Abstract
The Ras-related small GTPases Rac, Rho, Cdc42, and RalA bind filamin, an actin filament-crosslinking protein that also links membrane and other intracellular proteins to actin. Of these GTPases only RalA binds filamin in a GTP-specific manner, and GTP-RalA elicits actin-rich filopods on surfaces of Swiss 3T3 cells and recruits filamin into the filopodial cytoskeleton. Either a dominant negative RalA construct or the RalA-binding domain of filamin 1 specifically block Cdc42-induced filopod formation, but a Cdc42 inhibitor does not impair RalA’s effects, which, unlike Cdc42, are Rac independent. RalA does not generate filopodia in filamin-deficient human melanoma cells, whereas transfection of filamin 1 restores the functional response. RalA therefore is a downstream intermediate in Cdc42-mediated filopod production and uses filamin in this pathway.
Activation of the Rho family of Ras-related small GTPases causes the construction of particular actin-rich surface structures in cells. For example, forced expression of constitutively active forms of RhoA, Rac1, and Cdc42 in serum-starved Swiss 3T3 cells induces actin stress fibers, membrane ruffles, and filopodia, respectively (1). These GTPases do not appear to affect actin dynamics directly, but rather engage downstream intermediates. Such targets include the Rho-activated phosphorylation of myosin, which leads to cellular contractile activity (2), and the phosphorylation of proteins of the ezrin-moesin-radixin group to promote their ability to link actin filaments to cell membranes (3). Rac1-induced polyphosphoinositide synthesis causes cellular actin assembly by uncapping actin filament ends (4), and the actin filament severing and barbed end capping protein gelsolin participates in this process (5). N-WASP, a profilin-binding protein, is an intermediate in filopodial extension, possibly by acquiring actin filament depolymerizing activity when ligated by Cdc42 (6, 7).
We report two related connections in the pathway between Cdc42 and filopodial formation. One part of this connection involves the Ras-related small GTPase RalA (8). Expression of dominant negative RalA delays border cell migration in Drosophila oogenesis (9), and transfection of the guanine nucleotide releasing stimulator of RalA affects cell morphology (10), suggesting a linkage between RalA and the actin cytoskeleton. The second part of this connection is the actin filament crosslinking protein filamin. Three distinct filamin cDNAs have been reported, which originate from three different chromosomes, with additional variants ascribable to alternative mRNA splicing (11–14). Of these protein isoforms, filamin 1 (also called ABP-280) is the founding and most widely distributed (11, 15). Filamin efficiently crosslinks actin filaments and is a docking site for various cell surface receptors and certain intracellular proteins involved in signal transduction or endocytosis (16–22). Genetic evidence shows that filamin 1 in particular is an essential component for stabilization of the cell surface and for efficient translocational motility of melanocytic, neuronal, and other cells (23, 24). Although phosphorylation of filamin occurs in vitro and in cells and may affect its actin-binding properties (25), little information is available about filamin’s cellular regulation. Filamin was found previously to associate with unidentified GTP-binding proteins in human platelets (26, 27). We report here that filamin binds Rac, Rho, and Cdc42. We present evidence for the functional importance of the RalA-filamin interaction in vivo.
MATERIALS AND METHODS
Proteins and Plasmids.
Small GTPase proteins were expressed in Escherichia coli fused with glutathione S-transferase (GST) and incubated with glutathione-Sepharose beads. The GST fusion proteins were eluted from the beads with 5 mM reduced glutathione and dialyzed against 20 mM Tris⋅HCl, pH 7.5/150 mM NaCl (buffer A) containing 10% glycerol. For microinjection, the recombinant GTPases were released from the glutathione-Sepharose beads by cleavage with thrombin. Thrombin was removed by incubation with benzamidine-Sepharose 6B, and the proteins were dialyzed against a solution of buffer A with 5 mM MgCl2. Filamin (ABP-280) was purified from human uterine leiomyomas or from platelets as described (25). Filamin 1 is the predominant isotype of human platelets (12), and the actin filament gelation activities of platelet and leiomyoma filamins are equivalent (28). Nevertheless, isotype-specific antibodies have not been validated, and we cannot conclude that filamin 1 is the only isotype present in our binding experiments or reacting with the antibodies used. Therefore, we use the general term filamin unless genetic transfection studies permit specification of the filamin 1 isotype. The RalA-binding domain of filamin 1 (amino acids 2554–2647) was expressed in bacteria as a GST-fusion protein (GST-filamin 1–2554–2647), purified on glutathione-Sepharose beads, and dialyzed against a solution of buffer A with 0.5 mM phenylmethylsulfonyl fluoride. Wild-type RalA (subcloned into the pEFBos mammalian expression vector), PGEX2T-RalA, PGEX2T-V23RalA, PGEX2T-N28RalA, and Flag-tagged RalA constructs subcloned into pCMV5 vectors were gifts from Hiroshi Koide (Tokyo Institute of Technology, Yokohama) and Yoshito Kaziro (Tokyo Institute of Technology), and pCMV5-V12Cdc42 and pCMV5-N17Cdc42 were from Takaya Satoh, Tokyo Institute of Technology and Yoshito Kaziro. PGEX2T-Cdc42 and PGEX2T-Rac constructs were provided by Alan Hall (University College, London). The Cdc42 mutant cDNAs were subcloned into PGEX2T vectors.
Association of Filamin with Small GTPases.
GST-GTP-binding proteins (12 μg) were loaded with nucleotides by incubation with 1 mM EDTA and 200 μM gluanosine 5′-β-thiodiphosphate (GDPβS) or guanosine 5′-γ-thiotriphosphate (GTPγS) for 5 min at 30°C, mixed with 5 mM MgCl2 and placed on ice. The GST-GTP binding proteins (20 μl) were mixed with 20 μl of purified filamin (20 μg). Glutathione-Sepharose beads (50 μl, 50% vol/vol) were added, and the immobilized GST-fusion proteins were incubated in 0.4 ml of a solution containing 50 mM β-glycerophosphate (pH 7.5), 5 mM EGTA, 1 mM DTT, 0.1% Triton X-100, 5 mM MgCl2, and 0.1 mM Na3VO4 (buffer B) for 60 min at 4°C. The reacted beads were washed four times with buffer B, suspended in 60 μl of 1% SDS, boiled, and centrifuged. The supernatants were collected and subjected to PAGE. Bound filamin 1 was detected either by staining the gels with Coomassie brilliant blue or Western blotting probed with an antiplatelet filamin mAb (Chemicon).
The plasmids pCMV-myc-filamin 1 and pCMV-Flag-V23RalA or pCMV-Flag-N28RalA were transfected into Cos7 cells to express filamin 1 and Flag-tagged V23RalA and N28RalA. Forty-eight hours later, the cells were washed twice with 10 ml of PBS, suspended in 0.5 ml of buffer B with 10 μg/ml of leupeptin and antipain, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and homogenized in a Dounce homogenizer. The cell lysates were centrifuged at 100,000 × g for 15 min at 4°C by using Beckman TL-100 centrifuge. The supernatant fluid was subjected to immunoprecipitation by antibody to Flag (Eastman Kodak). Bound protein was detected by Western blotting probed with the antifilamin antibody for filamin or anti-Flag antibody for RalA. Subfragments derived from filamin 1 were constructed by using restriction enzyme digestion of filamin 1 cDNA (fragment 1, ScaI and BglII; fragment 2, BglII and NcoI; and fragment 3, NcoI and NotI) or the PCR (fragment 4), expressed in E. coli as GST fusion proteins by using the PGEX4T vector. Bacterial extracts were incubated with glutathione-Sepharose beads and eluted with 5 mM reduced glutathione. The purified proteins were dialyzed against buffer A with 0.5 mM PMSF. V23RalA was tagged with the recognition sequence (RRASV) for the catalytic subunit of cAMP-dependent protein kinase (cAMP kinase) by using the PGEX-2TK vector, expressed in E. coli and purified by glutathione-Sepharose affinity chromatography. The purified RalA proteins were phosphorylated by cAMP kinase in the presence of [γ32P]ATP for 5 min at 30°C, and the reaction was terminated by adding 10 μM of cAMP kinase inhibitor (PKI-tide). 32P-labeled RalA protein was loaded with either GTPγS or GDPβS (200 μM) and incubated with subfragments of filamin 1 (30 μg/ml) in buffer C containing 20 mM Tris⋅HCl (pH 7.5), 0.5% Nonidet P-40, 150 mM NaCl, and 10 mM MgCl2 for 1 h at 4°C and further incubated with glutathione-Sepharose beads (50 μl, 50% vol/vol) for 15 min at 4°C. The beads were washed four times with buffer C, and bound protein was released from the beads by boiling for 2 min in 30 μl of electrophoresis sample buffer and resolved by PAGE (15%) (29). The relative intensity of the 32P-labeled RalA band was determined by a Bioimage Analyzer BAS2000 (Fuji).
Cell Culture and Microinjection.
Swiss 3T3 cells were grown at 37°C in DMEM supplemented with 10% fetal calf serum. For microinjection, cells were cultured in 35-mm dishes containing 15-mm glass coverslips coated with poly-l-lysine (50 μg/ml). Confluent cells were cultured for 16 h in serum-free DMEM. Proteins were microinjected into the cytoplasm with fluorescein isothiocyanate (FITC)-coupled dextran (5 mg/ml) to localize microinjected cells. A filamin 1 cDNA, which lacks the carboxyl-terminal last 333 nt (filamin 1δCT112), was ligated to the mammalian expression vector pME18SNeo, which contains the neomycin resistance gene. The plasmid was transfected into M2 cells with liposomes (Lipofectamine, Gibco/BRL). After 1 day, the culture medium was replaced with medium containing G418 (1 mg/ml). Resistant colonies of cells [M2(δCT112)] were serially diluted into multiwell plates and cultured separately. The human melanoma cell lines M2, M2(δCT112), and A7 were grown in minimum essential medium (MEM) supplemented with 8% newborn calf serum and 2% fetal calf serum. Cells were allowed to spread for 4 days and serum-starved in MEM for 48 h. After microinjection, cells were rinsed in PBS and fixed in 3.7% paraformaldehyde at room temperature and permeabilized in 0.5% Triton X-100. Cells were rinsed and incubated with PBS containing 5% horse serum. For actin localization, cells were rinsed and incubated with 1 unit of Texas red-X phalloidin (Molecular Probes). For localization of filamin, cells were incubated with monoclonal antifilamin antibodies, rinsed in PBS, and further incubated with Texas red-conjugated sheep anti-mouse antibody (Amersham Pharmacia). Cells were rinsed, and the coverslips were mounted on slide glass with 10 μl of Perma Fluor. Cells were examined on a confocal scanning laser-beam microscope (Molecular Dynamics). Images were recorded and imported into Adobe Photoshop program for compilation.
Subcellular Fractionation.
Subconfluent COS7 cells were transfected by the DEAE-dextran method using 10 μg of plasmids per 10-cm Petri dish. The cells were grown in DMEM with 10% fetal calf serum. Forty-eight hours later, transfected cells were serum-starved in DMEM for 16 h. Triton-insoluble cytoskeletons were prepared at room temperature. Cells were washed twice with 10 ml of PBS and subjected to 1 ml of lysis buffer containing 20 mM Pipes (pH 6.8), 2 mM MgCl2, 50 mM KCl, 5 mM EGTA, 5 mM DTT, 1 mM ATP, 0.5% Triton X-100, 42 mM leupeptin, 1.0 mM benzamidine HCl, 12 mM aprotinin, and 1.0 mM phenylmethylsulfonyl fluoride. Cell suspensions were collected with a rubber policeman and centrifuged for 3 min at 8,730 × g in a microfuge. Supernatant fluids were removed, and the pellets were suspended in 1 ml of a solution containing 1% SDS. Fractions were subjected to PAGE, transferred to poly(vinylidine fluoride) PVF membranes, and probed with monoclonal antifilamin antibody or anti-RalA antibody (Transduction Laboratories, Lexington, KY). The relative amounts of filamin and RalA proteins in the cytoskeleton and membrane fractions were quantitated from digitized images of autoradiograms of immunoblots by using the National Institutes of Health image analysis program. Protein concentrations were determined by the method of Bradford (30) using BSA as the standard.
RESULTS
Filamin Binds Small GTPases and Preferentially to Activated RalA.
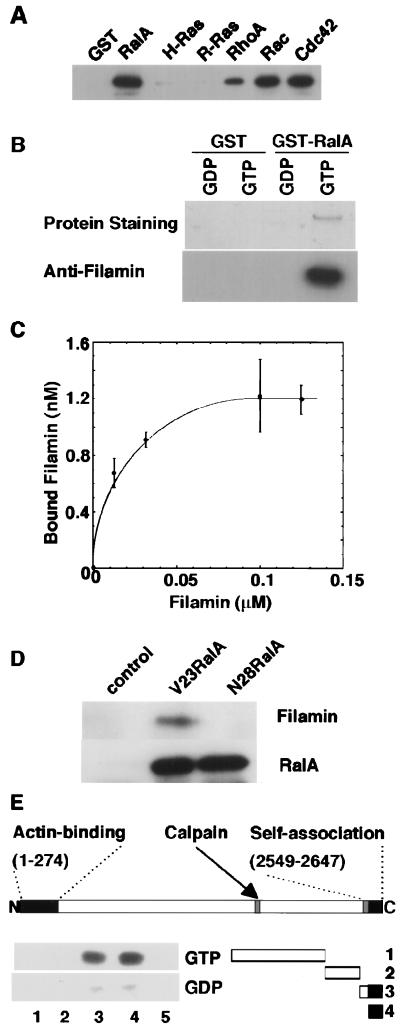
To identify the filamin-binding small GTPase(s), we used the GTPγS-bound form of a panel of GST-small GTPases as an affinity reagent. Human filamin precipitated with glutathione Sepharose-bound RhoA, Rac1, and Cdc42, although these associations were unaffected by whether the nucleotide bound was GTP or GDP. Filamin did not bind to H-Ras or R-Ras (Fig. 1A). On the other hand, filamin bound the GST-RalA containing GTPγS but not GDPβS (Fig. 1B). The association of filamin with GTP-ligated RalA is stable when the ionic strength is made physiologic (0.15 M) with NaCl and occurs rapidly, reaching saturation within 10 min at 4°C (data not shown). Purified filamin bound to the glutathione Sepharose-bound GST-RalA-GTPγS complex in a concentration-dependent manner with an apparent Kd of 12 nM (Fig. 1C). This affinity is equivalent to the interaction between GTP-bound Ras with the N-terminal region of Raf kinase (50 nM) and stronger than that between Ras and neurofibromin (250 nM) (31, 32). We confirmed qualitatively that activated RalA and filamin can form a stable complex in intact cells. Filamin bound activated RalA (V23RalA) immunoprecipitated from Cos7 cells transfected with DNAs encoding these proteins (Fig. 1D). Filamin did not associate with N28RalA, a mutant protein with preferential affinity for GDP. Little RalA precipitated with filamin from Cos7 transfectants (data not shown). This result suggests that only a small fraction of filamin binds RalA in vivo, a finding reminiscent of other filamin 1 binding partners with the exception of actin (20).
Figure 1.
Binding of filamin to RalA. (A) Various GST-small GTPase fusion proteins or GST protein were loaded with GTPγS and mixed with purified filamin. Bound filamin was visualized by immunoblot. (B) GTPγS, GDPβS RalA-GST, or GST were loaded with and mixed with purified human filamin, precipitated by using glutathione-Sepharose beads, washed, eluted in 1% SDS, and displayed by SDS/PAGE. (C) Dose-dependent binding of filamin to GTPγS-coupled RalA-GST. GTPγS-loaded RalA-GST protein was coupled to glutathione-Sepharose beads, and protein conjugates were mixed with increasing amounts of filamin for 30 min and washed, and bound filamin was visualized by SDS/PAGE and silver staining of the gel. Bound filamin was quantitated by using the National Institutes of Health image software program. Each value represents the mean ± SE from triplicate measurements. (D) V23RalA and N28RalA was immunoprecipitated from Cos7 cells overexpressing V23RalA or N28RalA and filamin 1. The washed immunoprecipitates were immunoblotted for the presence of filamin and RalA. (E) Schematic diagram of truncated filamin 1 constructs and mapping of the RalA binding domain of filamin 1. Fragments of filamin 1 were engineered, expressed in E. coli, and purified as GST fusion proteins. The GST fusion filamin 1 fragments shown in the diagram (lanes 1–4) or GST alone (lane 5) were incubated with the 32P-labeled V23RalA protein and precipitated with glutathione-Sepharose beads. Bound RalA proteins were resolved by PAGE and analyzed by a Bioimage Analyzer BAS2000. The N-terminal amino acid and carboxyl-terminal amino acid of each construct is 1, 1524–2283; 2, 2283–2490; 3, 2495–2647, and 4, 2554–2647.
Filamin 1 is an elongated dimer composed of identical 280-kDA subunits, each containing extended rod domains formed from 24 Ig-like β-sheet motifs that connect an amino-terminal actin-binding domain to a carboxyl-terminal self-association site. Sequence insertions of 23 and 35 aa, designated as hinges, precede repeats 16 and 24, and these hinges contain calpain cleavage sites (11). We localized the specific binding of RalA in vitro to the carboxyl-terminal calpain-cleaved fragment of filamin (data not shown) and then pursued this analysis by generating GST fusion constructs containing carboxyl-terminal residues of filamin 1. The RalA binding activity resided in filamin 1 fragments encompassing amino acid residues 2254–2647, which correspond to the carboxyl-terminal last repeat (repeat 24), the subunit dimerization domain, and part of the second hinge (Fig. 1E).
RalA Induces Filopodia in Serum-Starved Swiss 3T3 Cells.
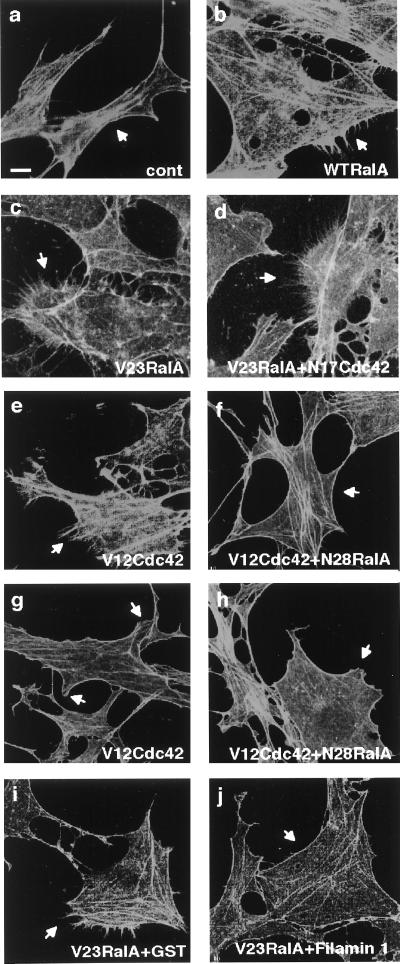
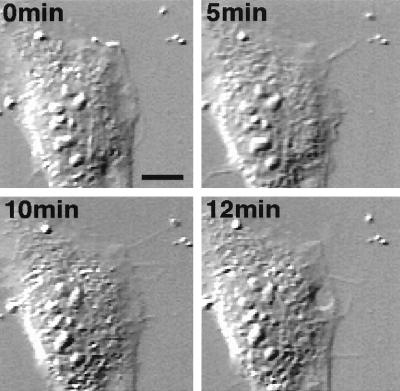
Having documented binding of filamin to RalA in vitro, we then microinjected RalA protein into Swiss 3T3 cells, which have the best characterized responses to small GTPases (1). RalA induced filopodia around the cell periphery, and the filopodia stained with phalloidin, indicating that RalA reorganized the actin cytoskeleton (Fig. 2). Constitutively active recombinant V23RalA also stimulated the formation of filopodia, whereas the dominant negative RalA inhibitor N28RalA did not. Observations with time-lapse differential interference contrast microscopy revealed that the filopodia induced by RalA resulted from protrusive activity of the cells and not cell retraction (Fig. 3). The induction of filopodia occurs within 5 min after the microinjection of RalA, reaches a maximal level at around 15 min, and declines to a basal level within 90 min (Fig. 3 and Table 1).
Figure 2.
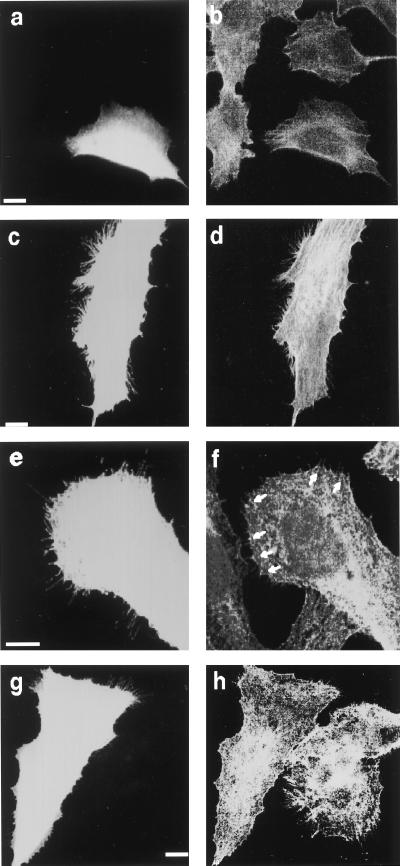
RalA induces filopodia in serum-starved Swiss 3T3 cells. Serum-starved, confluent Swiss 3T3 fibroblasts were fixed 15 min after injection with FITC-dextran alone (a), together with RalA (1.4 mg/ml) (b), V23RalA (0.4 mg/ml) (c), or coinjection with V23RalA (0.4 mg/ml) and N17Cdc42 (2.0 mg/ml) (d), V23RalA (0.7 mg/ml) and GST (0.8 mg/ml) (i), V23RalA (0.7 mg/ml) and GST-filamin 1 (2554–2647) (1.2 mg/ml) (j). Cells were fixed 5 min (e and f) or 90 min (g and h) after injection with V12 Cdc42 (0.46 mg/ml) (e and g), or coinjection with V12Cdc42 (0.46 mg/ml) and N28RalA (1.6 mg/ml) (f and h). Actin was localized by Texas red-X phalloidin. Injected cells were marked by FITC-labeled Dextran and indicated by arrows. (Scale bar represents 10 μm.)
Figure 3.
Time-lapse images of a cell injected with V23RalA. V23RalA protein (1.6 mg/ml) was microinjected into serum-starved subconfluent fibroblasts, which were recorded under differential interference contrast microscopy (Nikon TE300). Images were collected with a charge-coupled device camera and time-lapse controller (ARGUS-20, Hamamatsu Photonics, Hamamatsu City, Japan), transferred to a Macintosh computer with a frame grabber (Scion, Frederick, MD), and processed by using National Institutes of Health image analysis. Frames at selected times after injection were shown. (Scale bar represents 10 μm.)
Table 1.
Percentages of phenotypes observed after microinjection of small GTPase proteins into Swiss 3T3 cells
| Treatment | Time, min | Filopodia | Lamellipodia |
|---|---|---|---|
| Control | 15 | 13.8 ± 2.5 | 0.7 ± 0.7 |
| RalA (1.4 mg/ml) | 5 | 54.8 ± 1.6 | 0.0 ± 0.0 |
| RalA | 15 | 68.6 ± 3.2 | 0.3 ± 0.3 |
| RalA | 60 | 46.1 ± 4.0 | 1.3 ± 1.3 |
| RalA | 90 | 27.3 ± 1.0 | 4.3 ± 1.1 |
| V23RalA (0.4 mg/ml) | 15 | 62.2 ± 8.2 | 1.1 ± 1.1 |
| V12Cdc42 (1.0 mg/ml) | 5 | 60.0 ± 2.6 | 1.4 ± 1.4 |
| V12Cdc42 | 15 | 32.9 ± 7.3 | 29.2 ± 4.6 |
| V12Cdc42 | 60 | 26.3 ± 1.2 | 39.3 ± 6.0 |
| V12Cdc42 | 90 | 3.6 ± 2.6 | 64.7 ± 7.5 |
| N28RalA (1.4 mg/ml) | 5 | 16.4 ± 0.4 | 0.0 ± 0.0 |
| N17Cdc42 (2.0 mg/ml) | 15 | 16.7 ± 0.0 | 0.0 ± 0.0 |
| V23RalA (0.4 mg/ml) + N17Cdc42 (2.0 mg/ml) | 15 | 60.5 ± 1.4 | 2.4 ± 0.9 |
| V12Cdc42 (0.46 mg/ml) + N28RalA (1.6 mg/ml) | 5 | 15.2 ± 4.9 | 1.3 ± 1.3 |
| V12Cdc42 + N28RalA | 90 | 15.3 ± 2.1 | 62.9 ± 6.5 |
| V23RalA (0.7 mg/ml) + GST (0.82 mg/ml) | 15 | 51.0 ± 1.8 | 11.4 ± 2.3 |
| V23RalA (0.7 mg/ml) + GST-filamin 1 | 15 | 24.1 ± 6.4 | 8.0 ± 3.1 |
| (2554–2647) (1.2 mg/ml) |
The percentages of filopod- or lamellipod-positive cells were calculated and data are expressed as the mean ± SE. The GST-filamin 1 fragment did not affect the actions of 0.8 mg/ml Rac or 0.8 mg/ml RhoA, which produced respectively lamellipodia or stress fibers in 60–70% of injected cells.
The morphology of filopodia induced by RalA protein resembles the hairlike protrusions induced by Cdc42 or by treatment of cells with bradykinin (1). We therefore determined whether RalA’s effects involve Cdc42. The dominant negative Cdc42 inhibitor (N17Cdc42) not only failed to suppress filopodial formation induced by RalA but even enhanced it (Fig. 2). Microinjection of constitutively active Cdc42 (V12Cdc42) into serum-starved Swiss 3T3 cells induced the expected filopodia within 5 min (1) (Fig. 2 and Table 1), but microinjection of the dominant negative RalA inhibitor (N28RalA) blocked the Cdc42-dependent formation of filopodia (Fig. 2 and Table 1). Cdc42 also elicits ruffling lamellipodia because of a secondary activation of Rac (1). We occasionally observed small lamellipodia at the roots of RalA-induced filopodia, but the lamellipodia did not grow into ruffles up to 90 min after the injection of RalA protein (Fig. 3 and Table 1). Microinjection of the dominant negative RalA inhibitor (N28RalA) did not block the Cdc42-dependent formation of lamellipodia (Fig. 2 and Table 1).
Filamin 1 Is Required for the Formation of Filopodia Induced by RalA.
We examined the functional importance of RalA’s binding to filamin 1 in filopod protrusion with three sets of experiments. First, we expressed the RalA-binding domain of filamin 1 containing amino acids 2554–2647 in bacteria as a GST-fusion protein, purified it on glutathione beads, and injected it along with V23RalA into confluent serum-starved Swiss 3T3 cells. The fragment markedly inhibited filopod formation induced by RalA (Fig. 2 and Table 1). However, the fragment neither affected the baseline morphology of the injected cells nor impaired the customary induction of stress fibers or ruffling lamellae, respectively, by microinjection of RhoA or Rac (1). The fragment does not appear to cause dissociation of filamin’s dimeric subunits, because a 65-molar excess of the fragment did not change the actin gelling activity of filamin 1 in vitro.
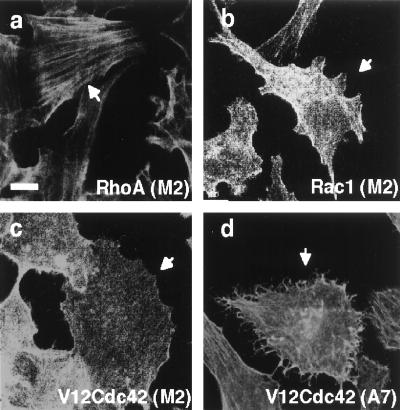
Second, we took advantage of a human melanoma cell line that does not transcribe filamin mRNA (M2 cells) to test the role of filamin in RalA-induced filopod formation. M2 cells exhibit prolonged membrane blebbing after plating and almost no translocational cell movement (23). Stable transfection of recombinant human filamin 1 in these cells, such that the molar ratio of filamin to actin (1:160) is comparable to that found in cells normally expressing filamin, corrects these defects (A7 cells). Blebbing M2 cells and cortically more stable A7 cells have abundant filopodia, and bradykinin increased the filopod density in both cell lines. However, filamin-deficient cells stop blebbing 3 days after plating because of a compensatory increase in their content of actin filaments compared with A7 cells (33). We therefore analyzed the regulation of filopod formation in the filamin-null and filamin-repleted cell lines at 6 days after plating when both cell lines more closely resemble Swiss 3T3 cells in morphology. The appearance of phalloidin-stainable actin filaments is similar between these two cell lines after serum depletion, and cortical filamentous actin is visible in the fluorescence microscope as a thick rim around the cell periphery. Filamin staining distributes with the actin filaments and concentrates at the cell periphery. As in Swiss 3T3 cells, microinjection of RhoA and Rac1 produced stress fibers and confluent peripheral actin collections, respectively in filamin-deficient M2 and filamin-1-transfected A7 cells (Fig. 4 and Table 2). On the other hand, microinjection of constitutively active Cdc42 (V12Cdc42) caused no significant change in the surface morphology of the filamin-deficient M2 cells whereas filamin-1-repleted cells formed numerous filopodia after Cdc42 microinjection (Fig. 4a and Table 2). In accordance with the above results, microinjection of constitutively active RalA (V23RalA) did not cause any change in surface morphology of the filamin-deficient M2 cells but filamin-1-transfected A7 cells produced abundant filopodia after RalA microinjection (Fig. 5 and Table 2). Microinjection of A7 melanoma cells with constitutively active RalA (V23RalA) caused filamin to concentrate at the roots of filopodia (Fig. 5).
Figure 4.
Distribution of actin in human melanoma cells. Serum-starved subconfluent filamin−1-deficient M2 (a–c) or filamin−1-repleted A7 (d) cells were injected with RhoA (1.0 mg/ml) (a), Rac1 (1.8 mg/ml) (b), or V12Cdc42 (1.8 mg/ml) (c and d). Cells were fixed at 180 (a), 150 (b), or 70 (c and d) min after injection and stained for actin filaments by using Texas red-X phalloidin. Injected cells were marked by FITC-labeled Dextran and indicated by arrows. (Scale bar represents 10 μm.)
Table 2.
Percentages of phenotypes observed after microinjection of small GTPase proteins into human melanoma cells
| Cells | Treatment | Time, min | F‡ | L | SF |
|---|---|---|---|---|---|
| M2 | Control | 60 | 4.8 ± 2.0 | 5.7 ± 1.2 | 10.3 ± 0.3 |
| V23RalA | 90 | 5.8 ± 3.9 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| V23RalA + N17Cdc42 | 60 | 0.0 ± 0.0 | N.D. | N.D. | |
| RhoA | 180 | 3.6 ± 1.8 | 7.8 ± 4.3 | 52.7 ± 6.4 | |
| Rac | 150 | 6.5 ± 4.1 | 31.2 ± 5.1 | 1.8 ± 1.8 | |
| V12Cdc42 | 70 | 8.1 ± 4.6 | 2.7 ± 1.3 | 1.3 ± 1.3 | |
| A7 | Control | 60 | 11.8 ± 2.5 | 9.0 ± 1.2 | 2.0 ± 0.3 |
| V23RalA | 90 | 52.7 ± 8.2 | 9.0 ± 4.3 | 0.0 ± 0.0 | |
| V23RalA + N17Cdc42 | 60 | 67.2 ± 4.5 | N.D. | N.D. | |
| RhoA | 180 | 5.9 ± 0.8 | 4.8 ± 2.1 | 64.6 ± 7.8 | |
| Rac | 150 | 9.1 ± 1.7 | 40.7 ± 9.4 | 0.0 ± 0.0 | |
| V12Cdc42 | 70 | 48.8 ± 6.1 | 3.2 ± 1.1 | 0.0 ± 0.0 | |
| M2 (δCT112) | |||||
| Control | 90 | 60.0 ± 7.0 | 4.0 ± 1.0 | 0.0 ± 0.0 | |
| V23RalA | 90 | 57.9 ± 4.9 | 0.0 ± 0.0 | 0.0 ± 0.0 |
Experimental conditions are described and the data are expressed as in Table 1. The concentration of microinjected RhoA and Racl are RhoA (1.0 mg/ml) and Racl (1.8 mg/ml), respectively. F, filopodia; L, lamellipodia, SF, stress fibers; N.D., not determined.
Figure 5.
Distribution of actin and filamin 1 in human melanoma cells. Serum-starved subconfluent filamin-deficient M2 (a and b), filamin−1-repleted A7 (c–f), or M2 cells transfected with RalA-binding site deletion mutant filamin 1 [M2(δ CT112)] (g and h) were injected with V23RalA (1.6 mg/ml). Cells were fixed 90 min after injection and stained for actin filaments by using Texas red-X phalloidin (b, d, and h) or filamin 1 by using antifilamin antibody (f). Injected cells were marked by FITC-labeled Dextran (a, c, e, and g). (f) Accumulation of filamin 1 at the roots of filopods is indicated by arrows. (Scale bar represents 10 μm.)
Third, we stably transfected filamin-deficient M2 cells with an filamin 1 truncate construct (filamin-1-δCT112) that fails to encode the carboxyl-terminal 112 aa encompassing the RalA binding and dimerization domain of filamin 1. This mutant filamin 1 binds but does not crosslink actin filaments (34). Cells expressing this construct exhibited numerous filopodia even 6 days after plating, but microinjection of constitutively active RalA (V23RalA) did not cause any change in surface morphology of this cell line (Fig. 5 and Table 2). Unlike A7 cells, these cells initially bleb like M2 cells and even after spreading, they have a comparatively thin cortical actin network (Fig. 5).
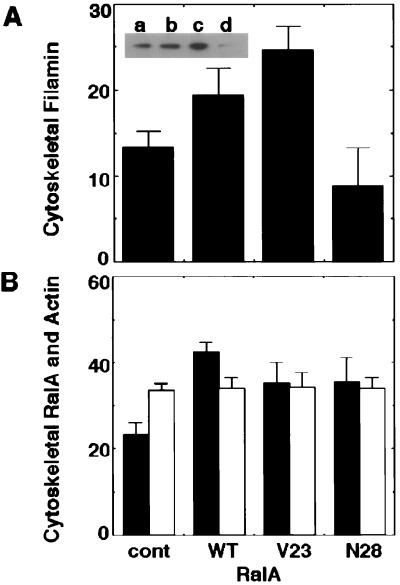
We did not detect an effect of RalA protein on the actin filament binding or crosslinking activity of purified filamin in vitro. However, active RalA changed the distribution of filamin among cellular compartments. Most of the filamin protein in serum-starved Cos7 cells was in a detergent-soluble fraction (Fig. 6A). Transfection of wild type, as well as constitutively active RalA (V23RalA), increased the fraction of cytoskeleton-associated filamin by 1.4- to 1.8-fold, whereas the dominant negative inhibitor of RalA (N28RalA) decreased the incorporation of filamin into the cytoskeleton (Fig. 6A). Both endogenous as well as forcibly expressed RalA reside in the Triton-insoluble cytoskeletal fractions (Fig. 6B). The amount of cytoskeletal actin in resting cells was about 34% and did not change after transfection of RalA (Fig. 6B).
Figure 6.
Fractionation of filamin, RalA, and actin proteins. (A) Cos7 cells were transfected with vector (control), wild-type RalA (WT), V23RalA (V23), or N28RalA (N28). Cells were serum-starved and Triton-solubilized, and insoluble cytoskeletal fractions were prepared after cell lysis. Samples of each fraction were immunoblotted with antifilamin antibody. The relative amounts of filamin in the cytoskeletons were quantitated from digitized images of autoradiograms of immunoblots by using a National Institutes of Health image analysis program. Total filamin protein represents the sum of the amounts of filamin in Triton-soluble and insoluble fractions. Each value represents the percentage of total and the mean ± SE (n = 5). Statistical significance was determined by the one-factor ANOVA (P < 0.05). (Inset) The representative autoradiograph of the cytoskeleton-associated filamin in cells transfected with control (a), wild-type RalA (b), V23RalA (c), and N28RalA (d) probed with antifilamin antibody. (B) Samples of Triton-soluble and insoluble fractions from RalA transfectants were immunoblotted with anti-RalA antibody. The relative amounts of RalA proteins were analyzed as in A and are presented as filled bars. The relative content of actin was determined by scanning of Coomassie brilliant blue-stained gel and National Institutes of Health image analysis program and are presented as open bars. Each value represents the mean ± SE (n = 4).
DISCUSSION
Our findings add RalA to the repertoire of signal transduction intermediates that regulate the actin cytoskeletal remodeling and identify filamin, a protein with known actin modulating, membrane binding, and intracellular adaptor functions as its target. Our results further suggest that Cdc42 acts through RalA to induce filopodia, because a dominant negative inhibitor of RalA (N28RalA) blocked the formation of filopodia induced by Cdc42. Some additional evidence is consistent with communication between Cdc42 and RalA. A proposed effector protein factor for RalA called Ral-binding protein 1 (RalBP1) or Ral-interacting protein 1 (RLIP1 or RIP1) binds specifically to GTP-RalA and has GTPase-activating protein activity in vitro for Rac and Cdc42 but not for RhoA (35–37). However, our results imply that RalA does not activate Rac. First, RalA elicited filopodia but not lamellipodia, whereas Cdc42 stimulated the formation of lamellipodia as a result of Rac activation (1). Second, microinjection of N28RalA did not suppress the formation of lamellipodia induced by Cdc42.
Filamin appears to participate in the formation of filopodia as a downstream target of RalA. First, filamin binds RalA in a GTP-dependent manner in vitro. Second, expression of the RalA-binding domain of filamin 1 in Swiss 3T3 cells specifically inhibited the formation of filopodia induced by RalA. Third, RalA did not induce filopodia in a human melanoma cell line that lacks expression of filamin, but an filamin 1-expressing subline of this line made filopodia in response to active RalA. We observed these phenomena reproducibly at a time when the two cell lines had been cultured long enough for filamin-deficient cells to polymerize sufficient actin to develop a stable plasma membrane (33). Under these conditions, filamin is necessary for Cdc42 and RalA to induce filopodia. Finally, human melanoma cells expressing mutant filamin 1, which lacks the RalA-binding domain, failed to respond to active RalA, although this construct gains the function of constitutively inducing filopodia.
The molecular mechanism of how RalA regulates filamin 1 deserves further analysis. Our results suggest that active RalA may recruit filamin into the cytoskeleton before filopodial protrusion. First, transfection of active RalA increased the amount of cytoskeletal filamin, whereas the dominant negative inhibitor of RalA decreased it. Second, filamin concentrates at the root of filopodia in cells microinjected with active RalA. In vitro low concentrations of filamin promote high angle branching of actin filaments, whereas higher amounts organize parallel bundles of actin filaments as exist in filopodia (38, 39). The fact that the RalA protein increased the cytoskeletal filamin, but not the total amount of crosslinked cytoskeletal actin filaments, suggests one possible mechanism for filopod formation in which active RalA increases the concentration of filamin at cellular sites where Cdc42 and N-WASP recruit actin filament fragments (6, 7) for the filamin to crosslink into bundles. The ability of M2 cells to make filopodia at certain times in the absence of RalA stimulation or filamin expression supports the growing appreciation that the same cells can use more than one set of actin regulating machinery to change shape and move (5). Although binding of other small GTPases to filamin was not GTP dependent, they also may participate in the regulation of filamin’s interactions with actin and other binding partners.
Acknowledgments
We thank Seisuke Hattori for helpful discussions, Yuko Niino for subcloning cDNAs, Akemi Takayama for technical assistance, Jim McGrath for video microscopy, and Ikuroh Ohsawa for help with confocal microscopy. N.S. is on leave from the Third Department of Medicine, Nippon Medical School, Tokyo 113 and thanks Ichiji Wakabayashi for support. Y.O. is supported by the Science and Technology Agency of Japan. J.H.H. is supported by National Institutes of Health Grants HL54145, HL56252, HL56949, and HL56993. T.P.S. is supported by National Institutes of Health Grant HL19429 and a Clinical Research Professorship of the American Cancer Society.
ABBREVIATIONS
- GST
glutathione S-transferase
- GTPγS
guanosine 5′-γ-thiotriphosphate
- FITC
fluorescein isothiocyanate
References
- 1.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 2.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 3.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartwig J H, Bokoch G M, Carpenter C L, Janmey P A, Taylor L A, Toker A, Stossel T P. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 5.Azuma T, Witke W, Stossel T P, Hartwig J H, Kwiatkowski D J. EMBO J. 1998;17:1362–1370. doi: 10.1093/emboj/17.5.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki H, Sasaki T, Takai Y, Takenawa T. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 7.Suetsugu S, Miki H, Takenawa T. EMBO J. 1998;22:6516–6536. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feig L A, Urano T, Cantor S. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 9.Lee T, Feig L, Montell D J. Development (Cambridge, UK) 1996;122:409–418. doi: 10.1242/dev.122.2.409. [DOI] [PubMed] [Google Scholar]
- 10.Albright C A, Giddings B W, Liu J, Vito M, Weinberg R A. EMBO J. 1993;12:339–347. doi: 10.1002/j.1460-2075.1993.tb05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlin J, Yamin R, Egan S, Stewart M, Stossel T, Kwiatkowski D, Hartwig J. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takafuta T, Wu G, Murphy G F, Shapiro S S. J Biol Chem. 1998;273:17531–17538. doi: 10.1074/jbc.273.28.17531. [DOI] [PubMed] [Google Scholar]
- 13.Xu W, Xie Z, Chung D W, Davie E W. Blood. 1998;92:1268–1276. [PubMed] [Google Scholar]
- 14.Xie Z, Xu W, Davie E W, Chung D W. Biochem Biophys Res Commun. 1998;251:914–919. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- 15.Maestrini E, Patrosso C, Mancini M, Rivella S, Rocchi M, Repetto M, Villa A, Frattini A, Zoppé M, Vezzoni P, Toniolo D. Hum Mol Genet. 1993;2:761–766. doi: 10.1093/hmg/2.6.761. [DOI] [PubMed] [Google Scholar]
- 16.Brotschi E, Hartwig J, Stossel T. J Biol Chem. 1978;253:8988–8993. [PubMed] [Google Scholar]
- 17.Fox J, Goll D, Reynolds C, Phillips D. J Biol Chem. 1985;260:1060–1066. [PubMed] [Google Scholar]
- 18.Ohta Y, Stossel T P, Hartwig J H. Cell. 1991;67:275–282. doi: 10.1016/0092-8674(91)90179-3. [DOI] [PubMed] [Google Scholar]
- 19.Edwards D N, Towb P, Wasserman S A. Development (Cambridge, UK) 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 20.Marti A, Luo Z, Cunningham C, Ohta Y, Hartwig J, Stossel T P, Kyriakis J M, Avruch J. J Biol Chem. 1997;272:2620–2628. doi: 10.1074/jbc.272.5.2620. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Thomas L, Warren R A, Enns C A, Cunningham C C, Hartwig J H, Thomas G. J Cell Biol. 1997;139:1719–1733. doi: 10.1083/jcb.139.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaff M, Liu S, Erle D J, Ginsberg M H. J Biol Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham C C, Gorlin J B, Kwiatkowski D J, Hartwig J H, Janmey P A, Byers R, Stossel T P. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 24.Fox J W, Lampert E D, Eksioglu Y, Hong S E, Scheffer I E, Dobyns W B, Hirsch B A, Radtke R A, Berkovic S F, Huttenlocher P R, Walsh C A. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 25.Ohta Y, Hartwig J H. J Biol Chem. 1996;271:11858–11864. doi: 10.1074/jbc.271.20.11858. [DOI] [PubMed] [Google Scholar]
- 26.Yada Y, Okano Y, Nozawa Y. Biochem Biophys Res Commun. 1990;172:256–261. doi: 10.1016/s0006-291x(05)80202-5. [DOI] [PubMed] [Google Scholar]
- 27.Ueda M, Oho C, Takisawa H, Ogihara S. Eur J Biochem. 1992;203:347–352. doi: 10.1111/j.1432-1033.1992.tb16556.x. [DOI] [PubMed] [Google Scholar]
- 28.Janmey P A, Hvidt S, Lamb J, Stossel T P. Nature (London) 1990;345:89–92. doi: 10.1038/345089a0. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Martin G A, Viskochil D, Bollag G, McCabe P C, Crosier W J, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon R M, et al. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 32.Warne P H, Viciana P R, Downward J. Nature (London) 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham C C. J Cell Biol. 1995;129:1589–1599. doi: 10.1083/jcb.129.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer S C, Zuerbig S, Cunningham C C, Hartwig J H, Bissell T, Gardner K, Fox J E B. J Biol Chem. 1997;272:2914–2919. doi: 10.1074/jbc.272.5.2914. [DOI] [PubMed] [Google Scholar]
- 35.Cantor S B, Urano T, Feig L A. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jullien-Flores V, Dorseuil O, Romeo F, Letoiurneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis J H. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 37.Park S-H, Weinberg R A. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 38.Hartwig J, Tyler J, Stossel T. J Cell Biol. 1980;87:841–848. doi: 10.1083/jcb.87.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartwig J, Stossel T. J Mol Biol. 1981;145:563–581. doi: 10.1016/0022-2836(81)90545-3. [DOI] [PubMed] [Google Scholar]