Abstract
The spatial and temporal regulation of lipid molecules in cell membranes is a hallmark of cellular signaling and membrane trafficking events. Lipid-mediated targeting provides for strict control and versatility, because cell membranes harbor a large number of lipid molecules with variation in head group and acyl chain structures. Signaling and trafficking proteins contain a large number of modular domains that exhibit specific lipid binding properties and play a critical role in their localization and function. Nearly 20 years of research including structural, computational, biochemical and biophysical studies have demonstrated how these lipid-binding domains recognize their target lipid and achieve subcellular localization. The integration of this individual lipid-binding domain data in the context of the full-length proteins, macromolecular signaling complexes, and the lipidome is only beginning to be unraveled and represents a target of therapeutic development. This review brings together recent findings and classical concepts to concisely summarize the lipid-binding domain field while illustrating where the field is headed and how the gaps may be filled in with new technologies.
Keywords: C1 domain, C2 domain, peripheral protein
Cellular membranes harbor receptors, ion channels, lipid domains, lipid signals, and scaffolding complexes, which function to maintain cellular growth, metabolism, and homeostasis. Moreover, abnormalities in lipid metabolism attributed to genetic changes among other causes are often associated with diseases such as cancer (1). Thus, there is a need to understand molecular events occurring within and on membranes as a means of grasping disease etiology and identifying viable targets for drug development (2). The lipid bilayer has a highly polarized structure that consists of a central hydrocarbon core and 2 flanking interfacial regions that are highly dynamic and may contain >1,000 different lipids (3). This dynamic variety of glycerophospholipids, sphingolipids, and sterols in the membrane organelles provides spatial and temporal architecture to direct signaling processes through target proteins. Because nearly one-half of all proteins are located in or on membranes, it is not surprising that there is a variety of conserved lipid binding domains in eukaryotes. Some of these domain families rank in the top 15 modular domains in the human genome and are most often found in signal transduction and membrane trafficking proteins (4).
Lipid-binding domain research can be traced back to the discovery of protein kinase C (PKC) in the late 1970s (5), which led to the identification of conserved regions among PKC isoforms, now termed C1 and C2 domains, which harbor distinct lipid binding properties. Later, a substrate of PKC termed pleckstrin was identified to possess the first phosphoinositide (PI) binding region termed the pleckstrin homology (PH) domain. This was followed by an array of structural data on C1 (6), C2 (7), and PH domains (8) and eventually the identification of more lipid-binding domain genomes with bioinformatic studies. Subsequently, these individual domains have been rigorously characterized to understand how they bind membranes, translocate to membrane docking sites in the cell, and function to regulate their parent protein. Yet, there is still a paucity of predictive data for lipid-binding domains, meaning they have to be studied in vitro and in cells on an individual basis to characterize their function. Nonetheless, the mechanisms by which these modular domains control host protein activity are beginning to be unraveled, and we should expect a rush of informative studies in the coming decade. The advent of high throughput methods geared toward studying these domains on a genomic, proteomic, and lipidomic scale will play a major role in this regard. My aim in this review is to introduce general principles governing the lipid binding and cellular localization of some of the lipid-binding domains. The description and examples used are selective, and I refer the reader to other compelling reviews (4, 9–11) or illustrative original manuscripts to fill in the gaps. The concise overview of the lipid-binding domains leads into a critical discussion of areas to be addressed in the lipid-binding field and potential ways of new discovery.
LIPID-BINDING DOMAINS
At least 11 lipid-binding domains have been identified to date, including: C1 (12), C2 (13), PH (14), FYVE (15), PX (15), ENTH (16), ANTH (16), BAR (16), FERM, PDZ, and tubby domains. Figure 1 summarizes some of the common properties of these lipid-binding domains. All intracellular membranes contain a varying degree of anionic lipids, and a majority of lipid-binding domains contain cationic surfaces, at least locally. Despite specific or nonspecific interactions primarily with anionic lipids, lipid-binding domains have distinct membrane binding mechanisms allowing control in cellular signaling mechanisms and a niche to be exploited in drug development. While some lipid-binding domains are highly specific in their coordination of a lipid head group, others are nonspecific and associate with a membrane based upon its physical property such as charge or curvature. Thus, spatial and temporal signals can be crucial to regulating targeting of lipid-binding domains to the intracellular membrane.
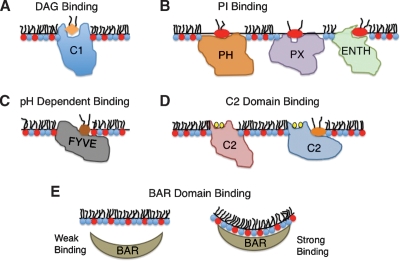
Fig. 1.
Membrane binding modes of the most common lipid-binding domains. A: C1 domain hydrophobic interactions facilitate the docking to the bilayer for efficient DAG or phorbol ester coordination. B: PH domains can bind a variety of PIs as described in the text. Some PH domains primarily associate with the membrane through electrostatic interactions with the PI, while a number of PH domains anchor to the membrane upon PI docking through penetration of hydrophobic residues adjacent to the PI binding site. PIs induce the membrane penetration of PX domains through a reduction in the desolvation penalty surrounding exposed hydrophobic residues adjacent to the PI binding site. The ENTH domain coordinates PIP2, which induces formation of a N-terminal α-helix from an unstructured region. The amphipathic α-helix then inserts into the membrane to induce changes in membrane curvature. C: FYVE domains selectively bind PtdIns(3)P at a site adjacent to the hydrophobic membrane protrusion loop. Following nonspecific electrostatic association, PtdIns(3)P docking to FYVE domains induces the membrane penetration through a reduction of the desolvation penalty. D: Two modes of binding are depicted for C2 domains. Ca2+ ions are shown in yellow. Ca2+-binding C2 domains can associate with anionic or zwitterionic membranes depending upon the residues present in their Ca2+-binding loops. Some C2 domains contain a patch of cationic residues in their β-groove that is able to bind lipids such as PIs and may function in coincidence detection. E: Many BAR domains resemble the shape of a crescent moon and are rich in cationic residues on their concave surface. Thus, many BAR domains selectively associate with membranes of high curvature due to the shape of their concave surface and the greater ability for facilitating electrostatic interactions with the highly curved membranes.
Spatial regulation can be mediated by degree of membrane curvature and inherent differences in bulk lipid compositions among membrane organelles. Temporal regulation can occur through metabolism of PIs and diacylglycerol (DAG), the second messenger Ca2+, and local membrane curvature changes induced by cell signaling processes. Moreover, a number of peripheral proteins are regulated in both a spatial and temporal fashion. For instance, proper targeting may require a certain degree of membrane curvature harboring a temporally regulated lipid or the presence of two different lipids with docking sites on a single protein. This dual mode of recognition by a single lipid-binding domain is referred to as coincidence detection (11) and has been observed for PX (17), PH (18), and C2 domains (13). Recently, it was also demonstrated that lipid-binding domains could be regulated by pH. The Kutateladze laboratory identified perhaps the most compelling feature among FYVE domains in their ability to target endosomes at acidic pH due to His protonation (19). This opened the door for them to demonstrate that other domains such as PH (20) and ENTH (21) can target membranes in a pH-dependent manner due to His protonation.
Because many proteins harboring lipid-binding domains are enzymes, the stereospecific recognition of lipid head group and hydrophobic membrane penetration have important functional consequences with regards to biological activity. Indeed, there are a number of cases where a disease is attributable to the abrogation of these properties (22). Thus, therapeutic intervention at the level of protein-lipid interactions may be invaluable. For instance, a recent computational structural analysis of several lipid-binding domains demonstrated that they have a druggable pocket, and several drug-like compounds have been developed that have demonstrated efficacy in inhibiting lipid binding (23). Bioinformatics and computational biology have also begun to play a major role in lipid-binding domain studies, eliciting predictions of potential lipid-binding proteins (24), the role of electrostatics in lipid binding (25), and assembly of databases that compile lipid-binding domain data and predictions of membrane binding properties (26, 27). The BAR domain field is one area where atomistic molecular dynamics simulations have excelled. Studies from the Voth (28) and Schulten (29) laboratories have been informative on how fast these domains induce membrane curvature, the curvature dependency of BAR-membrane associations, and how different arrangements of BAR domains on lipid surfaces lead to a variety of curvature changes.
C1 DOMAINS
The C1 domain was first identified as the interaction site for DAG and phorbol ester in PKCs. It has a well-conserved, cysteine-rich compact structure (∼50 amino acids) that contains five short β strands, a short α-helix, and two zinc ions (30). More than 30 different mammalian proteins contain the C1 domain, and most of them have been shown to bind DAG and phorbol ester. The X-ray crystal structure of the PKCδ C1B domain shows that it has unique structural features that are consistent with its membrane binding mechanism (6). The domain has a polar binding pocket for DAG/phorbol ester located at the tip of the molecule. This pocket is surrounded by hydrophobic and aromatic residues that penetrate the membrane (31) to anchor the domain for DAG binding. Surface plasmon resonance measurements indicate DAG binding increases the vesicle affinity of the PKC C1 domains by more than 2 orders of magnitude, mainly by reducing the dissociation rate constant. Because hydrophobic and aromatic residues surrounding the DAG binding pocket are exposed, isolated C1 domains typically have a tendency to aggregate in solution. Thus, the C1 domain in the full-length protein is often buried in the inactive form of the enzyme and becomes accessible to DAG or phorbol esters only after an interdomain conformational change in the case of PKCα (32) and chimaerins (33, 34).
Originally, it was thought DAG bound in the same mode as phorbol ester; however, minor variations in sequence homology are responsible for dramatic changes in affinity for DAG and phorbol ester, including abrogation of binding to one ligand and nanomolar affinity for the other. The structural basis of differential DAG and phorbol ester affinities of these C1 domains is not fully understood, partially due to lack of structural information on DAG coordination, which may stem from the difficulty associated with purifying these small hydrophobic domains. The recognition that the C1 domain is a drug target led to the development of a number of lead compounds, including several that are close to clinical trial. I refer you to the work of Peter Blumberg et al. (12) for critical insight.
C2 DOMAINS
Following its discovery in PKC, the C2 domain (∼130 residues) was identified in other proteins such as synaptotagmins and group IVA cytosolic phospholipase A2 (cPLA2α), which also bind membranes in a Ca2+-dependent fashion. The C2 domain represents the second-most abundant lipid-binding domain with at least 200 examples identified in the Pfam database. While most C2 domain proteins are peripheral and bind reversibly to membranes, some C2 parent proteins are transmembrane proteins involved in membrane trafficking. Although the parent protein is anchored to the membrane, these C2 domains can bind reversibly in a Ca2+-dependent manner (35). The lipid affinity as well as the Ca2+ affinity of the Ca2+-binding C2 domains can vary greatly. The general mechanism of how a number of C2 domains associate with membranes in a Ca2+-dependent mode is well established; however, physiological functions and Ca2+- and membrane-binding properties of a large portion of C2 domains still remain unknown. Structural studies have shown that C2 domains have a common fold of conserved eight-stranded antiparallel β-sandwich connected by surface loops (7). The specificity in C2 domain targeting arises in the surface loops, which are variable in amino acid sequence and conformation and most often involved in lipid binding. Also of functional consequence is a cationic patch in the concave face of the β-sandwich, termed the cationic β-groove, which varies in size and electrostatics among C2 domains (13). Cationic β-grooves have been shown to bind ceramide-1-phosphate (36) as well as PIs including PI(4,5)P2 (37). Thus, many C2 domains are able to coordinate multiple lipids in both a Ca2+-dependent or independent manner. The preliminary data available on the dual lipid recognition mode of C2 domains suggests C2 domains may be multiply regulated by different lipids and Ca2+ signals and in some cases may require coincidence detection (e.g., interaction with multiple lipid targets) to achieve high affinity and cellular localization.
PH DOMAINS
The PH domain is composed of ∼100 amino acids and is the most abundant lipid-binding domain with >225 examples identified (14). Of the seven PIs in the mammalian cell, the PH domain binds specifically to PIP3, PI(4,5)P2, or PI(3,4)P2 (14). Reports of binding PI(3)P and PI(4)P are well documented, but the reliability of this specificity is still controversial. The membrane binding of PH domains is initially driven by nonspecific electrostatic interactions, which is followed by specific PI binding to increase the membrane residence time. A recent report demonstrated some PH domains anchor to the membrane through aliphatic residues adjacent to the PI-binding site (38).
The majority of PH domains have a conserved basic motif [K-Xn-(K/R)-X-R] in which the basic lysines and arginines play an important role in forming H-bonds with the head group of the PI. Other basic residues located within the domain vary from domain to domain and can provide a stronger binding affinity and create a unique binding pocket. Two distinct members of the PH domain family (TIAM1 and ARHGAP9) (39) were recently discovered that bind membranes through a site on the opposite side of the β1-β2 loop, suggesting that there are still novel PH domains to be discovered within the genome.
The importance of PH domains and disease was recently highlighted in an elegant study demonstrating that an E17K mutation in the AKT1 PH domain causes cancer. The E17K mutant was constitutively active due to pathological localization of E17K to the plasma membrane (PM) (22). The Falke laboratory (40) has shed some light on this pathological mechanism, demonstrating that the PI specificity of the AKT1 PH domain is drastically altered by the E17K mutation. Their biophysical analysis demonstrated E17K binds PI(4,5)P2 with even greater affinity than PIP3, and the constitutive PM localization of E17K may be due to binding to pools of PI(4,5)P2 often found in high concentration on the inner leaflet of the PM.
CONCLUSIONS and FUTURE DIRECTIONS
The critical role membrane-protein interactions play in the execution and regulation of many cellular processes, including cell signaling and membrane trafficking, has become evident in the past decade. Since the first discovery of a lipid-binding domain in 1989, progress in our understanding of the membrane binding mechanisms of these small modular domains and their host proteins has been substantial thanks to rapid developments in structural biology, computational biology, in vitro biophysical studies, and microscopic cell imaging.
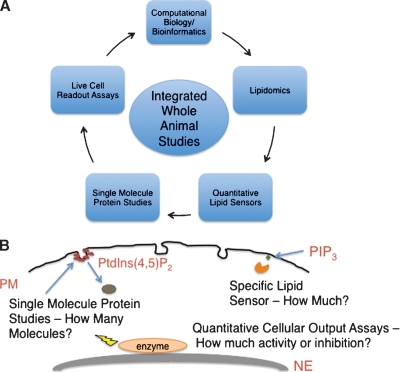
The main gaps in the field lie in understanding the detailed orchestration of cell signaling and membrane trafficking events mediated by lipid-binding domains. For instance, some questions in need of answers are: 1) How much of a particular signaling lipid is necessary to elicit a basal physiological response? 2) How many proteins (or lipid-binding domains) must associate with the biological membrane to obtain physiological activity? 3) How do different cell types or disease states influence the recruitment and activation of lipid binding proteins? For example, for enzymes with multiple binding sites such as PKC or cPLA2, are there different modes of activation where different membranes induce different patterns of activity? 4) How many lipid-binding domains or parent proteins are necessary for membrane remodeling in processes such as endocytosis? Figure 2 illustrates a multipronged approach to answering these questions, which should be possible with recently developed methodologies.
Fig. 2.
Illustration of future directions of lipid-binding domain research. A: Future directions will be geared toward integration of new technologies to perform whole-animal studies with translational capabilities. Combining single molecule imaging of protein dynamics with quantitative lipid sensing could lead to an unprecedented view of the molecular architecture of signaling complexes. Computational biology will serve to predict signaling cascades, drug targets, and the effects of drug compounds on the disease state of the cell. B: Depiction of how new technologies will be applied to live cell studies.
The lipidomics initiative has made significant strides in elucidating the different lipid structures among cell types and membrane organelles. Lipid sensors such as green fluorescent protein (GFP)-tagged lipid-binding domains have provided qualitative data on PI and DAG levels, and a recent DAG reporter shows promise as a more quantitative assessment of DAG levels in real-time (41). Certainly, even more quantitative lipid sensors should be possible such as those engineered to detect phosphatidylserine (42). Synthetic sensors may hold promise, as they can be engineered to harbor imaging capabilities such as those of Ca2+ indicators developed by Tsien et al. (43). Limitations in the lipid-binding realm will be sensitivity, selectivity, cellular uptake, and inhibition of downstream signaling events. These sensors will be less likely to interact with cellular proteins than GFP-domain fusions and with high sensor affinity may be able to overcome serving as a dominant negative in studying signaling events. Single molecule studies using fluorescence correlation spectroscopy (FCS) hold promise for investigating quantitative questions of lipid diffusion and lipid binding. The FCS studies should unveil the differences between ensemble and single molecule measurements and can answer questions such as: How many ENTH domains does it take to bend a locale region of the PM to initiate endocytosis? Combining the proposed studies such as in an elegant genome-wide study of PIP3 regulated PH domains by recursive-learning algorithm and rapid live imaging (44) will yield a quantitative picture of lipid-dependent events and should impact translation studies.
Acknowledgments
I thank Wonhwa Cho, Diana Murray, Tatiana Kutateladze, and Bradley Smith for helpful discussions. I apologize to authors whose works could not be cited due to significant space limitation.
Abbreviations
cPLA2, cytosolic phospholipase A2
DAG, diacylglycerol
FCS, fluorescence correlation spectroscopy
GFP, green fluorescent protein
PH, pleckstrin homology
PI, phosphoinositide
PIP3 or PtdIns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate
PIP2 or PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate
PKC, protein kinase C
PM, plasma membrane
PS, phosphatidylserine
This research was supported by grants from the American Heart Association (0735350N), the American Cancer Society (IRG-84-002-22), and the Indiana University School of Medicine.
Published, JLR Papers in Press, November 13, 2008.
References
- 1.Yuan T. L., and L. C. Cantley. 2008. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 27 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudhahar C. G., R. M. Haney, Y. Xue, and R. V. Stahelin. 2008. Cellular membranes and lipid-binding domains as attractive targets for drug development. Curr. Drug Targets. 9 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meer G. 2005. Cellular lipidomics. EMBO J. 24 3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho W., and R. V. Stahelin. 2005. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34 119–151. [DOI] [PubMed] [Google Scholar]
- 5.Takai Y., A. Kishimoto, Y. Iwasa, Y. Kawahara, T. Mori, and Y. Nishizuka. 1979. Calcium-dependent activation of a multifunctional protein kinase by membrane phospholipids. J. Biol. Chem. 254 3692–3695. [PubMed] [Google Scholar]
- 6.Zhang G., M. G. Kazanietz, P. M. Blumberg, and J. H. Hurley. 1995. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 81 917–924. [DOI] [PubMed] [Google Scholar]
- 7.Sutton R. B., B. A. Davletov, A. M. Berghuis, T. C. Sudhof, and S. R. Sprang. 1995. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 80 929–938. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson K. M., M. A. Lemmon, J. Schlessinger, and P. B. Sigler. 1995. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 83 1037–1046. [DOI] [PubMed] [Google Scholar]
- 9.DiNitto J. P., T. C. Cronin, and D. G. Lambright. 2003. Membrane recognition and targeting by lipid-binding domains. Sci. STKE. 2003 re16. [DOI] [PubMed] [Google Scholar]
- 10.Hurley J. H. 2006. Membrane binding domains. Biochim. Biophys. Acta. 1761 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemmon M. A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9 99–111. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg P. M., N. Kedei, N. E. Lewin, D. Yang, G. Czifra, Y. Pu, M. L. Peach, and V. E. Marquez. 2008. Wealth of opportunity: the C1 domain as a target for drug development. Curr. Drug Targets. 9 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho W., and R. V. Stahelin. 2006. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 1761 838–849. [DOI] [PubMed] [Google Scholar]
- 14.Lemmon, M. A. 2007. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 81–93. [DOI] [PMC free article] [PubMed]
- 15.Kutateladze T. G. 2007. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol 3-phosphate containing membranes. Prog. Lipid Res. 46 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh T., and P. De Camilli. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 1761 897–912. [DOI] [PubMed] [Google Scholar]
- 17.Karathanassis D., R. V. Stahelin, J. Bravo, O. Perisic, C. M. Pacold, W. Cho, and R. L. Williams. 2002. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao C., G. Du, K. Skowronek, M. A. Frohman, and D. Bar-Sagi. 2007. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9 706–712. [DOI] [PubMed] [Google Scholar]
- 19.Lee S. A., R. Eyeson, M. L. Cheever, J. Geng, V. V. Verkhusha, C. Burd, M. Overduin, and T. G. Kutateladze. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 102 13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J., R. M. Haney, M. Vora, V. V. Verkhusha, R. V. Stahelin, and T. G. Kutateladze. 2008. Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 49 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hom R. A., M. Vora, M. Regner, O. M. Subach, W. Cho, V. V. Verkhusha, R. V. Stahelin, and T. G. Kutateladze. 2007. pH-dependent binding of the epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P(2)-containing bilayers. J. Mol. Biol. 373 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpten J. D., A. L. Faber, C. Horn, G. P. Donoho, S. L. Briggs, C. M. Robbins, G. Hostetter, S. Boguslawski, T. Y. Moses, S. Savage, et al. 2007. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 448 439–444. [DOI] [PubMed] [Google Scholar]
- 23.Segers K., O. Sperandio, M. Sack, R. Fischer, M. A. Miteva, J. Rosing, G. A. Nicolaes, and B. O. Villoutreix. 2007. Design of protein membrane interaction inhibitors by virtual ligand screening, proof of concept with the C2 domain of factor V. Proc. Natl. Acad. Sci. USA. 104 12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj N., R. V. Stahelin, R. E. Langlois, W. Cho, and H. Lu. 2006. Structural bioinformatics prediction of membrane-binding proteins. J. Mol. Biol. 359 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulgrew-Nesbitt A., K. Diraviyam, J. Wang, S. Singh, P. Murray, Z. Li, L. Rogers, N. Mirkovic, and D. Murray. 2006. The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta. 1761 812–826. [DOI] [PubMed] [Google Scholar]
- 26.Bhardwaj N., R. V. Stahelin, G. Zhao, W. Cho, and H. Lu. 2007. MeTaDoR: a comprehensive resource for membrane targeting domains and their host proteins. Bioinformatics. 23 3110–3112. [DOI] [PubMed] [Google Scholar]
- 27.Lomize M. A., A. L. Lomize, I. D. Pogozheva, and H. I. Mosberg. 2006. OPM: orientations of proteins in membranes database. Bioinformatics. 22 623–625. [DOI] [PubMed] [Google Scholar]
- 28.Blood P. D., and G. A. Voth. 2006. Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 103 15068–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arkhipov A., Y. Yin, and K. Schulten. 2008. Four-scale description of membrane sculpting by BAR domains. Biophys. J. 95 2806–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colon-Gonzalez F., and M. G. Kazanietz. 2006. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta. 1761 827–837. [DOI] [PubMed] [Google Scholar]
- 31.Medkova M., and W. Cho. 1999. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem. 274 19852–19861. [DOI] [PubMed] [Google Scholar]
- 32.Stahelin R. V., J. Wang, N. R. Blatner, J. D. Rafter, D. Murray, and W. Cho. 2005. The origin of C1A–C2 interdomain interactions in protein kinase Calpha. J. Biol. Chem. 280 36452–36463. [DOI] [PubMed] [Google Scholar]
- 33.Canagarajah B., F. C. Leskow, J. Y. Ho, H. Mischak, L. F. Saidi, M. G. Kazanietz, and J. H. Hurley. 2004. Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin. Cell. 119 407–418. [DOI] [PubMed] [Google Scholar]
- 34.Colon-Gonzalez F., F. Coluccio Leskow, and M. G. Kazanietz. 2008. Identification of an autoinhibitory mechanism that restricts C1 domain-mediated activation of the Rac-GAP alpha 2-chimaerin. J. Biol. Chem. 283 35247–35257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis D. B., K. R. Doherty, A. J. Delmonte, and E. M. McNally. 2002. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 277 22883–22888. [DOI] [PubMed] [Google Scholar]
- 36.Stahelin R. V., P. Subramanian, M. Vora, W. Cho, and C. E. Chalfant. 2007. Ceramide-1-phosphate binds group IVA cytosolic phospholipase a2 via a novel site in the C2 domain. J. Biol. Chem. 282 20467–20474. [DOI] [PubMed] [Google Scholar]
- 37.Corbalan-Garcia S., J. Garcia-Garcia, J. A. Rodriguez-Alfaro, and J. C. Gomez-Fernandez. 2003. A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase Calpha. J. Biol. Chem. 278 4972–4980. [DOI] [PubMed] [Google Scholar]
- 38.Manna D., A. Albanese, W. S. Park, and W. Cho. 2007. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 282 32093–32105. [DOI] [PubMed] [Google Scholar]
- 39.Ceccarelli D. F., I. M. Blasutig, M. Goudreault, Z. Li, J. Ruston, T. Pawson, and F. Sicheri. 2007. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 282 13864–13874. [DOI] [PubMed] [Google Scholar]
- 40.Landgraf, K. E., C. Pilling, and J. J. Falke. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. Epub ahead of print. Oct 28, 2008. [DOI] [PMC free article] [PubMed]
- 41.Sato M., Y. Ueda, and Y. Umezawa. 2006. Imaging diacylglycerol dynamics at organelle membranes. Nat. Methods. 3 797–799. [DOI] [PubMed] [Google Scholar]
- 42.Lampkins A. J., E. J. O'Neil, and B. D. Smith. 2008. Bio-orthogonal phosphatidylserine conjugates for delivery and imaging applications. J. Org. Chem. 73 6053–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grynkiewicz G., M. Poenie, and R. Y. Tsien. 1985. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260 3440–3450. [PubMed] [Google Scholar]
- 44.Park W. S., W. D. Heo, J. H. Whalen, N. A. O'Rourke, H. M. Bryan, T. Meyer, and M. N. Teruel. 2008. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell. 30 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]