Abstract
In 2005, the International Lipid Classification and Nomenclature Committee under the sponsorship of the LIPID MAPS Consortium developed and established a “Comprehensive Classification System for Lipids” based on well-defined chemical and biochemical principles and using an ontology that is extensible, flexible, and scalable. This classification system, which is compatible with contemporary databasing and informatics needs, has now been accepted internationally and widely adopted. In response to considerable attention and requests from lipid researchers from around the globe and in a variety of fields, the comprehensive classification system has undergone significant revisions over the last few years to more fully represent lipid structures from a wider variety of sources and to provide additional levels of detail as necessary. The details of this classification system are reviewed and updated and are presented here, along with revisions to its suggested nomenclature and structure-drawing recommendations for lipids.
Keywords: lipidomics, nomenclature, structure, databases
In an effort to support the growing field of lipidomics and establish the importance of lipids as a major class of biomolecules, the International Lipid Classification and Nomenclature Committee (ILCNC) developed a “Comprehensive Classification System for Lipids” that was published in 2005 (1). For the purpose of classification, we define lipids as hydrophobic or amphipathic small molecules that may originate entirely or in part by carbanion-based condensations of thioesters (fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, saccharolipids, and polyketides) and/or by carbocation-based condensations of isoprene units (prenol lipids and sterol lipids). The comprehensive classification system organizes lipids into these eight well-defined categories (Table 1) that cover eukaryotic and prokaryotic sources. It has been adopted internationally and widely accepted by the lipidomics community. The system is also available online on the LIPID MAPS (2) website (http://www.lipidmaps.org). The comprehensive classification system has been under the guidance of the ILCNC,3 which meets periodically to propose changes and updates to classification, nomenclature, and structural representation.
TABLE 1.
Lipid categories of the comprehensive classification system and the number of structures in the LIPID MAPS database
| Category | Abbreviation | Structures in Database |
|---|---|---|
| Fatty acyls | FA | 2678 |
| Glycerolipids | GL | 3009 |
| Glycerophospholipids | GP | 1970 |
| Sphingolipids | SP | 620 |
| Sterol Lipids | ST | 1744 |
| Prenol Lipids | PR | 610 |
| Saccharolipids | SL | 11 |
| Polyketides | PK | 132 |
The initial version of the comprehensive classification system was more heavily focused on mammalian lipids, reflecting a bias toward the experimental interests of the LIPID MAPS Consortium (2). However, due to considerable attention and requests from lipid researchers in a variety of fields, the classification system has now been extended to more fully represent lipid structures from nonmammalian sources, such as plants, bacteria, and fungi. For example, two new main classes (Glycosyldiradylglycerols and Glycosylmonoradylglycerols) have been added to the Glycerolipids category to accommodate key plant structural lipids, such as the sulfoquinovosyldiacylglycerols (3) found in chloroplasts. Also, the list of subclasses under the Sterols main class has been expanded to include a set of 15 different core structures (Ergosterols, Gorgosterols, Furostanols, etc.), which provide a structure-based classification of these molecules that span multiple phyla.
Another key development has been the adoption of existing hierarchies (4) for the Polyketide category and Prenol Lipids/Isoprenoids subclasses where the majority of these molecules are derived from natural product sources and have been studied intensively from a pharmaceutical and ecological standpoint. This in turn has necessitated the expansion of the number of existing classification levels (category, main class, and subclass) to accommodate an additional level of stratification in the case of the C10 to C30 isoprenoid subclasses that now contain entries at a fourth level of detail. The “LM_ID” identifier, whose format provides a systematic means of assigning a unique identification to each lipid molecule, has accordingly been expanded in length in these particular cases, with an additional two characters being used to describe the fourth level.
A detailed overview of the changes and updates to the comprehensive classification system is presented below.
CLASSIFICATION UPDATES
Expansion of LM_ID identifier
As a consequence of adding an extra level of classification detail, the length of the LM_ID identifier was lengthened from 12 characters to 14 in cases where a lipid defined with four levels of classification is being described (Table 2). In this case, characters 9 and 10 specify the level-4 class. It should be emphasized that all lipids that do not require a fourth level of detail (i.e., the vast majority of them) still use a 12-digit LM_ID identifier.
TABLE 2.
Format of LIPID MAPS identifier (LM_ID) in the comprehensive classification system
| Characters | Description | Example | Comments |
|---|---|---|---|
| 1–2 | Fixed “LM” designation | LM | Always LM |
| 3–4 | Two-letter category code | PR | One of eight categories |
| 5–6 | Two-digit class code | 01 | — |
| 7–8 | Two-digit subclass code | 03 | “00” when no subclass |
| 9–10 | Two-digit fourth level code | 06 | Only used for lipids with four levels |
| Last four digits | Unique four-character identifier within subclass or within fourth level | 0002 | First two of the last four digits are letters in the case of the Glycosphingolipid subclasses |
Removal of classes based on lipid source
In keeping with the theme of having a classification system dictated by molecular structure and function, the sterol lipid subclasses Phytosterols, Marine sterols, and Fungal sterols were retired because these refer to the lipid source (marine) or biological kingdom (plants and fungi). It is possible to identify a particular sterol in more than one of these three sources. These subclasses have been replaced by a new set of subclasses based on the carbon skeleton of the sterol core structure (Ergosterols, Gorgosterols, Furostanols, etc.). The details are outlined under the Sterol Lipids section below, and the complete description of this category can be found on the LIPID MAPS website4 (http://www.lipidmaps.org).
Adoption of existing natural products classification hierarchies for isoprenoids and polyketides
The natural products chemistry and medicinal chemistry literature describes tens of thousands of molecules that fall under the scope of lipids, based on their biosynthetic origin. In particular, isoprenoids and polyketides from diverse sources, such as plant, fungi, algae, bacteria, and marine invertebrates, are well documented and have been reviewed and classified in detail. The Dictionary of Natural Products (4), a database available from Chapman and Hall/CRC (http://dnp.chemnetbase.com), has a classification hierarchy that covers polyketides and isoprenoids in depth. The LIPID MAPS comprehensive classification system has now incorporated some of these hierarchies relevant to natural products, with a view to covering both mammalian and nonmammalian lipids comprehensively.3
Expansion of classification levels
It was recognized that additional levels of stratification were required to classify certain types of lipids and that the current three-level system of category/main class/subclass needed to be expanded. For example, in the Prenol Lipids category,3 the Sesquiterpene C15 subclass contains ∼90 known variants based on their carbon skeletons (Bisabolanes, Germacranes, etc.). A fourth level of detail has been added to the LIPID MAPS comprehensive classification system to handle cases such as these.
Additional classes and subclasses
In response to worldwide interest in the comprehensive classification system for lipids, the scope has been expanded to cover lipids from nonmammalian sources, such as plants, bacteria, fungi, algae, and marine organisms. To accomplish this, several new lipid classes have been added, such as fatty acyl glycosides, glycosyldiradylglycerols, and various sterol skeletons. The Polyketide category has also been revised comprehensively.3
NOMENCLATURE UPDATES
Changes to glycerophospholipid abbreviations
The nomenclature of lipids falls into two main categories: systematic names and common or trivial names. The latter includes abbreviations that are a convenient way to define acyl/alkyl chains in acylglycerols, sphingolipids, and glycerophospholipids and synonyms such as “phosphatidyl” for “glycerophospho.” The generally accepted guidelines for lipid systematic names have been defined by the International Union of Pure and Applied Chemists and the International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) Commission on Biochemical Nomenclature (http://www.chem.qmul.ac.uk/iupac/) (5–8). In response to several requests from knowledgeable lipid experts, abbreviations for Glycerophospholipid classes (see http://www.lipidmaps.org for GP category3) have been changed now in the comprehensive classification system to the more universally used two-letter “PC/PE/PS/PA/PI” format. Consequently, glycerophospholipids in the LIPID MAPS structure database and LIPID MAPS standards database as well as all the Glycerophospholipids drawing tools and mass spectrometry prediction tools have been updated to conform to this new abbreviation format (Table 3).
TABLE 3.
Changes in abbreviations for Glycerophospholipids in the comprehensive classification system
| Class | Synonym | Old | New |
|---|---|---|---|
| Glycerophosphocholines | Phosphatidylcholines | GPCho | PCa |
| Glycerophosphoethanolamines | Phosphatidylethanolamines | GPEtn | PE |
| Glycerophosphoserines | Phosphatidylserines | GPSer | PS |
| Glycerophosphoglycerols | Phosphatidylglycerols | GPGro | PG |
| Glycerophosphoglycerophosphates | Phosphatidylglycerol phosphates | GPGroP | PGP |
| Glycerophosphoinositols | Phosphatidylinositols | GPIns | PI |
| Glycerophosphoinositol monophosphates | Phosphatidylinositol phosphates | GPInsP | PIP |
| Glycerophosphoinositol bis-phosphates | Phosphatidylinositol bis-phosphates | GPInsP2 | PIP2 |
| Glycerophosphoinositol tris-phosphates | Phosphatidylinositol tris-phosphates | GPInsP3 | PIP3 |
| Glycerophosphates | Phosphatidic acids | GPA | PA |
| Glyceropyrophosphates | GPP | PPA | |
| Glycerophosphoglycerophosphoglycerols | Cardiolipins | CL | CL |
| CDP-glycerols | GCDP | CDP-DG | |
| Glycosylglycerophospholipids | [glycan]GP | [glycan]GP | |
| Glycerophosphoinositolglycans | [glycan]GPIns | [glycan]PI | |
| Glycerophosphonocholines | GPnCho | PnC | |
| Glycerophosphonoethanolamines | GPnEtn | PnE |
For abbreviations of monoradyglycerophospholipids (lysophospholipids), LPX may be used, for example, LPC, LPE, LPA, etc.
LIPID STRUCTURE REPRESENTATION UPDATES
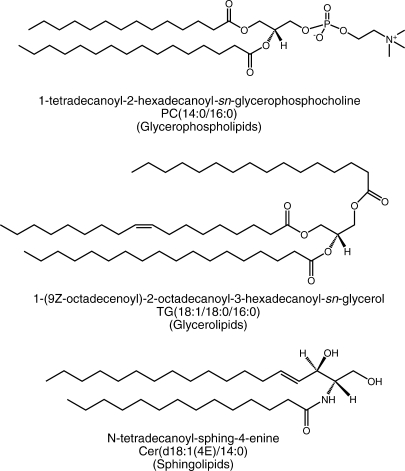
The LIPID MAPS Consortium has invested considerable effort to establish guidelines for drawing lipid structures in a clear and consistent fashion. Large and complex lipids are difficult to draw, which leads to the use of many unique formats that often generate more confusion than clarity among the lipid research community. Additionally, the structure-drawing step is often the most time-consuming process in creating molecular databases of lipids. However, many classes of lipids lend themselves well as targets for automated structure-drawing due to their consistent two-dimensional layout. A suite of structure-drawing tools has been developed and deployed that permits “on-demand” generation of structures, systematic names, and abbreviations (9). The structures may be viewed and exported in a variety of formats. Online versions of the structure-drawing tools for fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, and sterols are available in the “Tools” section of the LIPID MAPS website (http://www.lipidmaps.org). Some examples of the computationally drawn structures are shown in Fig. 1.
Fig. 1.
Examples of the computationally drawn structures with systematic name, abbreviation, and lipid category illustrating the stereochemical relationship between glycerolipids, glycerophospholipids, and sphingolipids and allowing one to visualize transformations between their components.
LIPID DATABASE UPDATES
Given the importance of these molecules in cellular function and pathology, it is essential to have a well-organized database of lipids with a defined ontology that is extensible, flexible, and scalable. The ontology of lipids must incorporate classification, nomenclature, structure representations, definitions, related biological/biophysical properties, cross-references, and structural features of all objects stored in the database. We have developed a large publicly available comprehensive object-relational database of lipid structures through integration of lipid molecules from existing repositories and from the LIPID MAPS project. This database (10, 11), in addition to serving as the most up to date and exhaustive catalog of lipid molecules, also contains systematic classification, nomenclature, ontology, and structure representations of lipids along with mass spectrometric characterization where available. More than 10,000 lipid molecules are now available on the LIPID MAPS website, and these have been adopted by the National Center for Biotechnology Information (NCBI) PubChem site (http://pubchem.ncbi.nlm.nih.gov/) as well as the Kyoto Encyclopedia of Genes and Genomes database (KEGG; http://www.genome.jp/kegg/). All structures have been classified and redrawn according to LIPID MAPS guidelines. A number of different molecular viewing formats, such as GIF image, Chemdraw CDX, and the Java-based Marvin and JMol interfaces, are offered. Where applicable, stereochemical representation and standardized nomenclature of these molecules are also provided. The database is accessible through any web browser (http://www.lipidmaps.org/data/structure/index.html) and the interfaces include text-based and structure-based query tools.
UPDATES BY LIPID CATEGORY
Within the Fatty acyls category, the Eicosanoid subclasses Hydroxyeicosatrienoic acids, Hydroxyeicosatetraenoic acids, and Hydroxyeicosapentaenoic acids have been changed to Hydroxy/hydroperoxyeicosatrienoic acids, Hydroxy/hydroperoxyeicosatetraenoic acids, and Hydroxy/hydroperoxyeicosapentaenoic acids to accommodate hydroperoxides. The names of the fatty amide subclasses N-acyl amides and N-acyl ethanolamides have been corrected to N-acyl amines and N-acyl ethanolamines. A new “Fatty acyl glycosides” main class has been added to cover the great number of simple glycolipids found in bacteria, yeast, and lower marine invertebrates (12, 13). New subclasses include Fatty acyl glycosides of mono- and disaccharides, Sophorolipids, and Rhamnolipids.
The Glycerolipids category was reorganized to include two new main classes (Glycosyldiradylglycerols and Glycosylmonoradylglycerols) that contain key plant structural lipids, such as the Sulfoquinovosyldiacylglycerols found in chloroplasts. The existing Glycerolglycoside subclasses were removed.
Due to the fact that it is extremely difficult to experimentally determine the exact position of radyl groups on the glycerol backbone for diradylglycerols and triradylglycerols, most of glycerolipid structures in the LIPID MAPS structure database have been computationally generated. For diradylglycerols with two different radyl groups, two different structural isomers are possible, whereas for triradylglycerols with three different radyl groups, six different isomers exist. Instead of drawing all possible structural isomers explicitly, an isomeric specification is used as a descriptor. A suffix containing “iso” along with the number of possible isomers is appended to the abbreviation (e.g., [iso2] and [iso6]) and a single unique LM_ID is assigned. An example of this format is the triacylglycerol TG(16:0/17:0/17:1(9Z))[iso6]. The structure assigned to the LM_ID on the LIPID MAPS website corresponds to the radyl substitution shown in the abbreviation, and the option is provided to explicitly display all isomers in the group. There are also cases within the diradyglycerol and monoradylglycerol classes where enantiomeric mixtures are obtained due to hydrolysis by certain lipases of both the sn1 or sn3 ester bond to yield 1,2-diacylglycerols[S] and 2,3-diacylglycerols[R], where the stereochemical designation of the chiral center at sn2 is reversed by the Prelog-Ingold-Cahn rules. Furthermore, acyl migration from the sn2 position of diacylglycerols to the primary hydroxyl group at either sn1 or sn2 can form an isomeric diacylglycerol, viz. 1,3-diacylglycerol. In such cases when a racemic mixture is formed, it can be identified with a “[rac]” suffix, for example, DG(16:0/16:0/0:0)[rac], or the stereochemistry is left undefined with a “[U]” suffix, for example, DG(18:1,18:2,0:0)[U]. It should be noted that the two-letter abbreviation (MG, DG, or TG) encompasses all possible types of lipid chains, for example, those having constituent acyl, alkyl, and (1Z)-alkenyl groups. The alkyl ether linkage is represented by the “O-” prefix, for example, DG(O-16:0/18:1(9Z)/0:0), and the (1Z)-alkenyl ether (neutral Plasmalogen) species by the “P-” prefix, for example, DG(P-14:0/18:1(9Z)/0:0). The same rules apply to the headgroup classes within the Glycerophospholipids category. In cases where glycerolipid total composition is known, but side chain regiochemistry and stereochemistry is unknown, abbreviations such as TG(52:1) and DG(34:2) may be used, where the numbers within parentheses refer to the total number of carbons and double bonds of all the chains.
For the Glycerophospholipids category, the subclass 1-alkyl glycerophosphocholines has been replaced by the more generic Monoalkylglycerophosphocholines due to the fact that 1-acyl-2-alkyl-glycerophosphocholines exist in nature. Examples are the ladderane phospholipids in anammox bacteria (14). Similar updates have been made for the other Glycerophospholipid headgroups. The Glycerophosphoglucose lipids class has been replaced by the Glycosylglycerophospholipids class to allow coverage of glycerolipids with sugar groups other than glucose.
As mentioned above, we have changed to two-letter abbreviations (PC, PE, etc.) to describe glycerophospholipids in shorthand form. They are generic abbreviations for all molecular species of their respective classes. These shorthand names lend themselves to fast, efficient, text-based searches and are used widely in lipid research as compact alternatives to systematic names. This “Headgroup(sn1/sn2)” format specifies one or two radyl side chains where the structures of the side chains are indicated within parentheses [e.g., PC(16:0/18:1(9Z)]. By default, R stereochemistry is implied at the C-2 carbon of glycerol, and the headgroup is attached at the sn3 position. In rare cases of molecules with opposite (S) stereochemistry at C-2 of the glycerol group and attachment of the headgroup at the sn1 position, the stereochemistry specification of [S] is appended to the abbreviation and the Headgroup (sn3/sn2) abbreviation format is used. Finally, for molecules with unknown stereochemistry at the C-2 carbon of the glycerol group, the stereochemistry specification of [U] is appended to the abbreviation, and the structure is drawn with C-2 stereochemistry unspecified.
In cases where glycerophospholipid total composition is known, but side chain regiochemistry and stereochemistry is unknown, abbreviations, such as PE(36:1), may be used to indicate total numbers of carbons and double bonds for all chains. Alkyl ether and 1Z-alkenyl ether (Plasmalogen) species are similarly represented by an “O-” or “P-” identifier, as in PC(O-36:2) and PC(P-36:1). In the latter case, the “P-” denotes an alkyl ether linkage (typically at sn1 of glycerol) and a double bond at the 1Z position. Monoradylglycerophospholipids or lysophospholipids may be specified with a letter “L” in the abbreviation, for example, LPC(16:0). The synonym “phosphatidyl” (e.g., phosphatidylcholine) is nowadays used to refer to glycerophospholipid classes containing all types of radyl chains and not just acyl groups as was originally inferred by IUPAC-IUBMB guidelines (5).
The Sphingolipids classification is essentially unchanged from that reported in the original publication in this journal (2). One item worthy of note is that for most of the Glycosphingolipid subclasses, the structure of the glycan chain is known but the exact structure of the N-acyl side chain is unknown. In these cases, the last two digits of the LIPID MAPS LM_ID identifier are assigned as “00” to signify an unspecified N-acyl side chain, and the third and fourth last digits are given a different two-letter identifier for every unique glycan chain within that subclass. For example, in the Gangliosides subclass (LM_ID: LMSP0601), the GM1 generic structure is assigned an LM_ID of LMSP0601AP00, where the “AP” digits specify the unique Galβ1-3GalNAcβ1-4(NeuAcα2-3)Galβ1-4Glcβ glycan chain, and the terminal “00” digits indicate a generic ceramide structure. By contrast, the ganglioside GM1(12:0) has a specific N-acyl chain and is assigned a LM_ID of LMSP0601AP01.
The Sterol lipids subclasses Phytosterols, Marine sterols, and Fungal sterols have been removed and replaced with a set of 13 subclasses (Ergosterols, Stigmasterols, C24-propyl sterols, Gorgosterols, Furostanols, Spirostanols, Furospirostanols, Cycloartanols, Calysterols, Cardanolides, Bufanolides, Brassinolides, and Solanidines) that differ in the nature of their sterol core structures and cover multiple phyla (15, 16). The vitamin D class has been appended with the D4, D5, D6, and D7 subclasses. Similarly, the Bile acid class now includes C22, C23, C25, and C29 subclasses. The Hopanoids class has been relocated to the Prenol Lipids category since the six-carbon D ring of the hopane core structure is at odds with the five-carbon D ring of the other members of the Sterol Lipids category.
The Retinoids subclass of the Prenol lipids category has been added to the Isoprenoids class. Retinoids are a group of C20 prenols that are related chemically to vitamin A. They have many important functions, including roles in vision, regulation of cell proliferation, and immunity.
As mentioned above, the C10 to C30 isoprenoid subclasses now contain entries at a fourth level of detail. The corresponding LM_ID identifiers contain an extra two digits that specify the fourth level class, for example, the Bisabolane sesquiterpenoid Zingiberene is assigned an LM_ID of LMPR0103060002.
The Hopanoids class has been relocated to the Prenol Lipids category (from the Sterol Lipids category).
For the Saccharolipids, the main class “Other acyl sugars” has been added to cover a variety of metabolites from plants, bacteria, and fungi. An example is the diacyl sugar 2,3-di-0-hexanoyl-α-glucopyranose from the plant species Datura metel (17). It should be noted that this category only covers structures in which fatty acyl/alkyl groups are linked directly to a sugar backbone. All lipids linked to sugars via a glycosidic bond are found in their respective lipid-centered categories.
The Polyketides category was completely revised and modeled on the classification hierarchy used by the Dictionary of Natural Products (4). Polyketides are secondary metabolites from bacteria, fungi, plants, and invertebrates and have been heavily studied by natural products chemists and pharmacologists for many years. The new classification format provides a better representation of the great structural diversity within this category.
DISCUSSION
The first step toward classification of lipids is the establishment of an ontology that is extensible, flexible, and scalable. One must be able to classify, name, and represent these molecules in a logical manner that is amenable to databasing and computational manipulation. The ILCNC proposed the comprehensive classification system in 2005 and has been actively involved in enhancing and refining it on a continuous basis. Due to considerable attention and requests from lipid researchers in a variety of fields, the classification system has been extended to more fully represent lipid structures from nonmammalian sources, such as plants, bacteria, and fungi. This universal system has been internationally accepted and is now widely used in research and for teaching purposes. The LIPID MAPS classification system has also been adopted by KEGG, where functional hierarchies involving lipids, reactions, and pathways have been constructed (http://www.genome.ad.jp/brite/), and by the knowledgebase in Wiki format of the EU Framework Project “LipidomicNet” (http://www.lipidomicnet.org). In an effort to increase public availability, LIPID MAPS lipid structures are now available on NCBI's PubChem website (http://pubchem.ncbi.nlm.nih.gov), where they have been assigned PubChem Substance IDs.
The classification system is available online (http://www.lipidmaps.org), where it has been integrated with an object-relational database of >10,000 lipids. This database, in addition to serving as the most up-to-date and exhaustive catalog of lipid molecules, also contains systematic classification, nomenclature, ontology, and structure representations of lipids along with mass spectrometric characterization where available. All structures have been classified and redrawn according to LIPID MAPS guidelines. The format of the LM_ID identifier (Table 2) provides a systematic means of simultaneously encapsulating the classification hierarchy and assigning a unique identification to each lipid molecule. It also allows for the addition of new classification elements in the future. The database is accessible through any web browser, and the interfaces include text-based and structure-based query tools. This database is described in detail elsewhere (10, 11).
A suite of lipid structure-drawing tools (available in the Tools section of the LIPID MAPS website) has been developed to enable rapid structure generation consistent with LIPID MAPS guidelines. These tools are also capable of generating systematic names and detailed ontologies, enabling rapid and efficient databasing of lipid molecules. Unix-based and web-based software has been created to register and edit database records and to automatically classify and assign LIPID MAPS IDs. These tools will be expanded and refined as the scope of the classification system and databases evolves over the coming years.
Acknowledgments
The authors appreciate the input of numerous lipid researchers around the world who have made valuable suggestions and brought to our attention anomalies in the original Comprehensive Classification System for Lipids, which to be useful must constantly adapt to new information and advances in the lipid field. The authors are also grateful to the entire LIPID MAPS Consortium for their input and especially to Dr. Jean Chin, Program Director at the National Institutes of General Medical Sciences, for her valuable input to this effort.
Abbreviations
ILCNC, International Lipid Classification and Nomenclature Committee
IUPAC-IUBMB, International Union of Pure and Applied Chemists and the International Union of Biochemistry and Molecular Biology
KEGG, Kyoto Encyclopedia of Genes and Genomes
NCBI, National Center for Biotechnology Information
This work was supported by the LIPID MAPS Large Scale Collaborative Grant number GM-069338 from the National Institutes of Health.
Published, JLR Papers in Press, December 19, 2008.
Footnotes
Guest editor for this article was Trudy M. Forte, Children's Hospital Oakland Research Institute.
The ILCNC currently consists of Dr. Edward A. Dennis, Chair, (US), Dr. Robert C. Murphy (US), Dr. Masahiro Nishijima (Japan), Dr. Christian R. H. Raetz (US), Dr. Takao Shimizu (Japan), Dr. Friedrich Spener (Austria), Dr. Gerrit van Meer (The Netherlands), and Dr. Michael Wakelam (UK). Dr. Shankar Subramaniam serves as Informatics Advisor, and Dr. Eoin Fahy serves as Director. Meetings were held May 7, 2006 and May 4, 2008 in La Jolla, CA.
Supplementary tables that provide the complete list of the classes, subclasses, and fourth class level (where applicable) of each of the eight categories of lipids are available on the LIPID MAPS website at http://www.lipidmaps.org.
References
- 1.Fahy E., S. Subramaniam, H. A. Brown, C. K. Glass, A. H. Merrill, Jr., R. C. Murphy, C. R. Raetz, D. W. Russell, Y. Seyama, W. Shaw, et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46 839–862. [DOI] [PubMed] [Google Scholar]
- 2.Schmelzer K., E. Fahy, S. Subramaniam, and E. A. Dennis. 2007. The lipid maps initiative in lipidomics. Methods Enzymol. 432 171–183. [DOI] [PubMed] [Google Scholar]
- 3.Norman H. A., C. F. Mischke, B. Allen, and J. S. Vincent. 1996. Semi-preparative isolation of plant sulfoquinovosyldiacylglycerols by solid phase extraction and HPLC procedures. J. Lipid Res. 37 1372–1376. [PubMed] [Google Scholar]
- 4.Buckingham, J., editor. 1998. Dictionary of Natural Products on CD-ROM, Version 6.1. Chapman & Hall, London.
- 5.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). The nomenclature of lipids (Recommendations 1976). 1977. Eur. J. Biochem. 79: 11–21; 1977. Hoppe-Seylers Z. Physiol. Chem. 358: 617–631; 1977. Lipids 12: 455–468; 1977. Mol. Cell. Biochem. 17: 157–171; 1978. Chem. Phys. Lipids 21: 159–173; 1978. J. Lipid Res. 19: 114–128; 1978. Biochem. J. 171: 21–35. (http://www.chem.qmul.ac.uk/iupac/lipid/).
- 6.I. U. P. A. C-I. U. B. Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids. (Recommendations 1997) 2000. Adv. Carbohydr. Chem. Biochem. 55: 311–326; 1988. Carbohydr. Res. 312: 167–175; 1998. Eur. J. Biochem. 257: 293–298; 1999. Glycoconjugate J. 16:1–6; 1999. J. Mol. Biol. 286: 963–970; 1997. Pure Appl. Chem. 69: 2475–2487. (http://www.chem.qmul.ac.uk/iupac/misc/glylp.html).
- 7.I. U. P. A. C-I. U. B. Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of prenols. (Recommendations 1987) 1987. Eur. J. Biochem. 167: 181–184. (http://www.chem.qmul.ac.uk/iupac/misc/prenol.html). [PubMed]
- 8.I. U. P. A. C-I. U. B. Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of steroids (Recommendations 1989) 1989. Eur. J. Biochem. 186: 429–458. (http://www.chem.qmul.ac.uk/iupac/steroid/). [PubMed]
- 9.Fahy E., M. Sud, D. Cotter, and S. Subramaniam. 2007. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35 W606–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sud M., E. Fahy, D. Cotter, A. Brown, E. A. Dennis, C. K. Glass, A. H. Merrill, Jr., R. C. Murphy, C. R. Raetz, D. W. Russell, et al. 2007. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35 D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahy E., D. Cotter, R. Byrnes, M. Sud, A. Maer, J. Li, D. Nadeau, Y. Zhau, and S. Subramaniam. 2007. Bioinformatics for lipidomics. Methods Enzymol. 432 247–273. [DOI] [PubMed] [Google Scholar]
- 12.Dembitsky V. M. 2004. Astonishing diversity of natural surfactants: 1. Glycosides of fatty acids and alcohols. Lipids. 39 933–953. [DOI] [PubMed] [Google Scholar]
- 13.Dembitsky V. M. 2004. Chemistry and biodiversity of the biologically active natural glycosides. Chem. Biodivers. 1 673–781. [DOI] [PubMed] [Google Scholar]
- 14.Boumann H. A., E. C. Hopmans, I. van de Leemput, H. J. Op den Camp, J. van de Vossenberg, M. Strous, M. S. Jetten, J. S. Sinninghe Damsté, and S. Schouten. 2006. Ladderane phospholipids in anammox bacteria comprise phosphocholine and phosphoethanolamine headgroups. FEMS Microbiol. Lett. 258 297–304. [DOI] [PubMed] [Google Scholar]
- 15.Kanazawa A. 2001. Sterols in marine invertebrates. Fish. Sci. 67 997–1007. [Google Scholar]
- 16.Moreau R. A., B. D. Whitaker, and K. B. Hicks. 2002. Phytosterols, phytostanols, and their conjugates in foods: structural diversity, quantitative analysis, and health-promoting uses. Prog. Lipid Res. 41 457–500. [DOI] [PubMed] [Google Scholar]
- 17.King R. R., and L. A. Calhoun. 1988. 2,3-di-O- and 1,2,3-tri-O-acylated glucose esters from the glandular trichomes of Datura metel. Phytochemistry. 27 3761–3763. [Google Scholar]