Abstract
The field of inositol signaling has expanded greatly in recent years. Given the many reviews on phosphoinositide kinases, we have chosen to restrict our discussion to inositol lipid hydrolysis focused on the phosphatases and a brief mention of the lipase isoforms. We also discuss recent discoveries that link mutations in phosphoinositide phosphatases to disease.
Keywords: membrane trafficking, lipid code, kinase, myotubularin, Lowe syndrome, Charcot-Marie-Tooth disease, Jobert syndrome
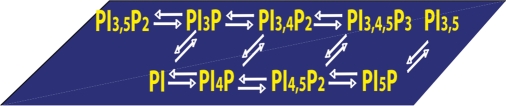
There are eight known phosphoinositides (also known as phosphatidylinositol phosphates or PIPs) with phosphate linked to diacylglycerol (DAG) and monoester phosphates in every possible combination of the 3, 4, and 5 positions of the inositol ring as shown in Fig. 1. Budding yeast have most of the PIPs, including PI(3)P, PI(4)P, PI(3,5)P2, and PI(4,5)P2, consistent with their evolutionarily conserved functions. The inositol phospholipids are interconverted by kinases and phosphatases as well as cleaved by phospholipase C (PLC) enzymes.
Fig. 1.
Inositol lipid regulatory pathways. Seven individual phosphoinositides comprise a “lipid code”. Forward and reverse arrows indicate kinase and phosphatase reactions. Nomenclature of PIxPn: x designates the inositol ring positions that are phosphorylated, whereas n refers to the total number phosphomonoesters.
A concept in the field has emerged in which each PIP has a distinct role: a so-called “lipid code” hypothesis. The lipid code postulates that distinct lipids mark each of the cellular membranes to maintain an orderly flow required for the complexities of membrane trafficking and the spatio-temporal signaling reaction. For example, Golgi membranes are enriched in PI(4)P, endosomal membranes are decorated with PI(3)P, and the plasma membrane with PI(4,5)P2.
Among the most well-studied codes are the PIPs harboring a D-3 phosphate, including PI(3,4,5)P3 (commonly referred to as PIP3), PI(3,4)P2, PI(3,5)P2 and PI3P. Importantly, the D-3 class of lipids are not substrates for PLC enzymes. This, along with the work showing that stimulation of cells by many growth factors activates receptor-linked phosphatidylinositol 3-kinases to transiently generate PIP3, helped secure the roles of inositol lipids as signaling molecules in their own right. While PI(3,4)P2 was considered as the breakdown product of PI(3,4,5)P3, evidence points to a distinct role for this lipid in cell signaling. Remarkably, interpretation of the inositol lipid code has rested on hundreds of recently discovered lipid binding protein domains, which are found attached to numerous signaling proteins. There are about a half-dozen classes of lipid binding domains, for example PH, FYVE, and PX domains. Overall, the complexity of lipid signaling has exceeded expectations, and its importance is underscored by the identification of disease states that arise from mutations in these enzymes. Many of these diseases result from defects in inositol lipid hydrolysis. In the following pages, we will outline the diseases and describe the hydrolytic enzymes responsible for regulating these important chemical messengers within cells.
PATHOLOGY RESULTING FROM DERANGED INOSITOL SIGNALING
The significance of inositol signaling with respect to human health is highlighted by the fact that mutations in enzymes of inositol signaling cause numerous diseases and pathologies (Table 1). Mutation of the gene encoding a PIP2 5-phosphatase (5-ptase) (OCRL) in humans causes a severe X-linked disorder called Lowe syndrome, while the OCRL knockout mouse has no phenotype (1). Mutation of myotubularin (MTM) causes a severe and fatal disorder associated with a failure of skeletal muscle development. Subsequently, it has been shown that MTM is the founding member of a large family of phosphatidylinositol 3-phosphate phosphatases, designated now as MTM-related proteins (MTMR1-13). MTMR1-8 have 3-phosphatase activity, while MTMR9-13 lack the catalytic active site cysteine residue. The inactive subunits partner with active MTM proteins to form heteromers, which increase enzyme activity and in some cases alter cellular location. The importance of the inactive subunits is illustrated by the finding that mutations of MTMR13, the inactive partner of MTMR2, cause the same disorder as mutation of the active partner. Mutations in factor-induced gene 4 (FIG4) in mice cause the pale tremor mouse syndrome, while mutation of the human ortholog causes a form of Charcot Marie Tooth disease. Similarly, different types of mutation of the type IV inositol polyphosphate 5-ptase produce discrete disorders. Mutations that truncate the last 18 amino acids result in MORM syndrome, while mutations in the catalytic domain cause Jobert's syndrome.
TABLE 1.
Disorders and pathologies associated with mutations in inositol lipid phosphatases
| Enzyme | Disorder | Species | Symptoms | Reference |
|---|---|---|---|---|
| PTEN | Cancer | Human | Many tumor types | (10) |
| Inositolpolyphosphate 5-ptase (OCRL) | Lowe Syndrome | Human | X-linked renal failure, mental retardation, blindness | (4) |
| Inositol polyphosphate 4-phosphatase I | Weeble mouse | Mouse | Ataxia, cerebellar degeneration, early death | (7, 8) |
| Phosphatidyl inositol 3-phosphate phosphatase (MTM) | Myotubular myopathy | Human | Lack of muscle development, respiratory failure | (50) |
| MTMR2 | Charcot Marie Tooth disease type 4B | Human | Neurodegeneration | (20) |
| MTMR13 | Charcot Marie Tooth disease type 4B | Human | Neurodegeneration | (20) |
| FIG4 5-ptase on phosphatidyl inositol 3,5-bisphosphate | Pale tremor mouse | Mouse | Neuronal degeneration, early death | (19) |
| FIG4 5-ptase on phosphatidyl inositol 3,5-bisphosphate | Charcot Marie Tooth disease type 4 J | Human | Neurodegeneration | (19) |
| Inositolpolyphosphate 5-ptase type IV | MORM syndrome | Human | Mental retardation, micropenis | (51) |
| Inositolpolyphosphate 5-ptase type IV | Jobert's syndrome | Human | Hypotonia, ataxia, retardation | (52, 53) |
ENZYMES THAT REGULATE LEVELS OF CELLULAR PIPS
PIP Selective Phospholipases
PLC enzymes are phosphodiesterases that upon stimulation cleave PIPs to produce soluble inositol phosphates and DAG. A number of PLCs have been identified in mammalian cells. These PLCs differ from a bacterial phosphatidylinositol (PI)-specific PLC in their absolute requirement for calcium and in their ability to hydrolyze phosphorylated forms of PI (PIP and PIP2). Based on comparison of their sequences and structural studies, mammalian PLCs are divided into five families: PLCβ, PLCγ, PLCδ, PLCɛ, and PLCζ (2). The PLCδ class is conserved from yeast to man, whereas the others appear primarily in metazoans. The PLCγ-isoforms are activated by receptor tyrosine kinases, whereas the PLCβ-class are activated by trimeric G-protein coupled receptors, typically Gαq.
PIP 5-ptase
The most abundant and perhaps best studied of the inositol lipid phosphatases are the 5-ptase family (3). 5-Ptases were first identified based on their ability to terminate I(1,4,5)P3-mediated calcium release, because the product I(1,4)P2 is unable to mobilize calcium. As more gene products were discovered, it became apparent that many, including all those in Saccharomyces cerevisiae, have a preference for breaking down PIPs rather than their soluble counterparts. Over 10 distinct mammalian and 4 yeast 5-ptases have been identified. All share magnesium-dependent phosphomonoesterase activities and contain two signature motifs: (F/I)WxGDxN(F/Y)R and (R/N)xP(S/A)(W/Y)(C/T)DR(I/V)(L/I). These motifs are critical for substrate binding and catalysis.
5-Ptases cay be divided into four groups according to their substrate selectivity. Group I 5-ptases hydrolyze only the water-soluble substrates I(1,4,5)P3 and I(1,3,4,5)P4. These enzymes are membrane-associated through isoprenylation and function to terminate calcium signaling. Group II enzymes hydrolyze both water-soluble and lipid substrates and they are also membrane-associated proteins. OCRL, the protein mutated in Lowe (oculocerebrorenal) Syndrome, is a member in this group (4). Another family member, synaptojanin (discussed below), is involved in synaptic vesicle trafficking (5). Group III enzymes hydrolyze mainly lipid substrates. SHIP1 and SHIP2 are members in this group and have been shown to regulate cytokine signaling in hematopoietic cells and insulin signaling (6). Group IV 5-ptases hydrolyze only PIP3 and PI(4,5)P2 and form complexes with PI 3-kinase.
PIP 4-Phosphatases
These enzymes commonly referred to as 4-ptases were originally identified as hydrolases for inositol 1,3,4-trisphosphate and inositol 3,4-bisphosphate. Upon cloning and further biochemical characterization, it became apparent that the 4-ptase enzymatic activity may be most relevant to hydrolysis of the D-4 position phosphate from the second messenger PI(3,4)P2, yielding PI(3)P as a product (7, 8). Therefore, it is now thought that a major function for 4-ptases is to terminate the PI(3,4)P2 signal. Enzymes in the 4-ptase family, types I and II, all have a Cx5R(S/T) motif where the cysteine is critical for catalysis. All these enzymes are metal independent. Loss of 4-ptase activity results in neurological phenotypes highlighted by the discovery that the “weeble” mouse harbors a causative mutation in the type I gene (8).
The PTEN Class of Lipid Phosphatases
PTEN, a tumor suppressor, is found mutated in a variety of human cancers, including breast, prostate, and brain cancer. PTEN contains the Cx5R(S/T) and was originally thought to be a dual-specificity serine/threonine and tyrosine protein phosphatase. Later it was discovered that PTEN catalyzes the dephosphorylation of PIP3 at the D-3 position to produce PI(4,5)P2 as product (9). In this way, PTEN functions as a PIP3 3-phosphatase to negatively control the PI 3-kinase signaling pathway. The Cx5R(S/T) active site motif shared with the 4-ptase family provides a strong evolutionary link to other lipid phosphatases and, as described below, the MTM and Sac1-like proteins. The biology and role of PTEN in human cancers has been reviewed in detail elsewhere (9, 10).
Promiscuous PIP Phosphatases: the Sac1-Like Proteins
The Sac1-like class of enzymes encode “multi” phosphatase activities that cleave the D-3, D-4, and D-5 position phosphates from PI(3)P, PI(4)P, PI(3,5)P2, and PI(3,4)P2 to produce PI (11). The promiscuity of the Sac1-like activities has confounded the interpretation of which lipid substrate is most physiologically relevant. However, the observed changes in specific PIPs in Sac1-like mutants, for example, selective elevations in PI(4)P and PI(3,5)P2 in sac1 and fig4 mutants, indicates that individual proteins may prefer distinct lipid substrates in cells (11–14). Alternatively, it has also been proposed that the broad specificity of Sac1-like enzymes functions as a general mechanism to clear multiple signaling PIPs, thereby enabling lipid recycling, neutrality of membranes, and/or termination of many distinct signals.
There are several gene products designated to be Sac1-like, including three in mammals and four in budding yeast (11, 15).
The active site motif of the 400-amino acid Sac1-like domains is Cx5R and they fall into two groups based on their domain structure. Group I gene products, including Sac1 and Fig4 and their homologs in higher eukaryotes, have a single Sac1-like domain (15). Group II proteins, including the synaptojanin class of proteins in mammals and yeast Sjl2/Inp52 and Sjl3/Inp53, are dual-functional lipid phosphatases that harbor both a Sac1-like domain and 5-ptase domain (discussed in more detail below).
Functional studies of the Sac1-like proteins suggest roles in a number of cellular processes. Sac1 is a transmembrane protein enriched in endoplasmic reticulum (ER)-Golgi fractions and is involved in ATP transport into the ER, growth factor signaling, and cell shape (13). Loss of sac1 in yeast genetically links to overcoming defects in both actin and secretory mutants (16, 17) and in mice results in early embryonic lethality (18). Mutations in FIG4 in mice cause the pale tremor mouse syndrome, while mutation of the human ortholog causes a form of Charcot Marie Tooth disease (19, 20) (Table 1). Budding yeast Fig4 collaborates with the Fab1 lipid kinase and the regulatory proteins Vac7 and Vac14 to regulate lipid synthesis, vacuole biogenesis/dynamics, and endosomal transport (12, 14).
The PIP substrates hydrolyzed by Sac1-like domains have been implicated in the regulation of membrane trafficking (reviewed in (20–23). PI(4)P is synthesized by the PI 4-kinases, Pik1 in yeast and PI4Kβ in mammals, and is required for the formation of secretory vesicles from the late-Golgi (23, 24). Pik1 mutants are defective for protein secretion from the Golgi, vacuolar protein maturation, endocytosis, and lipid homeostasis. Pik1 also controls an essential nuclear pool of PIPs (23). A distinct non-Golgi pool of PI(4)P is produced by the PI 4-kinase, Stt4. Production of this PI(4)P is required for actin polarity, cell wall integrity, and phosphatidylserine metabolism (23, 25). Stt4 also fuels PI(4,5)P2 production at the plasma membrane through the Stt4-Mss4 [a PI(4)P 5-kinase] pathway. The pleckstrin homology (PH) domain found in FAPP1 specifically binds PI(4)P and, along with other lipid binding domains, participates in decoding this messenger (26).
Components of the PI(3)P pathway are found in many eukaryotes, suggesting a conserved role of PI(3)P in protein transport from Golgi to the endosomal system (27). Interpretation of these signals occurs through FYVE domain containing proteins, which recognize PI(3)P, and the endosomal sorting complex required for transport (ESCRT) (22, 28). Additionally, the lipid binding properties of certain PX and PH domains mediate interactions of proteins with PI(3)P (29).
Another important role for PI(3)P is to serve as the precursor for PI(3,5)P2, an osmotic stress-induced lipid regulator of multivesicular body endosomal pathways (14, 30) produced through the action of PI(3)P 5-kinases, Fab1 in yeast and PIKfyve in mammalian cells (31, 32).
The MTM 3-Phosphatases
The MTMs are a large family of 13 proteins, 8 of which have catalytic activity. These enzymes act to dephosphorylate PI(3)P and PI(3,5)P2 (20, 33). Six members of the family are not active, because they lack the cysteine residue in the Cx5R(S/T) PTP motif. Despite having nearly identical catalytic activity, biochemical and genetic evidence supports the hypothesis that the MTMs are not redundant and have unique functions within cells. Current evidence suggests that each MTM protein regulates a specific pool of PI(3)P and/or PI(3,5)P2 that serves a variety of cellular functions.
The functions of MTM proteins are altered by the formation of heteromers between active and inactive subunits. The initial purification of MTM1 demonstrated the presence of MTM1 homodimers and MTM1/3-ptase adaptor protein (3PAP) heteromers (34). 3PAP was later designated as MTMR12. MTMR2 partners with MTMR5, and MTMR9 forms heteromers with MTMR6, MTMR7, and MTMR8. Heteromer formation has two major effects: 1) increased activity of the enzymatic unit; and 2) altered subcellular location. Both PI(4)P and PI(5)P, nonsubstrate inositides, cause an allosteric changes in the enzyme that increase activity (35).
Dual-Functional Lipid Phosphatases: the Synaptojanin Family
Synaptojanin was originally identified as a synaptic vesicle-localized PIP2 5-ptase involved in synaptic vesicle recycling (5). Its name was derived from Janus, the God of two faces, based on links of the N-terminal Sac1-like domain to actin/secretion and a 5-ptase domain to PIPs. Subsequent studies showed that the synaptojanin-class of proteins had the ability to hydrolyze a variety of PIPs to PI (11, 36). The molecular basis for the “multi” phosphatase activity was revealed to be autonomously encoded by the 5-ptase and Sac1-like domains (11), which function independently to dephosphorylate PI(4,5)P2 and PI(3,4,5)P3 (5-ptase substrates), as well as PI(3)P, PI(4)P, PI(3,4)P2, and PI(3,5)P2 (Sac1-like substrates).
Two isoforms of mammalian synaptojanin, synaptojanin-1 and -2, along with several splice variants have been characterized (21). Genetic studies in mice, worms, and yeast have established roles for synaptojanin-enzymes in vesicle recycling, endocytosis, and actin cytoskeleton. The PI(4,5)P2 5-ptase activity of synaptojanin has been implicated in modulating the clathrin uncoating process during synaptic vesicle recycling (21, 37) and in control of lipid/actin regulatory protein interactions (38). A broader role for synaptojanin in multiple steps of synaptic vesicle recycling, budding, uncoating, and tethering to the cytoskeleton has been identified from studies in worms (37). A recent study has indicated a role for coordinated regulation of the two activities providing a basis for their tethering on a single peptide chain (39).
PI(4,5)P2 4-Phosphatases
The most recently discovered of the seven known PIPs is phosphatidylinositol 5-ptase, PI(5)P. Analysis of the changes in the cellular levels of PI(5)P suggested that it is synthesized by the action of a phosphatase rather than by a kinase (40). The discovery of two 4-phosphatases that convert PI(4,5)P2 to PI(5)P provides a molecular route for the synthesis of this lipid (41).
Recently, it was suggested that PI(5)P specifically interacts with a plant homeodomain (PHD) finger of inhibitor of growth protein-2 (ING2) protein and that this interaction is required for ING2-dependent acetylation of p53, which leads to increased apoptosis (42). This suggestion was based on the finding that RNAi of ING2 or overexpression of the phosphatidylinositol phosphate kinase (PIPK) type IIβ, an enzyme that converts PI(5)P to PI(4,5)P2, decreases apoptosis. Thus, it was presumed that both ING2 and PI(5)P were required for acetylation of p53, although cellular PI(5)P was not measured in that study. Cells overexpressing type I 4-ptase have elevated levels of PI(5)P (43). When HeLa cells are treated with the DNA-damaging agents etoposide or doxorubicin, type I 4-ptase translocate to the nucleus and nuclear levels of PI(5)P increase. Overexpression of type I 4-ptase increased apoptosis, which is inhibited by cotransfection of PIPk IIβ. This enzyme therefore controls levels of PI(5)P and thereby p53-dependent apoptosis.
PIPS AND THEIR EFFECTOR PROTEINS
The decoding of PIP signals occurs through hundreds of lipid binding domains that are typically found attached to a variety of signaling modules, such as protein kinases, protein phosphatases, adaptor molecules, and small GTPases. Lipid binding domains are typically 50–120 amino acids in length and fall into a few common protein folds (reviewed in (29). Among the modules that have been characterized to bind inositol lipids are: pleckstrin-homology (PH), PX, C2, SH2, protein tyrosine binding, FYVE, PHD, GRAM, BAR, and ENTH/ANTH domains that collectively represent some of the largest families of domains within the proteome (29). While some of these domains are highly selective for a particular lipid, others bind a broad range of phospholipids with relatively weak affinity. Several examples exist in which various modules are fused, thereby generating a coincidence detector capable of being activated by combinations of lipids.
It is noteworthy that some of the lipid binding modules also bind the water-soluble inositol phosphates. For example, the PH domain of PLCδ binds PI(4,5)P2 and its head group I(1,4,5)P3 with high specificity. It has been proposed that such properties would enable another layer of regulation and/or termination by a competition model in which production of soluble molecules may compete the PH-containing protein off membranes. Lastly, when some modules dimerize, such as the BAR domains, they form a concave surface that may provide a template for membrane curvature. Different BAR domains have different curvature radii, possibly explaining in part their ability to discriminate among different sized vesicles or cellular membranes.
The lipid binding domain serves to impart a membrane-targeting signal that in many cases depends on the transient synthesis of a PIP, thereby enabling recruitment to specific locations within the cell and assuring a spatio-temporal mode to signaling biology. Two well-characterized lipid binding domains are the PH domain of Akt/PKB and the so-called FYVE-finger, named after the first four proteins identified to contain this structure: Fab1p, YOTB, Vac1p, and EEA1. The FYVE domains consist of 60–80 amino acids and harbor 2 zinc-finger motifs that specifically recognize PI(3)P [reviewed in (44)]. Several proteins involved in the ESCRT pathway that is required for multivesicular body formation utilize FYVE domains (22, 28).
Akt/PKB is critical for cell growth and survival through its binding to both PI(3,4)P2 and PIP3 in vitro, independent of PI(4,5)P2 [reviewed in (45, 46)]. PI(3,4)P2 and PIP3 activate PKB/Akt by spatio-temporal synthesis of 3-phosphorylated lipids that leads to the translocation of Akt/PKB from the cytosol to the membrane in stimulated cells that primes Akt/PKB for its phosphorylation and activation by PIP-dependent kinase 1. Akt/PKB may also require intrinsic binding of these 3-phosphorylated lipids to be fully active.
Recent biochemical, electrophysiological, and molecular work has also shown that PIPs play a key role in regulating the intrinsic properties of transmembrane proteins, such as ion channels and vesicle fusion machinery (47, 48). Using molecular approaches, specific lipid phosphatases have been spatio-temporally activated to manipulate the levels of PIPs at specific membranes within cells. For example, methods in which PI(4,5)P2 selective 5-ptase proteins have been inducibly targeted to cellular membranes have highlighted roles for PI(4,5)P2 in membrane adhesion, ion channel gating activity, membrane trafficking, receptor internalization, and many other cellular processes (47, 49).
A technological benefit of the discovery of lipid binding domains has been their use as illuminators of lipid synthesis in living cells when fused to green fluorescent protein (GFP). GFP-tagged PKB/Akt PH and GFP-PH (PLCδ) are useful probes for marking PIP3 and PI(4,5)P2 levels in real time. One note of caution when using these probes is that some lipid binding modules appear to require more than PIP binding to translocate to membranes, such as protein-protein interactions. Nonetheless, the ability to monitor lipids in response to cellular signals has been a major advance.
CONCLUDING REMARKS
Overall, the complexity of PIP signaling has exceeded expectations. The link of in-born errors in PIP metabolism to vertebrate disease states highlights the relevance and significance of the field to human health. Studies of proteins dedicated to the regulation and interpretation of PIP signals have provided several new insights into fundamental questions in biology related to signaling specificity. While this review covers only a small portion of the field, we hope the readers glean a sense that inositol lipid metabolism and signaling remains a tremendously active area of research full of many surprises.
Abbreviations
DAG, diacylglycerol
ER, endoplasmic reticulum
ESCRT, endosomal sorting complex required for transport
GFP, green fluorescent protein
ING2, inhibitor of growth protein-2
MTM, myotubularin
MTMR, myotubularin-related protein
3PAP, 3-ptase adaptor protein
PHD, plant homeodomain
PIP, phosphoinositide
PLC, phospholipase C
5-ptase, 5-phosphatase
This work is supported by funds from the Howard Hughes Medical Institute (J.D.Y.), and from the National Institutes of Health HL-55672 (J.D.Y.) and HL-016634 (P.W.M.)
Published, JLR Papers in Press, November 11, 2008.
References
- 1.Janne P. A., S. F. Suchy, D. Bernard, M. MacDonald, J. Crawley, A. Grinberg, A. Wynshaw-Boris, H. Westphal, and R. L. Nussbaum. 1998. Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J. Clin. Invest. 101 2042–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suh P. G., J. I. Park, L. Manzoli, L. Cocco, J. C. Peak, M. Katan, K. Fukami, T. Kataoka, S. Yun, and S. H. Ryu. 2008. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41 415–434. [DOI] [PubMed] [Google Scholar]
- 3.Astle, M. V., K. A. Horan, L. M. Ooms, and C. A. Mitchell. 2007. The inositol polyphosphate 5-phosphatases: traffic controllers, waistline watchers and tumour suppressors? Biochem. Soc. Symp. 161–181. [DOI] [PubMed]
- 4.Attree O., I. M. Olivos, I. Okabe, L. C. Bailey, D. L. Nelson, R. A. Lewis, R. R. McInnes, and R. L. Nussbaum. 1992. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 358 239–242. [DOI] [PubMed] [Google Scholar]
- 5.McPherson P. S., E. P. Garcia, V. I. Slepnev, C. David, X. Zhang, D. Grabs, W. S. Sossin, R. Bauerfeind, Y. Nemoto, and P. De Camilli. 1996. A presynaptic inositol-5-phosphatase. Nature. 379 353–357. [DOI] [PubMed] [Google Scholar]
- 6.Backers K., D. Blero, N. Paternotte, J. Zhang, and C. Erneux. 2003. The termination of PI3K signalling by SHIP1 and SHIP2 inositol 5-phosphatases. Adv. Enzyme Regul. 43 15–28. [DOI] [PubMed] [Google Scholar]
- 7.Norris F. A., R. C. Atkins, and P. W. Majerus. 1997. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splicing in the 4-phosphatase family. J. Biol. Chem. 272 23859–23864. [DOI] [PubMed] [Google Scholar]
- 8.Nystuen A., M. E. Legare, L. D. Shultz, and W. N. Frankel. 2001. A null mutation in inositol polyphosphate 4-phosphatase type I causes selective neuronal loss in weeble mutant mice. Neuron. 32 203–212. [DOI] [PubMed] [Google Scholar]
- 9.Maehama T., G. S. Taylor, and J. E. Dixon. 2001. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70 247–279. [DOI] [PubMed] [Google Scholar]
- 10.Yuan T. L., and L. C. Cantley. 2008. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 27 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S., L. E. Stolz, S. M. Lemrow, and J. D. York. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274 12990–12995. [DOI] [PubMed] [Google Scholar]
- 12.Duex J. E., F. Tang, and L. S. Weisman. 2006. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 172 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blagoveshchenskaya A., F. Y. Cheong, H. M. Rohde, G. Glover, A. Knodler, T. Nicolson, G. Boehmelt, and P. Mayinger. 2008. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 180 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botelho R. J., J. A. Efe, D. Teis, and S. D. Emr. 2008. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol. Biol. Cell. 19 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes W. E., F. T. Cooke, and P. J. Parker. 2000. Sac phosphatase domain proteins. Biochem. J. 350 337–352. [PMC free article] [PubMed] [Google Scholar]
- 16.Cleves A. E., P. J. Novick, and V. A. Bankaitis. 1989. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novick P., B. C. Osmond, and D. Botstein. 1989. Suppressors of yeast actin mutations. Genetics. 121 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., M. Boukhelifa, E. Tribble, E. Morin-Kensicki, A. Uetrecht, J. E. Bear, and V. A. Bankaitis. 2008. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol. Biol. Cell. 19 3080–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow C. Y., Y. Zhang, J. J. Dowling, N. Jin, M. Adamska, K. Shiga, K. Szigeti, M. E. Shy, J. Li, X. Zhang, et al. 2007. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 448 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicot A. S., and J. Laporte. 2008. Endosomal phosphoinositides and human diseases. Traffic. 9 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Paolo G., and P. De Camilli. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443 651–657. [DOI] [PubMed] [Google Scholar]
- 22.Hurley J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strahl T., and J. Thorner. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1771 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heck J. N., D. L. Mellman, K. Ling, Y. Sun, M. P. Wagoner, N. J. Schill, and R. A. Anderson. 2007. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit. Rev. Biochem. Mol. Biol. 42 15–39. [DOI] [PubMed] [Google Scholar]
- 25.Voelker D. R. 2005. Protein and lipid motifs regulate phosphatidylserine traffic in yeast. Biochem. Soc. Trans. 33 1141–1145. [DOI] [PubMed] [Google Scholar]
- 26.Dowler S., R. A. Currie, D. G. Campbell, M. Deak, G. Kular, C. P. Downes, and D. R. Alessi. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odorizzi G., M. Babst, and S. D. Emr. 2000. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25 229–235. [DOI] [PubMed] [Google Scholar]
- 28.Williams R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8 355–368. [DOI] [PubMed] [Google Scholar]
- 29.Lemmon M. A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9 99–111. [DOI] [PubMed] [Google Scholar]
- 30.Dove S. K., F. T. Cooke, M. R. Douglas, L. G. Sayers, P. J. Parker, and R. H. Michell. 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 390 187–192. [DOI] [PubMed] [Google Scholar]
- 31.Cooke F. T., S. K. Dove, R. K. McEwen, G. Painter, A. B. Holmes, M. N. Hall, R. H. Michell, and P. J. Parker. 1998. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr. Biol. 8 1219–1222. [DOI] [PubMed] [Google Scholar]
- 32.Sbrissa D., O. C. Ikonomov, and A. Shisheva. 1999. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274 21589–21597. [DOI] [PubMed] [Google Scholar]
- 33.Robinson F. L., and J. E. Dixon. 2006. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16 403–412. [DOI] [PubMed] [Google Scholar]
- 34.Caldwell K. K., D. L. Lips, V. S. Bansal, and P. W. Majerus. 1991. Isolation and characterization of two 3-phosphatases that hydrolyze both phosphatidylinositol 3-phosphate and inositol 1,3-bisphosphate. J. Biol. Chem. 266 18378–18386. [PubMed] [Google Scholar]
- 35.Clague M. J., S. K. Dove, and F. A. Barr. 2004. I-proteins–a proposed switch in myotubularin function. Trends Biochem. Sci. 29 58–61. [DOI] [PubMed] [Google Scholar]
- 36.Chung J. K., F. Sekiya, H. S. Kang, C. Lee, J. S. Han, S. R. Kim, Y. S. Bae, A. J. Morris, and S. G. Rhee. 1997. Synaptojanin inhibition of phospholipase D activity by hydrolysis of phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 272 15980–15985. [DOI] [PubMed] [Google Scholar]
- 37.Harris T. W., E. Hartwieg, H. R. Horvitz, and E. M. Jorgensen. 2000. Mutations in synaptojanin disrupt synaptic vesicle recycling. J. Cell Biol. 150 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakisaka T., T. Itoh, K. Miura, and T. Takenawa. 1997. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol. Cell. Biol. 17 3841–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mani M., S. Y. Lee, L. Lucast, O. Cremona, G. Di Paolo, P. De Camilli, and T. A. Ryan. 2007. The dual phosphatase activity of synaptojanin1 is required for both efficient synaptic vesicle endocytosis and reavailability at nerve terminals. Neuron. 56 1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts H. F., J. H. Clarke, A. J. Letcher, R. F. Irvine, and K. A. Hinchliffe. 2005. Effects of lipid kinase expression and cellular stimuli on phosphatidylinositol 5-phosphate levels in mammalian cell lines. FEBS Lett. 579 2868–2872. [DOI] [PubMed] [Google Scholar]
- 41.Ungewickell A., C. Hugge, M. Kisseleva, S. C. Chang, J. Zou, Y. Feng, E. E. Galyov, M. Wilson, and P. W. Majerus. 2005. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc. Natl. Acad. Sci. USA. 102 18854–18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gozani O., P. Karuman, D. R. Jones, D. Ivanov, J. Cha, A. A. Lugovskoy, C. L. Baird, H. Zhu, S. J. Field, S. L. Lessnick, et al. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 114 99–111. [DOI] [PubMed] [Google Scholar]
- 43.Zou J., J. Marjanovic, M. V. Kisseleva, M. Wilson, and P. W. Majerus. 2007. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. USA. 104 16834–16839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kutateladze T. G. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta. 1761 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brazil D. P., J. Park, and B. A. Hemmings. 2002. PKB binding proteins. Getting in on the Akt. Cell. 111 293–303. [DOI] [PubMed] [Google Scholar]
- 46.Toker A., and M. Yoeli-Lerner. 2006. Akt signaling and cancer: surviving but not moving on. Cancer Res. 66 3963–3966. [DOI] [PubMed] [Google Scholar]
- 47.Suh B. C., T. Inoue, T. Meyer, and B. Hille. 2006. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 314 1454–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James D. J., C. Khodthong, J. A. Kowalchyk, and T. F. Martin. 2008. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J. Cell Biol. 182 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stauffer T. P., S. Ahn, and T. Meyer. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8 343–346. [DOI] [PubMed] [Google Scholar]
- 50.Taylor G. S., T. Maehama, and J. E. Dixon. 2000. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA. 97 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hampshire D. J., M. Ayub, K. Springell, E. Roberts, H. Jafri, Y. Rashid, J. Bond, J. H. Riley, and C. G. Woods. 2006. MORM syndrome (mental retardation, truncal obesity, retinal dystrophy and micropenis), a new autosomal recessive disorder, links to 9q34. Eur. J. Hum. Genet. 14 543–548. [DOI] [PubMed] [Google Scholar]
- 52.Kisseleva M. V., M. P. Wilson, and P. W. Majerus. 2000. The isolation and characterization of a cDNA encoding phospholipid-specific inositol polyphosphate 5-phosphatase. J. Biol. Chem. 275 20110–20116. [DOI] [PubMed] [Google Scholar]
- 53.Zaki M. S., A. Abdel-Aleem, G. Abdel-Salam, S. E. Marsh, J. L. Silhavy, A. J. Barkovich, M. E. Ross, S. N. Saleem, W. B. Dobyns, and J. G. Gleeson. 2008. The molar tooth sign: a new Joubert syndrome and related cerebellar disorders classification system tested in Egyptian families. Neurology. 70 556–565. [DOI] [PubMed] [Google Scholar]