natl. Acad. Sci. USA. 101 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rainier S., M. Bui, E. Mark, D. Thomas, D. Tokarz, L. Ming, C. Delaney, R. J. Richardson, J. W. Albers, N. Matsunami, et al. 2008. Neuropathy target esterase gene mutations cause motor neuron disease. Am. J. Hum. Genet. 82 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kienesberger P. C., A. Lass, K. Preiss-Landl, H. Wolinski, S. D. Kohlwein, R. Zimmermann, and R. Zechner. 2008. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J. Biol. Chem. 283 5908–5917. [DOI] [PubMed] [Google Scholar]
- 36.Mancuso D. J., C. M. Jenkins, and R. W. Gross. 2000. The genomic organization, complete mRNA sequence, cloning, and expression of a novel human intracellular membrane-associated calcium-independent phospholipase A(2). J. Biol. Chem. 275 9937–9945. [DOI] [PubMed] [Google Scholar]
- 37.Mancuso D. J., X. Han, C. M. Jenkins, J. J. Lehman, N. Sambandam, H. F. Sims, J. Yang, W. Yan, K. Yang, K. Green, et al. 2007. Dramatic accumulation of triglycerides and precipitation of cardiac hemodynamic dysfunction during brief caloric restriction in transgenic myocardium expressing human calcium-independent phospholipase A2gamma. J. Biol. Chem. 282 9216–9227. [DOI] [PubMed] [Google Scholar]
- 38.Yan W., C. M. Jenkins, X. Han, D. J. Mancuso, H. F. Sims, K. Yang, and R. W. Gross. 2005. The highly selective production of 2-arachidonoyl lysophosphatidylcholine catalyzed by purified calcium-independent phospholipase A2gamma: identification of a novel enzymatic mediator for the generation of a key branch point intermediate in eicosanoid signaling. J. Biol. Chem. 280 26669–26679. [DOI] [PubMed] [Google Scholar]
- 39.Mancuso D. J., H. F. Sims, X. Han, C. M. Jenkins, S. P. Guan, K. Yang, S. H. Moon, T. Pietka, N. A. Abumrad, P. H. Schlesinger, et al. 2007. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J. Biol. Chem. 282 34611–34622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turk J., and S. Ramanadham. 2004. The expression and function of a group VIA calcium-independent phospholipase A2 (iPLA2beta) in beta-cells. Can. J. Physiol. Pharmacol. 82 824–832. [DOI] [PubMed] [Google Scholar]
- 41.Bao S., D. J. Miller, Z. Ma, M. Wohltmann, G. Eng, S. Ramanadham, K. Moley, and J. Turk. 2004. Male mice that do not express group VIA phospholipase A2 produce spermatozoa with impaired motility and have greatly reduced fertility. J. Biol. Chem. 279 38194–38200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J., R. W. Kriz, N. Wolfman, M. Shaffer, J. Seehra, and S. S. Jones. 1997. A novel cytosolic calcium-independent phospholipase A2 contains eight ankyrin motifs. J. Biol. Chem. 272 8567–8575. [DOI] [PubMed] [Google Scholar]
- 43.Ramanadham S., K. E. Yarasheski, M. J. Silva, M. Wohltmann, D. V. Novack, B. Christiansen, X. Tu, S. Zhang, X. Lei, and J. Turk. 2008. Age-related changes in bone morphology are accelerated in group VIA phospholipase A2 (iPLA2beta)-null mice. Am. J. Pathol. 172 868–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao S., D. A. Jacobson, M. Wohltmann, A. Bohrer, W. Jin, L. H. Philipson, and J. Turk. 2008. Glucose homeostasis, insulin secretion, and islet phospholipids in mice that overexpress iPLA2beta in pancreatic beta-cells and in iPLA2beta-null mice. Am. J. Physiol. Endocrinol. Metab. 294 E217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik I., J. Turk, D. J. Mancuso, L. Montier, M. Wohltmann, D. F. Wozniak, R. E. Schmidt, R. W. Gross, and P. T. Kotzbauer. 2008. Disrupted membrane homeostasis and accumulation of ubiquitinated proteins in a mouse model of infantile neuroaxonal dystrophy caused by PLA2G6 mutations. Am. J. Pathol. 172 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory A., S. K. Westaway, I. E. Holm, P. T. Kotzbauer, P. Hogarth, S. Sonek, J. C. Coryell, T. M. Nguyen, N. Nardocci, G. Zorzi, et al. 2008. Neurodegeneration associated with genetic defects in phospholipase A(2). Neurology. 71 1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaloske R. H., and E. A. Dennis. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 1761 1246–1259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.