Abstract
Apolipoprotein (apo) E has roles beyond lipoprotein metabolism. The detrimental effects of apoE4 in cardiovascular, neurological, and infectious diseases correlate with its structural features (e.g., domain interaction) that distinguish it from apoE3 and apoE2. Structure/function studies revealed that apoE2 is severely defective in LDL receptor binding because of a structural difference that alters the receptor binding region and helped unravel the mechanism of type III hyperlipoproteinemia. ApoE4 is the major genetic risk factor for Alzheimer's disease and sets the stage for neuropathological disorders precipitated by genetic, metabolic, and environmental stressors. ApoE also influences susceptibility to parasitic, bacterial, and viral infections. In HIV-positive patients, apoE4 homozygosity hastens progression to AIDS and death and increases susceptibility to opportunistic infections. The next phase in our understanding of apoE will be characterized by clinical intervention to prevent or reverse the detrimental effects of apoE4 by modulating its structure or blocking the pathological processes it mediates.
Keywords: cholesterol, neurodegeneration, HIV, coronary heart disease, LDL receptor, dysbetalipoproteinemia, heparan sulfate proteoglycans, infectious diseases, evolution
HISTORICAL BACKGROUND
Structural differences in apolipoprotein (apo) E isoforms impact cardiovascular, neurological, and infectious diseases (1–4). Discovered in the 1970s, this 34-kDa, 299-amino-acid protein was identified in triglyceride-rich lipoproteins and induced by cholesterol feeding in animal models and humans (1, 3, 5, 6). The three common isoforms (apoE2, apoE3, and apoE4) are encoded by a gene on chromosome 19. The three alleles differ in their frequencies: ɛ4 (15–20%), ɛ3 (65–70%), and ɛ2 (5–10%) and give rise to three homozygous and three heterozygous phenotypes. The nomenclature arose by consensus among key investigators (7).
Utermann, Hees, and Steinmetz (8) recognized differences in the apoE isoform patterns that distinguished normolipidemic subjects from patients with the genetic lipid disorder type III hyperlipoproteinemia (HLP) (dysbetalipoproteinemia). Studies by Zannis, Breslow, Havel, and others (9, 10) helped unravel the isoform pattern. Ultimately, apoE2 was associated with type III HLP. Gladstone investigators elucidated the structural basis for the polymorphism of apoE and showed that apoE isoforms differ at two sites: apoE3 has Cys-112 and Arg-158, whereas apoE4 has arginines at both sites, and apoE2 has cysteines (11, 12). They also determined the structure of apoE mRNA and the 3.6-kb gene encoding apoE and elucidated the regulatory elements that control its expression (13–15).
Plasma apoE (∼40–70 μg/ml) arises primarily from hepatic synthesis (>75%). The second most common site of synthesis is the brain (16). Although astrocytes produce a large proportion of cerebrospinal fluid apoE (∼3–5 μg/ml), neurons synthesize apoE when stressed (17). Macrophages and other cell types also synthesize apoE (1, 3, 16).
Initially, apoE was shown to be involved in lipid transport and cardiovascular disease (1–3). It is the critical ligand in the plasma clearance of triglyceride- and cholesterol-rich lipoproteins (chylomicron remnants, VLDL, intermediate density lipoproteins, and a subclass of HDL). After Goldstein and Brown (18) identified the LDL receptor, Gladstone investigators showed that apoE is the major ligand (1, 3, 19, 20). It is also the ligand for other members of the LDL receptor family, including the LDL-receptor-related protein, which contributes to remnant lipoprotein clearance. ApoE binds to heparan sulfate proteoglycans (HSPG), which also clear apoE-containing remnant lipoproteins (3, 21). Studies of type III HLP were pivotal in elucidating the roles of apoE in remnant lipoprotein metabolism and atherosclerosis (3, 22).
ApoE also has a key role in neurobiology and Alzheimer's disease (AD) (4). It is critical for lipid transport in the brain and contributes to neuronal maintenance and repair. ApoE4 is the major genetic risk factor for AD, and 60–80% of AD patients have at least one apoE4 allele. ApoE also modulates susceptibility to infectious disease and possibly immunoregulation (1, 3). ApoE4 enhances the infectivity of HIV in vitro and hastens progression to AIDS and death in HIV-positive subjects (23).
ApoE isoforms: structural differences predict function
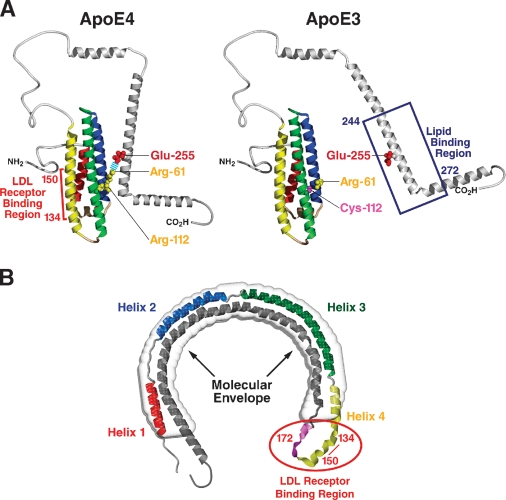
Structural analyses provided insight into the mechanisms of apoE's involvement in cardiovascular, neurological, and infectious diseases (1–4). ApoE has two structural domains separated by a hinge region (Fig. 1A). The N-terminal domain (amino acids 1–191) contains the receptor binding region (amino acids 134–150 and Arg-172) (1–3, 24) and forms a four-helix antiparallel bundle (3, 25). The C-terminal domain (amino acids ∼225–299) contains the major lipid binding region (amino acids ∼244–272) (2, 3). The amino acid differences among the isoforms profoundly affect their structures and roles in disease.
Fig. 1.
Structure of apoE. A: Model of domain interaction. In apoE4, Arg-112 orients the side chain of Arg-61 into the aqueous environment where it can interact with Glu-255, resulting in interaction between the N- and C-terminal domains. In apoE3, Arg-61 is not available to interact with glutamic acid–255. [Reproduced from (56).] B: Model of apoE bound to DMPC. X-ray analysis at 10Å resolution revealed that the molecular envelope of apoE bound to DMPC is in the shape of a circular horseshoe (gray), indicating that apoE undergoes extensive conformational change in binding to lipid. ApoE was modeled into the dimensions of the molecular envelope and is colored to show the known secondary structure of the N-terminal domain. red, helix 1; blue, helix II; green, helix III; yellow, helix IV and the connecting loop. Residues that contribute to the LDL receptor binding site are in pink, showing the juxtaposition of basic residues 134–150 with Arg-172. Residues in the C-terminal domain are shown in gray.
ApoE AND CARDIOVASCULAR DISEASE
ApoE2 and apoE4 increase the number of atherogenic lipoproteins and accelerate atherogenesis (1, 3, 6). Understanding structural differences in apoE isoforms helped establish the molecular mechanism responsible for the associated pathology. First, the altered structure and impaired function of the receptor binding region of apoE2 increase triglyceride and cholesterol levels caused by delayed clearance of hepatic and intestinal remnant lipoproteins (β-VLDL), resulting in type III HLP (3, 22). Cys-158 in apoE2 affects the receptor binding region by altering salt bridges and lowering the positive potential (25). Second, the increase in plasma cholesterol, LDL, and apoB associated with apoE4 appears to reflect the influence of Arg-112 (1–3). Arg-112 alters the lipid binding region of apoE4 and changes the lipid binding preference from small phospholipid-rich HDL (apoE2 and apoE3) to large triglyceride-rich VLDL (apoE4). This difference is due to apoE4 domain interaction, in which the N- and C-terminal domains interact, resulting in a more compact structure.
ApoE2: insights into the lipoprotein receptor and HSPG binding and type III HLP
The receptor and proteoglycan binding regions were identified through structural studies of apoE2 and its role in type III HLP (3, 22). Before the structure was delineated, arginine and lysine basic residues were shown to be critical for binding to the LDL receptor (26, 27). In addition, binding to phospholipids was required for high-affinity binding activity (20). Mutagenesis studies identified the critical basic residues within the 134–150 region of apoE as well as Arg-172 (24, 28). Modeling of apoE into the molecular envelope of apoE bound to phospholipid revealed why lipid binding is required for high-affinity binding to LDL receptors (29). To fit the molecular envelope, apoE folded into a helical horseshoe, bringing the critical residues for binding, amino acids 134–150 and Arg-172, into close proximity (Fig. 1B).
ApoE2, which is severely defective in LDL receptor binding activity (<2% LDL receptor binding activity compared with apoE3), differs structurally from apoE3 and apoE4, which bind avidly to LDL receptors. In apoE3, the 134–150 region is largely solvent exposed and forms a 20Å field of positive potential, likely available for receptor binding. In apoE2, the presence of Cys-158 rather than arginine alters the conformation and size of the positive potential (3, 25).
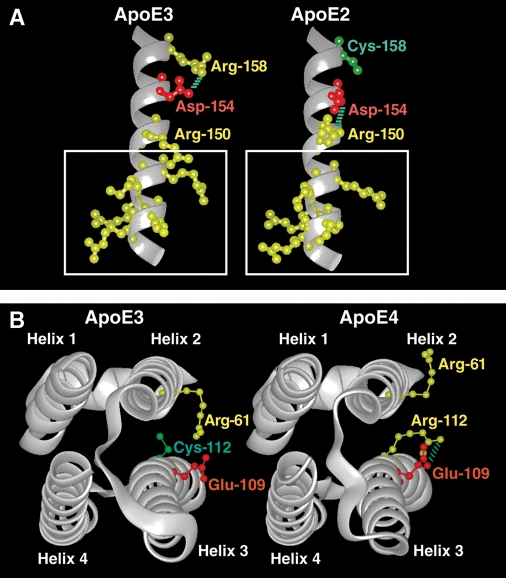
Residue 158 lies outside the 134–150 region, so how does it influence binding activity? In apoE3, Arg-158 forms a salt bridge with Asp-154. However, in apoE2, Cys-158 disrupts the salt bridge, causing Asp-154 to interact with Arg-150 (30) (Fig. 2A). This alters the size of the positively charged domain and disrupts LDL receptor binding. Nevertheless, apoE2 can still mediate lipoprotein clearance through binding HSPG (21).
Fig. 2.
Three-dimensional structure of regions of apoE highlighting isoform differences. A: The region of helix 4 where a critical salt bridge rearrangement in apoE2 reduces the positive potential of the LDL receptor binding site (boxed). Asp-154 changes its ionic interaction to Arg-150 in apoE2 because of the Cys-158 substitution, pulling the side chain of Arg-150 out of the positive potential cloud, reducing its potential, and causing a 100-fold reduction in apoE receptor binding. (Courtesy of Karl H. Weisgraber.) B: Four-helix bundle of apoE showing rearrangement of the Arg-61 side chain. The substitution of Arg-112 in apoE4 leads to an ionic interaction with Glu-109 that excludes the Arg-61 side chain from its usual position, causing Arg-61 to be more exposed at the side of helix 2 and allowing it to become available for interaction with Glu-255 in the C-terminal domain of apoE (data not shown). [Reproduced from (57) and used by permission of Lippincott Williams & Wilkins.]
Although apoE2 homozygosity is essential for type III HLP, the disorder is precipitated by genetic or environmental factors that result in saturated or impaired normal clearance pathways. These factors include estrogen deficiency, which can impair lipolytic processing; hypothyroidism, associated with decreased expression of LDL receptors; and obesity and diabetes, characterized by increased lipoprotein production. Transgenic mouse models expressing apoE2 provided proof of concept for how second “hits” induce type III HLP and how the hyperlipidemia is influenced by overproduction of apoB-containing lipoproteins and decreased numbers of LDL receptors (31). In summary, understanding the structural differences among apoE2, apoE3, and apoE4 helped to unravel the complexity of type III HLP and, at the same time, defined the receptor binding region and lipoprotein clearance pathways.
ApoE4: insights into lipid binding, domain interaction, and atherosclerosis
The C-terminal domain of apoE (amino acids 225–299) is predicted to form amphipathic α-helices (2). These helices are responsible for lipid binding in different apolipoproteins, and amino acids 244–272 constitute the lipid binding region of apoE (2, 3). ApoE3 and apoE2 preferentially bind to small, phospholipid-rich HDL, whereas apoE4 binds to large, triglyceride-rich VLDL. The difference is determined by how the N- and C-terminal domains interact (32). X-ray crystallographic analyses of apoE3 and apoE4 revealed differences in side chain orientation and rearrangements of salt bridges. In apoE4, Arg-112 forms a salt bridge with Glu-109 and causes the Arg-61 side chain to extend away from the four-helix bundle. In apoE3, this side chain is buried (25) (Fig. 2B).
The orientation of Arg-61 in apoE4 promotes domain interaction by interacting with Glu-255 within the lipid binding region, causing apoE4 to have a more compact conformation than apoE3 (Fig. 1A) (32, 33). Domain interaction is an important structural property of apoE4 (34, 35) that may be responsible for several of its pathogenic effects. ApoE3 and apoE2 are less likely to demonstrate domain interaction. Mutation of Arg-61 to threonine, or Glu-255 to alanine, abolishes domain interaction, causing the mutated apoE4 to function similarly to apoE3 (36). Domain interaction appears to alter the lipid binding region and, thus, lipoprotein preference. ApoE4 with Thr-61 corrects not only lipid binding preference but also several of the detrimental effects of apoE4 in neurobiology (4).
In most animals, including great apes, apoE has Arg-112 (like human apoE4) but has threonine at a site equivalent to Arg-61 in human apoE (2). Lacking domain interaction, it behaves like apoE3. Gene targeting to replace Thr-61 with arginine in mouse Apoe (37) produced a model of domain interaction (Arg-61 apoE mouse) that does not display other apoE4 structural properties. It established that domain interaction was responsible for the preference of apoE4 for VLDL.
ApoE4 increases plasma LDL levels and risk for atherosclerosis and is overrepresented in hyperlipidemic and cardiovascular disease patients (3). Because apoE4 binds preferentially to VLDL and remnants, it may accelerate their clearance, leading to downregulation of LDL receptors and increased LDL levels. Alternatively, remnants could compete for LDL receptors, retarding LDL clearance.
ApoE: roles in regulating VLDL production, lipolysis, and triglyceride levels
Plasma apoE levels are determinants of triglyceride-rich lipoprotein metabolism and explain 20–40% of variability in triglyceride levels in humans. Overexpression and accumulation of apoE appear to cause hypertriglyceridemia by stimulating VLDL triglyceride production and impairing VLDL lipolysis (38). In apoE-deficient mouse hepatocytes, VLDL triglyceride secretion is impaired, suggesting a physiological role for apoE in VLDL assembly secretion (39). In fact, hepatic overexpression of apoE stimulates VLDL production, increasing VLDL triglyceride secretion in transgenic mice and transfected hepatocytes and in hypertriglyceridemic patients. ApoE inhibits lipolysis in vitro, and plasma from hypertriglyceridemic patients has decreased lipoprotein lipase cofactor activity for apoCII, suggesting that the apoE:apoCII ratio in VLDL is critical for VLDL lipolysis by lipoprotein lipase (38). Thus, optimal expression of apoE is crucial for normal metabolism of triglyceride-rich lipoproteins. Too little apoE impairs plasma clearance of triglyceride-rich lipoproteins and their remnants. Too much apoE stimulates hepatic VLDL triglyceride production and impairs lipolysis, leading to hypertriglyceridemia.
ApoE AND NEURODEGENERATIVE DISORDERS
ApoE is the major known genetic risk factor for AD (40) and has a potential role in other neurological diseases and traumatic brain injury (4). In AD, apoE4 contributes to neuropathology by interacting with the amyloid pathway to modulate amyloid β (Aβ) peptide synthesis or clearance. ApoE4 can also cause neuropathology directly (4). ApoE4 sets the stage for “second hits” to precipitate neuropathology. These second hits may be genetic (α-synuclein in Parkinson's disease, SOD-1 in amyotrophic lateral sclerosis, and presenilin in AD), metabolic (ischemia and oxidative stress), or environmental (aging, inflammation, and central nervous system trauma). The neuropathologic effects of apoE4 are mediated by its structural features (e.g., domain interaction) (4).
ApoE4 increases of Aβ production
In cultured neuronal cells expressing amyloid precursor protein, exogenous apoE4 enhances Aβ production more than apoE3 (36). ApoE4 with Thr-61, which lacks domain interaction, behaves like apoE3. When domain interaction is blocked by a small molecule that docks with apoE near Arg-61, apoE4 no longer enhances Aβ production (36). Thus, altering a structural feature of apoE4 also converts its biological activity to one resembling apoE3.
ApoE4 destabilizes membranes and increases apoptotic cell death
In neuronal cells cultured with Aβ1−42, exogenous apoE4 causes lysosomal leakage and apoptosis, whereas apoE3 is protective (41, 42). ApoE4 causes greater membrane disruption than apoE3; and apoE4 with Thr-61, lacking domain interaction, behaves much more like apoE3. A small-molecule structure corrector also blocked the detrimental effects of apoE4. These studies provide proof of concept that such effects can be prevented with a structure corrector that blocks domain interaction and converts apoE4 to a molecule that is structurally and functionally similar to apoE3 (4).
ApoE synthesis by neurons leads to neuropathology
ApoE synthesis by neurons can be induced by various stressors or stimuli (17, 43), possibly including age, oxidative stress, trauma, Aβ deposition, and ischemia. Neuronal expression of apoE is likely induced to protect the cells from noxious stimuli or to repair damage, possibly by redistributing lipids. However, there is a marked difference in how apoE3 and apoE4 are handled by neurons.
ApoE4 synthesized by neurons undergoes proteolytic cleavage to a much greater extent than apoE3 (44). The resulting fragments with C-terminal truncations escape the secretory pathway and enter the cytosol. Most are neurotoxic (45). Similar apoE4 fragments are seen in the brains of transgenic mice expressing apoE4 in neurons (NSE-apoE) and in AD brains but not in transgenic mice expressing apoE4 in astrocytes. The initial fragment is produced by a chymotrypsin-like serine protease that primarily cleaves apoE at amino acids 268 and 272. The fragments accumulate with age in NSE-apoE4 mice, reaching a peak concentration at 6–8 months of age when these mice exhibit apoE4-associated neuropathology (reduced synaptophysin and microtubule-associated protein 2 immunoreactivity) and impaired learning and memory (water maze tests) (44).
The susceptibility of apoE4 to neuron-specific proteolysis is mediated by domain interaction. ApoE4 with Thr-61 is resistant to proteolysis by mouse brain lysate as the source of the cleaving enzyme. Here again, a structure corrector might protect apoE4 from proteolysis. Inhibition of the apoE-cleaving enzyme might be an additional therapeutic strategy (4).
In cultured neurons, the apoE fragments are primarily localized to mitochondria and to a lesser extent to filamentous neurofibrillary-tangle-like structures composed of phosphorylated tau (46). We suspect that the mitochondrial dysfunction characteristic of neurons synthesizing apoE4 is related to the translocation of the fragments to the mitochondria. The cytoskeletal abnormalities associated with apoE4 might relate to fragment localization within the filamentous inclusions.
Again, the pathology can be correlated with the apoE structure (4). Studies of truncated and mutated apoE4 forms expressed in cultured neurons demonstrated how the apoE4 fragments lacking the C-terminal 27 amino acids escape the secretory pathway, translocate into the cytosol, interact with mitochondria, and cause neurotoxicity (46). The minimal structure of apoE required for these detrimental effects includes the receptor binding region (amino acids 134–150) and the lipid binding region (amino acids 244–272).
Translocation of the fragments is mediated by positively charged amino acids in the receptor binding region (46). This sequence resembles the protein translocation domain of many viral proteins. Mutation of critical lysines or arginines in this domain abolishes translocation of the fragments to the cytosol.
The hydrophobic lipid binding region is responsible for mitochondrial targeting and neurotoxicity (46). Mutation of conserved residues in this region blocks the interaction with mitochondria. Mitochondrial dysfunction can profoundly alter neurobiology in many ways. For example, mitochondria are important in synaptogenesis, and their localization to the site of synaptic spine formation is essential. Neuronal cells incubated with apoE4 (especially the 29-kDa fragment) have fewer mitochondria located to the spines and many fewer spines than neurons incubated with apoE3 (47). These findings suggest an additional therapeutic strategy: preventing the interaction between apoE fragments and mitochondria with so-called mitochondrial protectors (4).
ApoE, INFECTIOUS DISEASE, AND IMMUNOREGULATION
ApoE also plays a role in susceptibility to infection (1, 3), including malaria (48) and bacterial infections (49, 50). However, isoform specificity has not been explored.
Evidence suggests that apoE modulates viral infections, and there are isoform-specific effects related to herpes simplex virus (HSV) and HIV. HSV1 infection is associated with increased risk of AD, and apoE4 is overrepresented in HSV-infected subjects (51). In HIV-positive patients, apoE4 homozygosity hastens progression to AIDS and death and increases susceptibility to opportunistic organisms (23). Cultured cells are more susceptible to infection with HIV in the presence of apoE4 than apoE3, reflecting enhancement of viral attachment and fusion. The structural differences between apoE4 and apoE3 may shed light on the mechanism by which apoE4 modulates infectivity and fusion.
Amphipathic helices of apolipoproteins interact with similar helices of the HIV gp41 protein and inhibit viral fusion (52). In apoE, these helices encompass the lipid binding region. Since domain interaction modulates lipid binding, isoform-specific differences in HIV infectivity and fusion may be explained by the structure of apoE4. Alternatively, both apoE and HIV bind to cell-surface HSPG, whose interaction with viral particles may be rate limiting in HIV infection. Another potential explanation is the differential effects of apoE on cholesterol content or other lipids in infected cells or the HIV envelope.
Do infectious diseases drive apoE evolution?
The source of differences in the apoE allele frequencies among population groups is unknown. ApoE3 is the most common, but apoE4 may be the ancestral allele (1, 3, 53, 54). Many animals, including all the great apes, have an apoE4-like allele (Arg-112) and do not display multiple isoforms (2). It is unlikely that the detrimental effects of apoE4 in cardiovascular or neurological disease provided the evolutionary pressure, as these effects are postreproductive. Any genetic drift from apoE4 to apoE3 to apoE2 most likely results from the selective pressures of infectious diseases.
For example, apoE4 is more frequent in Africa, perhaps because it afforded protection against specific endemic infectious diseases (e.g., parasitic diseases). However, 50,000 years ago, when populations migrated from Africa to Europe and beyond, new infectious diseases (e.g., viral infections) may have arisen in historical centers of agriculture, where population density might maintain viral diseases, resulting in selection against carriers of apoE4. Thus, apoE3 might have provided a selective advantage over apoE4. A cataclysmic event in human history driving the evolution of apoE4 to apoE3 to apoE2 could have been an infectious disease, such as the Great Plague, which killed 30–50% of Europeans in the 14th century, or smallpox. Much remains to be learned about the evolution of this fascinating protein and the selective pressures that account for the population differences in the frequency of the apoE alleles (3, 53–55).
Acknowledgments
The authors thank Sylvia Richmond for manuscript preparation and Stephen Ordway and Gary Howard for editorial assistance. The authors acknowledge the generous support of The J. David Gladstone Institutes.
Abbreviations
Aβ, amyloid β
AD, Alzheimer's disease
apo, apolipoprotein
HLP, hyperlipoproteinemia
HSPG, heparan sulfate proteoglycans
HSV, herpes simplex virus
This work was supported in part by grant R01 AG028793 and program project grant 2P01AG02207 from the National Institutes of Health.
Published, JLR Papers in Press, December 22, 2008.
References
- 1.Mahley R. W. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240 622–630. [DOI] [PubMed] [Google Scholar]
- 2.Weisgraber K. H. 1994. Apolipoprotein E: structure–function relationships. Adv. Protein Chem. 45 249–302. [DOI] [PubMed] [Google Scholar]
- 3.Mahley R. W., and S. C. Rall, Jr. 2000. Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1 507–537. [DOI] [PubMed] [Google Scholar]
- 4.Mahley R. W., K. H. Weisgraber, and Y. Huang. 2006. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 103 5644–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shore V. G., and B. Shore. 1973. Heterogeneity of human plasma very low density lipoproteins. Separation of species differing in protein components. Biochemistry. 12 502–507. [DOI] [PubMed] [Google Scholar]
- 6.Mahley R. W. 1983. Development of accelerated atherosclerosis. Concepts derived from cell biology and animal model studies. Arch. Pathol. Lab. Med. 107 393–399. [PubMed] [Google Scholar]
- 7.Zannis V. I., J. L. Breslow, G. Utermann, R. W. Mahley, K. H. Weisgraber, R. J. Havel, J. L. Goldstein, M. S. Brown, G. Schonfeld, W. R. Hazzard, et al. 1982. Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J. Lipid Res. 23 911–914. [PubMed] [Google Scholar]
- 8.Utermann G., M. Hees, and A. Steinmetz. 1977. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 269 604–607. [DOI] [PubMed] [Google Scholar]
- 9.Zannis V. I., P. W. Just, and J. L. Breslow. 1981. Human apolipoprotein E isoprotein subclasses are genetically determined. Am. J. Hum. Genet. 33 11–24. [PMC free article] [PubMed] [Google Scholar]
- 10.Havel R. J., and J. P. Kane. 1973. Primary dysbetalipoproteinemia: predominance of a specific apoprotein species in triglyceride-rich lipoproteins. Proc. Natl. Acad. Sci. USA. 70 2015–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisgraber K. H., S. C. Rall, Jr., and R. W. Mahley. 1981. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem. 256 9077–9083. [PubMed] [Google Scholar]
- 12.Rall S. C., Jr., K. H. Weisgraber, and R. W. Mahley. 1982. Human apolipoprotein E. The complete amino acid sequence. J. Biol. Chem. 257 4171–4178. [PubMed] [Google Scholar]
- 13.Taylor J. M., W. S. Simonet, S. J. Lauer, G. Zhu, and D. Walker. 1993. Regulation and expression of the human apolipoprotein E gene in transgenic mice. Curr. Opin. Lipidol. 4 84–89. [Google Scholar]
- 14.Grehan S., E. Tse, and J. M. Taylor. 2001. Two distal downstream enhancers direct expression of the human apolipoprotein E gene to astrocytes in the brain. J. Neurosci. 21 812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allan C. M., S. Taylor, and J. M. Taylor. 1997. Two hepatic enhancers, HCR.1 and HCR.2, coordinate the liver expression of the entire human apolipoprotein E/C-I/C-IV/C-II gene cluster. J. Biol. Chem. 272 29113–29119. [DOI] [PubMed] [Google Scholar]
- 16.Elshourbagy N. A., W. S. Liao, R. W. Mahley, and J. M. Taylor. 1985. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc. Natl. Acad. Sci. USA. 82 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Q., A. Bernardo, D. Walker, T. Kanegawa, R. W. Mahley, and Y. Huang. 2006. Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J. Neurosci. 26 4985–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein J. L., and M. S. Brown. 1976. The LDL pathway in human fibroblasts: a receptor-mediated mechanism for the regulation of cholesterol metabolism. Curr. Top. Cell. Regul. 11 147–181. [DOI] [PubMed] [Google Scholar]
- 19.Innerarity T. L., and R. W. Mahley. 1978. Enhanced binding by cultured human fibroblasts of apo-E-containing lipoproteins as compared with low density lipoproteins. Biochemistry. 17 1440–1447. [DOI] [PubMed] [Google Scholar]
- 20.Innerarity T. L., R. E. Pitas, and R. W. Mahley. 1979. Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. J. Biol. Chem. 254 4186–4190. [PubMed] [Google Scholar]
- 21.Mahley R. W., and Y. Huang. 2007. Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. J. Clin. Invest. 117 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahley R. W., Y. Huang, and S. C. Rall, Jr. 1999. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): questions, quandaries, and paradoxes. J. Lipid Res. 40 1933–1949. [PubMed] [Google Scholar]
- 23.Burt T. D., B. K. Agan, V. C. Marconi, W. He, H. Kulkarni, J. E. Mold, M. Cavrois, Y. Huang, R. W. Mahley, M. J. Dolan, et al. 2008. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE ɛ4/ɛ4 genotype accelerates HIV disease progression. Proc. Natl. Acad. Sci. USA. 105 8718–8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow J. A., K. S. Arnold, J. Dong, M. E. Balestra, T. L. Innerarity, and K. H. Weisgraber. 2000. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J. Biol. Chem. 275 2576–2580. [DOI] [PubMed] [Google Scholar]
- 25.Wilson C., M. R. Wardell, K. H. Weisgraber, R. W. Mahley, and D. A. Agard. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 252 1817–1822. [DOI] [PubMed] [Google Scholar]
- 26.Mahley R. W., T. L. Innerarity, R. E. Pitas, K. H. Weisgraber, J. H. Brown, and E. Gross. 1977. Inhibition of lipoprotein binding to cell surface receptors of fibroblasts following selective modification of arginyl residues in arginine-rich and B apoproteins. J. Biol. Chem. 252 7279–7287. [PubMed] [Google Scholar]
- 27.Weisgraber K. H., T. L. Innerarity, and R. W. Mahley. 1978. Role of the lysine residues of plasma lipoproteins in high affinity binding to cell surface receptors on human fibroblasts. J. Biol. Chem. 253 9053–9062. [PubMed] [Google Scholar]
- 28.Lalazar A., K. H. Weisgraber, S. C. Rall, Jr., H. Giladi, T. L. Innerarity, A. Z. Levanon, J. K. Boyles, B. Amit, M. Gorecki, R. W. Mahley, et al. 1988. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. J. Biol. Chem. 263 3542–3545. [PubMed] [Google Scholar]
- 29.Peters-Libeu C. A., Y. Newhouse, D. M. Hatters, and K. H. Weisgraber. 2006. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J. Biol. Chem. 281 1073–1079. [DOI] [PubMed] [Google Scholar]
- 30.Dong L-M., S. Parkin, S. D. Trakhanov, B. Rupp, T. Simmons, K. S. Arnold, Y. M. Newhouse, T. L. Innerarity, and K. H. Weisgraber. 1996. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat. Struct. Biol. 3 718–722. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y., S. C. Rall, Jr., and R. W. Mahley. 1997. Genetic factors precipitating type III hyperlipoproteinemia in hypolipidemic transgenic mice expressing human apolipoprotein E2. Arterioscler. Thromb. Vasc. Biol. 17 2817–2824. [DOI] [PubMed] [Google Scholar]
- 32.Dong L-M., and K. H. Weisgraber. 1996. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 271 19053–19057. [DOI] [PubMed] [Google Scholar]
- 33.Dong L-M., C. Wilson, M. R. Wardell, T. Simmons, R. W. Mahley, K. H. Weisgraber, and D. A. Agard. 1994. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 269 22358–22365. [PubMed] [Google Scholar]
- 34.Xu Q., W. J. Brecht, K. H. Weisgraber, R. W. Mahley, and Y. Huang. 2004. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J. Biol. Chem. 279 25511–25516. [DOI] [PubMed] [Google Scholar]
- 35.Hatters D. M., M. S. Budamagunta, J. C. Voss, and K. H. Weisgraber. 2005. Modulation of apolipoprotein E structure by domain interaction. Differences in lipid-bound and lipid-free forms. J. Biol. Chem. 280 34288–34295. [DOI] [PubMed] [Google Scholar]
- 36.Ye S., Y. Huang, K. Müllendorff, L. Dong, G. Giedt, E. C. Meng, F. E. Cohen, I. D. Kuntz, K. H. Weisgraber, and R. W. Mahley. 2005. Apolipoprotein (apo) E4 enhances amyloid β peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc. Natl. Acad. Sci. USA. 102 18700–18705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raffaï R. L., L-M. Dong, R. V. Farese, Jr., and K. H. Weisgraber. 2001. Introduction of human apolipoprotein E4 “domain interaction” into mouse apolipoprotein E. Proc. Natl. Acad. Sci. USA. 98 11587–11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y., X. Q. Liu, S. C. Rall, Jr., J. M. Taylor, A. von Eckardstein, G. Assmann, and R. W. Mahley. 1998. Overexpression and accumulation of apolipoprotein E as a cause of hypertriglyceridemia. J. Biol. Chem. 273 26388–26393. [DOI] [PubMed] [Google Scholar]
- 39.Kuipers F., M. C. Jong, Y. Lin, M. van Eck, R. Havinga, V. Bloks, H. J. Verkade, M. H. Hofker, H. Moshage, T. J. C. van Berkel, et al. 1997. Impaired secretion of very low density lipoprotein–triglycerides by apolipoprotein E–deficient mouse hepatocytes. J. Clin. Invest. 100 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corder E. H., A. M. Saunders, W. J. Strittmatter, D. E. Schmechel, P. C. Gaskell, G. W. Small, A. D. Roses, J. L. Haines, and M. A. Pericak-Vance. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 261 921–923. [DOI] [PubMed] [Google Scholar]
- 41.Ji Z-S., R. D. Miranda, Y. M. Newhouse, K. H. Weisgraber, Y. Huang, and R. W. Mahley. 2002. Apolipoprotein E4 potentiates amyloid β peptide-induced lysosomal leakage and apoptosis in neuronal cells. J. Biol. Chem. 277 21821–21828. [DOI] [PubMed] [Google Scholar]
- 42.Ji Z-S., K. Müllendorff, I. H. Cheng, R. D. Miranda, Y. Huang, and R. W. Mahley. 2006. Reactivity of apolipoprotein E4 and amyloid β peptide: lysosomal stability and neurodegeneration. J. Biol. Chem. 281 2683–2692. [DOI] [PubMed] [Google Scholar]
- 43.Xu Q., D. Walker, A. Bernardo, J. Brodbeck, M. E. Balestra, and Y. Huang. 2008. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 28 1452–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris F. M., W. J. Brecht, Q. Xu, I. Tesseur, L. Kekonius, T. Wyss-Coray, J. D. Fish, E. Masliah, P. C. Hopkins, K. Scearce-Levie, et al. 2003. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. USA. 100 10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brecht W. J., F. M. Harris, S. Chang, I. Tesseur, G-Q. Yu, Q. Xu, J. D. Fish, T. Wyss-Coray, M. Buttini, L. Mucke, et al. 2004. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 24 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang S., T. R. Ma, R. D. Miranda, M. E. Balestra, R. W. Mahley, and Y. Huang. 2005. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl. Acad. Sci. USA. 102 18694–18699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodbeck J., M. E. Balestra, A. M. Saunders, A. D. Roses, R. W. Mahley, and Y. Huang. 2008. Rosiglitazone increases dendritic spine density and rescues spine loss caused by apolipoprotein E4 in primary cortical neurons. Proc. Natl. Acad. Sci. USA. 105 1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinnis P., T. E. Willnow, M. R. S. Briones, J. Herz, and V. Nussenzweig. 1996. Remnant lipoproteins inhibit malaria sporozoite invasion of hepatocytes. J. Exp. Med. 184 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roselaar S. E., and A. Daugherty. 1998. Apolipoprotein E-deficient mice have impaired innate immune responses to Listeria monocytogenes in vivo. J. Lipid Res. 39 1740–1743. [PubMed] [Google Scholar]
- 50.de Bont N., M. G. Netea, P. N. M. Demacker, I. Verschueren, B. J. Kullberg, K. W. van Dijk, J. W. M. van der Meer, and A. F. H. Stalenhoef. 1999. Apolipoprotein E knock-out mice are highly susceptible to endotoxemia and Klebsiella pneumoniae infection. J. Lipid Res. 40 680–685. [PubMed] [Google Scholar]
- 51.Itzhaki R. F., W-R. Lin, D. Shang, G. K. Wilcock, B. Faragher, and G. A. Jamieson. 1997. Herpes simplex virus type 1 in brain and risk of Alzheimer's disease. Lancet. 349 241–244. [DOI] [PubMed] [Google Scholar]
- 52.Martin I., M-C. Dubois, T. Saermark, and J-M. Ruysschaert. 1992. Apolipoprotein A-1 interacts with the N-terminal fusogenic domains of SIV (simian immunodeficiency virus) GP32 and HIV (human immunodeficiency virus) GP41: implications in viral entry. Biochem. Biophys. Res. Commun. 186 95–101. [DOI] [PubMed] [Google Scholar]
- 53.Finch C. E., and R. M. Sapolsky. 1999. The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol. Aging. 20 407–428. [DOI] [PubMed] [Google Scholar]
- 54.Mahley R. W., and S. C. Rall, Jr. 1999. Is ɛ4 the ancestral human apoE allele? Neurobiol. Aging. 20 429–430. [DOI] [PubMed] [Google Scholar]
- 55.Gerdes L. U. 2003. The common polymorphism of apolipoprotein E: geographical aspects and new pathophysiological relations. Clin. Chem. Lab. Med. 41 628–631. [DOI] [PubMed] [Google Scholar]
- 56.Zhong, N., and K. H. Weisgraber. Understanding the association of apolipoprotein E4 with Alzheimer's disease: clues from its structure. J. Biol. Chem. Epub ahead of print. October 22, 2008. 10.1074/jbc.R800009200. [DOI] [PMC free article] [PubMed]
- 57.Mahley R. W., and Y. Huang. 1999. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 10 207–217. [DOI] [PubMed] [Google Scholar]