Abstract
The sphingolipidome is the portion of the lipidome that encompasses all sphingoid bases and their derivatives. Whereas the most studied sphingoid base is sphingosine [(2S,3R,4E)-2-aminooctadecene-1,3-diol], mammals have dozens of structural variants, and hundreds of additional types have been found in other eukaryotic organisms and some bacteria and viruses. Multiplying these figures by the N-acyl-derivatives (“ceramides”) and the more than 500 phospho- and glyco- headgroups places the number of discrete molecular species in the tens of thousands or higher. Structure-specific, quantitative information about a growing fraction of the sphingolipidome can now be obtained using various types of chromatography coupled with tandem mass spectrometry, and application of these methods is producing many surprises regarding sphingolipid structure, metabolism, and function. Such large data sets can be difficult to interpret, but the development of tools that display results from genomic and lipidomic studies in a pathway relational, nodal, context can make it easier for investigators to deal with this complexity.
Keywords: glycosphingolipids, ceramides, sphingoid bases, metabolism, lipidomics, systems biology
An “-omic” analysis encompasses all the members of that category of biomolecules, or at least a subfraction that provides a comprehensive picture of the pertinent biological system (1). Its immediate deliverables are to reduce the likelihood of erroneous conclusions regarding a biological phenomenon because they have been based on too few components (for which the parable of the blind men describing an elephant is an oft cited analogy) and to accelerate the discovery of new relationships. Both of these are appealing for those who study sphingolipids because the “sphingolipidome” is comprised of many thousands of compounds (2) that can have synergistic or antagonistic biologic effects, as exemplified by the cytotoxicity of sphingosine versus the inhibition of cell death by sphingosine 1-phosphate (3). It is fitting to review this subject as part of the 50th anniversary issue of the Journal of Lipid Research because this journal has played an important role in the evolution of our understanding of this category of compounds by publishing over 500 articles on the chemistry and biochemistry of sphingolipids.
COMPONENTS OF THE SPHINGOLIPIDOME
The sphingolipidome is comprised of all sphingoid bases and their derivatives (2). Sphingoid bases have been defined as “… long-chain aliphatic amino alcohols... represented by the compound originally called ‘dihydrosphingosine' (2S,3R)-2-aminooctadecane-1,3-diol…[and]… imply a chain length of 18 carbon atoms” (4, 5). Dihydrosphingosine (also called sphinganine) and its N-acyl-derivatives [dihydroceramides (DHCer)] are intermediates in the de novo biosynthesis of ceramide (N-acyl-sphingosine), which has the sphingosine backbone (Fig. 1) (6). Mammalian sphingolipids have over a dozen other types of sphingoid base backbones that differ in the number of double bonds, hydroxyls such as 4-hydroxysphinganine, “phytosphingosine,” and 1-deoxysphinganine (7), longer or shorter chain lengths, branching methyl groups, and other modifications. Hundreds of additional variations are found in other organisms (8).
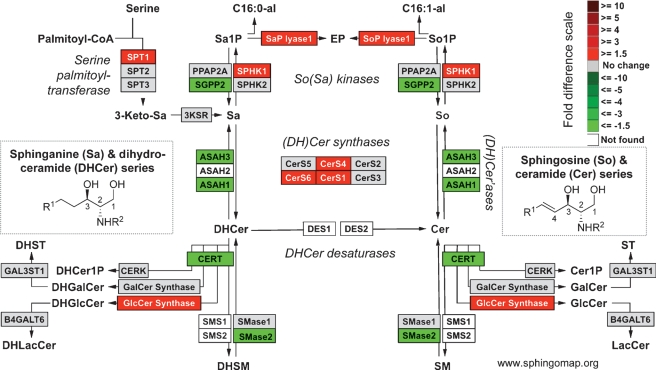
Fig. 1.
Illustration of differences in the expression of genes for early steps of sphingolipid metabolism using a KEGG-based microarray analysis tool (GenMapp v2.1) that has been modified to display this pathway more completely (www.sphingomap.org). The sphingolipid metabolites and genes (with the gene abbreviations shown in boxes, or enzyme names where gene names are ambiguous) are given for the condensation of serine and palmitoyl-CoA to form 3-ketosphinganine (3-ketoSa) by serine palmitoyltransferase, which is reduced to sphinganine (Sa), acylated to dihydroceramides, DHCer, by (DH)Cer synthases, and incorporated into more complex DH-sphingolipids (the 1-phosphate, DHCerP, sphingomyelins, DHSM, glucosylceramides, DHGlcCer, galactosylceramides, DHGalCer, lactosylceramides, DHLacCer, and sulfatides, or desaturated to Cer followed by headgroup addition. Also included are a number of the catabolic genes, e.g., sphingomyelinases, SMases, ceramidases, ASAH, sphingosine kinases, for the formation of sphinganine 1-phosphate (Sa1P) and sphingosine 1-phosphate (So1P), and phosphatases for the reverse reaction and the lyase that cleaves sphingoid base 1-phosphates to ethanolamine phosphate (EP), hexadecanal (C16:0al) and hexadecenal (C16:1al). For the purpose of illustration, the coloration is for the log2-fold difference in expression of these genes from a published microarray study (30) invasive lobular carcinoma cells versus normal lobular cells.
If one multiplies the number of known sphingoid bases by the dozens of different types of N-acyl-derivatives and the more than 500 phospho- and glyco- headgroups, the size of the mammalian sphingolipidome potentially numbers in the tens of thousands or higher. Data bases and bioinformatics tools to deal with this complexity are being developed by a number of organizations, such as the LIPID MAPS Consortium (www.lipidmaps.org), the Consortium for Functional Glycomics (http://www.functionalglycomics.org), the Japanese Lipid Bank (www.lipidbank.jp), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/glycan/) (9).
SPHINGOLIPIDOMIC ANALYSIS
Sphingolipids can be analyzed by a variety of traditional methods (reviewed in volumes 311 and 312 of Methods in Enzymology); however, for analysis of large numbers of compounds with a high degree of structural accuracy, the method of choice is mass spectrometry (MS) and tandem MS (MS/MS). Lipid MS uses various ionization methods [e.g., electrospray ionization, (ESI) and matrix-assisted laser-desorption ionization (MALDI)] combined with triple quadrupole or tandem quadrupole-linear ion trap mass analyzers for MS/MS and MSn (10), respectively, or for higher mass accuracy, time-of-flight (11) or Fourier transform instruments (12, 13). The combination of MS with liquid chromatography (10, 14), thin-layer chromatography (12, 15), and automated microfluidic “chip”-based devices (16) has been successful in meeting difficult analytical challenges posed by the presence of contaminants that suppress ionization, isomeric species, etc. Unfortunately, a major limitation for quantitative analysis is that certified internal standards are only available for the “core” sphingolipids (Avanti Polar Lipids, Alabaster, AL).
Additional challenges for sphingolipidomic analysis include the need for information about metabolic flux, which could be approached using stable isotope precursors (17, 18) and isotopomer analysis (19); and to know the location of these molecules, which is beginning to be addressed using bio-imaging MS. Bio-imaging MS (sometimes called tissue-imaging MS) generates images of tissue slices analogous to those using classical stains (such as H and E) but highlighting specific molecules (i.e., ions). The type of ions that can be seen and the resolution depends on the ionization method used, such as MALDI (20, 21), secondary ion MS (SIMS) (22–25), nanoSIMS (26), and desorption ESI (27). This technology is evolving rapidly, so tissue- and subcellular-imaging MS will undoubtedly become major adjuncts to traditional MS in the analysis of the sphingolipidome.
APPLICATION OF SPHINGOLIPIDOMICS TO SYSTEMS BIOLOGY
When “omic” methods are used, the amounts of data rapidly become unwieldy and tools are needed to facilitate visualization and interpretation of the results. Gene expression data can be displayed in the context of a pathway or biological process, as has been done in a recent comprehensive analysis of glycosyltransferase gene expression (28). Gene expression data are often represented using KEGG pathway tools, and this type of representation is shown in Fig. 1 using the pathway browser display tool GenMapp v2.1 (29) that has been expanded to include most of the currently known genes for sphingolipid biosynthesis (the download of this GenMapp pathway template is available at www.sphingomap.org). One of the key features of this depiction is that Fig. 1 reminds the viewer that DHCer are not merely intermediates in the biosynthesis of ceramides (Cer) and downstream sphingolipids, but are also utilized for biosynthesis of dihydrosphingolipids.
To illustrate the output of this tool, it was applied to data from a recently published study (30) that compared mRNA from invasive lobular carcinoma cells versus normal lobular cells (Fig. 1). This allows one to see readily that carcinoma cells have higher expression2 of CerS1, CerS4 and CerS6, which code for the ceramide synthases that make C16-, C18-, C20-, and C22-Cer (31); therefore, one would predict that they will have higher proportions of these Cer subspecies. Other interesting differences are elevations in serine palmitoyltransferase, sphingosine kinase, S1P lyase, and GlcCer synthase, any of which could alter the amounts of bioactive sphingolipids. Thus, this type of tool can be used to mine libraries of gene expression data to predict where interesting changes in metabolites might occur, which would then be followed up by analysis of the sphingolipids of the cells. When successful, this would represent a powerful bridging of “omic” technologies that could lead not only to a better understanding of sphingolipids in a systems biology context, but also, to discovery of new disease biomarkers or therapeutic targets.
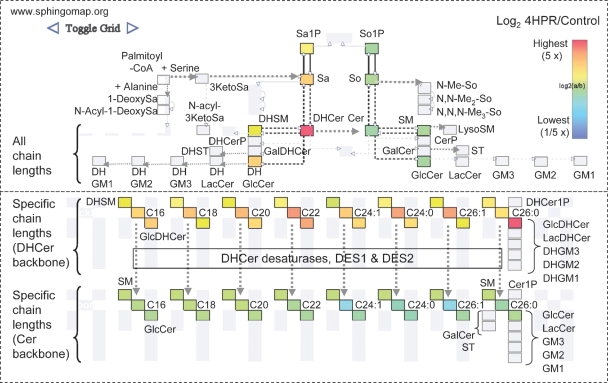
Similar types of data visualization tools have been created to display changes in the metabolites. Figure 2 was created using SphingoVisGrid (available on www.sphingomap.org)3 to compare the amounts of the sphingolipids in the breast cancer cell line MCF7 with and without treatment with fenretinide (4-hydroxyphenylretinamide, 4HPR), an anti-cancer drug currently undergoing clinical trials that has been found to affect sphingolipid metabolism (32, 33). The layout of the upper half of the display is similar to the GenMapp pathway (c.f., Fig. 1 and 2)4 with DHCer and downstream metabolites on the left and Cer and downstream metabolites on the right, and the lower layout depicts the specific chain length subspecies. In both of these formats, it is evident that fenretinide elevates the amounts of all of the sphingolipids with a sphinganine backbone (DHCer and other dihydro-sphingolipids as well as sphinganine and sphinganine 1-phosphate) whereas all of the compounds with a sphingosine backbone (Cer, SM, etc.) decrease, which is due to inhibition of DHCer desaturase (32, 33).
Fig. 2.
Display of the relative amounts of sphingolipids for early steps of sphingolipid metabolism and backbone turnover. This depicts the relative amounts of sphingolipids in MCF7 breast cancer cells treated with 10 μM fenretinide (4HPR) for 24 h versus the control cells minus the drug using the SphingoVisGrid tool (www.sphingomap.org). The pathway layout in the upper panel is similar to the gene display in Fig. 1 wherein all of the N-acyl-chain lengths of the complex sphingolipids are summed in one box. It also includes the additional metabolites (from upper left): 1-deoxysphinganine (1-deoxySa) and related species (such as N-acyl-1-deoxysphinganine); N-acyl-3-ketosphinganine (N-acyl3KetoSa); N-methyl (mono-, di- and tri-) derivatives of the sphingoid bases (N-MeSo, etc.); and gangliosides GM1, GM2, and GM3 with distinction of Cer and DHCer backbones. The lower box distinguishes the complex sphingolipids by the specific sphingoid base backbone and the chain length and number of double bonds of the N-acyl-fatty acid (e.g., C16 = palmitic acid, C24:1 = nervonic acid, etc.). The coloring of each subspecies reflects the fold difference (log2 as shown by the scale) for 4HPR-treated cells versus the control; gray boxes are for analytes that were not measured in this experiment.
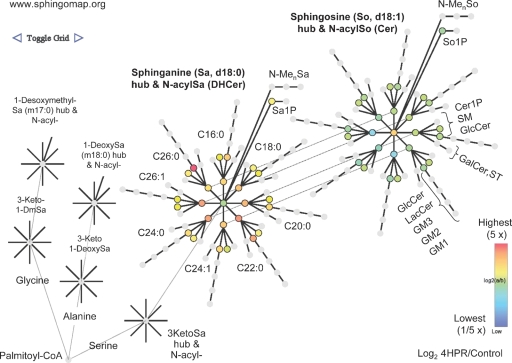
By toggling the link shown in the upper left of Fig. 2 online, SphingoVisGrid also displays this data in a pathway related, nodal, context (Fig. 3) that is similar to the interaction diagrams used in systems biology. This depiction includes nodes for the additional sphingoid bases (i.e., 1-deoxysphinganine and 1-desoxymethylsphinganine, on the left) that have been found in mammalian cells (7), illustrating how future expansions of SphingoVisGrid could add nodes for other sphingoid bases and downstream metabolites (2) when the availability of analytical data for them warrants.
Fig. 3.
Display of the relative amounts of sphingolipids for early steps of sphingolipid metabolism and backbone turnover in a pathway related, nodal, context. This depiction is obtained by toggling from Fig. 2 in SphingoVisGrid (www.sphingomap.org) and shows the same data set comparison (sphingolipids from MCF7 cells with versus without treatment with fenretinide). Beginning at the lower left corner is the initial condensation of palmitoyl-CoA with serine (or alanine or glycine for formation of 1-deoxy- and 1-desoxymethyl-Sa, respectively) and proceeding to the right are the formation of 3-ketosphinganine (3KSa), sphinganine (Sa) and downstream metabolites: (DH)Cer, (DH)SM, etc. The lines that radiate from the sphingoid base nodes reflect the different amide-linked fatty acids (shown by acyl chain length and number of double bonds). Also shown are the additional derivatives of sphingoid bases, the 1-phosphates (Sa1P and S1P) and N-methyl-metabolites (N-MenSa, etc.). Only a few of the nodes have been labeled because the rest fit the same pattern.
Investigators will eventually build computational models for the pathway, as has been reported in an elegant study of the simpler pathway in yeast (34, 35); however, modeling of the mammalian sphingolipid metabolism is still in its infancy (36).
CONCLUDING REMARKS
Because sphingolipids are involved in so many aspects of cell regulation as modulators of cell-cell and cell-substratum interactions, receptor regulation, and cell signaling (2, 3, 6, 37–40), the application of sphingolipidomic methods will help clarify the roles of these compounds in cell behavior and disease by revealing the ensemble of bioactive molecules that are playing important roles rather than limiting the focus to just a few species. Sphingolipidomic approaches have already found many interesting and often surprising new facets of sphingolipid biology, such as that sphingosine is a ligand for a nuclear receptor (41), and associations with disease, such as a role for sulfatides in Alzheimers disease (42), C18-Cer in head and neck cancer (43), and sphigolipid biosynthesis in insulin resistance and metabolic disease (44).
Abbreviations
1-deoxySa, 1-deoxysphinganine
3-ketoSa, 3-ketosphinganine
Cer, ceramides
Cerase, ceramidase
Cer1P, ceramide 1-phosphate
DHCer, dihydroceramide
EP, ethanolamine phosphate
ESI, electrospray ionization
GalCer, galactosylceramide
GlcCer, glucosylceramide
GM, ganglioside (GM1, GM2, GM3)
KEGG, Kyoto Encyclopedia of Genes and Genomes
LacCer, lactosylceramide
MS/MS, tandem mass spectrometry
Sa, sphinganine
Sa1P, sphinganine 1-phosphate
So, sphingosine
So1P, sphingosine 1-phosphate
SM, sphingomyelin
SMase, sphingomyelinase
ST, sulfatide
This work was supported by the Lipid MAPS Consortium grant (GM069338) (A.H.M.) and in part by funds from Microsoft Research, the National Institutes of Health (Bioengineering Research Partnership R01CA108468, P20GM072069, the Center for Cancer Nanotechnology Excellence U54CA119338), and the Georgia Cancer Coalition (M.D.W.).
Published, JLR Papers in Press, November 21, 2008.
Footnotes
With “higher” contingent on confirmation of the microarray results with a more quantitative method such as quantitative RT-PCR.
The labels shown in Figs. 2 and 3 have been added to the diagram to orient the reader; these are not shown on the original data display to make it easier for the viewer to see patterns.
SphingoVisGrid can display additional metabolites but these are colored grey when they have not been measured in a particular experiment.
References
- 1.Auffray C., S. Imbeaud, M. Roux-Rouquie, and L. Hood. 2003. From functional genomics to systems biology: concepts and practices. C. R. Biol. 326 879–892. [DOI] [PubMed] [Google Scholar]
- 2.Merrill A. H., Jr., M. D. Wang, M. Park, and M. C. Sullards. 2007. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 32 457–468. [DOI] [PubMed] [Google Scholar]
- 3.Hait N. C., C. A. Oskeritzian, S. W. Paugh, S. Milstien, and S. Spiegel. 2006. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim. Biophys. Acta. 1758 2016–2026. [DOI] [PubMed] [Google Scholar]
- 4.Chester M. A. 1998. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids–recommendations 1997. Eur. J. Biochem. 257 293–298. [DOI] [PubMed] [Google Scholar]
- 5.Fahy E., S. Subramaniam, H. A. Brown, C. K. Glass, A. H. Merrill, Jr., R. C. Murphy, C. R. Raetz, D. W. Russell, Y. Seyama, W. Shaw, et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46 839–861. [DOI] [PubMed] [Google Scholar]
- 6.Bartke, N., and Y. A. Hannun. 2009. Bioactive sphingolipids: metabolism and function. J. Lipid Res. S91–S96. [DOI] [PMC free article] [PubMed]
- 7.Zitomer N. C., T. Mitchell, K. A. Voss, G. S. Bondy, S. T. Pruett, E. C. Garnier-Amblard, L. S. Liebeskind, H. Park, E. Wang, M. C. Sullards, et al. 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxy-sphinganine: A novel category of bioactive 1-deoxy-sphingoid bases and 1-deoxy-dihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruett S. T., A. Bushnev, K. Hagedorn, M. Adiga, C. A. Haynes, M. C. Sullards, D. C. Liotta, and A. H. Merrill, Jr. 2008. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J. Lipid Res. 49 1621–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto K., S. Kawano, S. Goto, K. F. Aoki-Kinoshita, M. Kawashima, and M. Kanehisa. 2005. A global representation of the carbohydrate structures: a tool for the analysis of glycan. Genome Inform. 16 214–222. [PubMed] [Google Scholar]
- 10.Sullards M. C., J. C. Allegood, S. Kelly, E. Wang, C. A. Haynes, H. Park, Y. Chen, and A. H. Merrill, Jr. 2007. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol. 432 83–115. [DOI] [PubMed] [Google Scholar]
- 11.Kirsch S., M. Zarei, M. Cindric, J. Muthing, L. Bindila, and J. Peter-Katalinic. 2008. On-line nano-HPLC/ESI QTOF MS and tandem MS for separation, detection, and structural elucidation of human erythrocytes neutral glycosphingolipid mixture. Anal. Chem. 80 4711–4722. [DOI] [PubMed] [Google Scholar]
- 12.Ivleva V. B., Y. N. Elkin, B. A. Budnik, S. C. Moyer, P. B. O'Connor, and C. E. Costello. 2004. Coupling thin-layer chromatography with vibrational cooling matrix-assisted laser desorption/ionization Fourier transform mass spectrometry for the analysis of ganglioside mixtures. Anal. Chem. 76 6484–6491. [DOI] [PubMed] [Google Scholar]
- 13.Vukelic Z., A. D. Zamfir, L. Bindila, M. Froesch, J. Peter-Katalinic, S. Usuki, and R. K. Yu. 2005. Screening and sequencing of complex sialylated and sulfated glycosphingolipid mixtures by negative ion electrospray Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 16 571–580. [DOI] [PubMed] [Google Scholar]
- 14.Sommer U., H. Herscovitz, F. K. Welty, and C. E. Costello. 2006. LC-MS-based method for the qualitative and quantitative analysis of complex lipid mixtures. J. Lipid Res. 47 804–814. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K., Y. Suzuki, N. Goto-Inoue, C. Yoshida-Noro, and A. Suzuki. 2006. Structural characterization of neutral glycosphingolipids by thin-layer chromatography coupled to matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight MS/MS. Anal. Chem. 78 5736–5743. [DOI] [PubMed] [Google Scholar]
- 16.Vukelic Z., M. Zarei, J. Peter-Katalinic, and A. D. Zamfir. 2006. Analysis of human hippocampus gangliosides by fully-automated chip-based nanoelectrospray tandem mass spectrometry. J. Chromatogr. A. 1130 238–245. [DOI] [PubMed] [Google Scholar]
- 17.Merrill A. H., Jr., M. C. Sullards, J. C. Allegood, S. Kelly, and E. Wang. 2005. Sphingolipidomics: high throughput, structure specific and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36 207–224. [DOI] [PubMed] [Google Scholar]
- 18.Tserng K. Y., and R. Griffin. 2004. Studies of lipid turnover in cells with stable isotope and gas chromatograph-mass spectrometry. Anal. Biochem. 325 344–353. [DOI] [PubMed] [Google Scholar]
- 19.Bederman I. R., T. Kasumov, A. E. Reszko, F. David, H. Brunengraber, and J. K. Kelleher. 2004. In vitro modeling of fatty acid synthesis under conditions simulating the zonation of lipogenic [13C]acetyl-CoA enrichment in the liver. J. Biol. Chem. 279 43217–43226. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., J. Allegood, Y. Liu, E. Wang, B. Cachon-Gonzalez, T. M. Cox, A. H. Merrill, Jr., and M. C. Sullards. 2008. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 80 2780–2788. [DOI] [PubMed] [Google Scholar]
- 21.Hankin J. A., R. M. Barkley, and R. C. Murphy. 2007. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sjovall P., J. Lausmaa, and B. Johansson. 2004. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 76 4271–4278. [DOI] [PubMed] [Google Scholar]
- 23.Roy S., D. Touboul, A. Brunelle, D. P. Germain, P. Prognon, O. Laprevote, and P. Chaminade. 2006. [Imaging mass spectrometry: a new tool for the analysis of skin biopsy. Application in Fabry's disease] Ann. Pharm. Fr. 64 328–334. [DOI] [PubMed] [Google Scholar]
- 24.Borner K., H. Nygren, B. Hagenhoff, P. Malmberg, E. Tallarek, and J. E. Mansson. 2006. Distribution of cholesterol and galactosylceramide in rat cerebellar white matter. Biochim. Biophys. Acta. 1761 335–344. [DOI] [PubMed] [Google Scholar]
- 25.Baker M. J., L. Zheng, N. Winograd, N. P. Lockyer, and J. C. Vickerman. 2008. Mass spectral imaging of glycophospholipids, cholesterol, and glycophorin a in model cell membranes. Langmuir. 24 11803–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraft M. L., P. K. Weber, M. L. Longo, I. D. Hutcheon, and S. G. Boxer. 2006. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science. 313 1948–1951. [DOI] [PubMed] [Google Scholar]
- 27.Wiseman J. M., D. R. Ifa, Q. Song, and R. G. Cooks. 2006. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl. 45 7188–7192. [DOI] [PubMed] [Google Scholar]
- 28.Nairn A. V., W. S. York, K. Harris, E. M. Hall, J. M. Pierce, and K. W. Moremen. 2008. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 283 17298–17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlquist K. D., N. Salomonis, K. Vranizan, S. C. Lawlor, and B. R. Conklin. 2002. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 31 19–20. [DOI] [PubMed] [Google Scholar]
- 30.Turashvili G., J. Bouchal, K. Baumforth, W. Wei, M. Dziechciarkova, J. Ehrmann, J. Klein, E. Fridman, J. Skarda, J. Srovnal, et al. 2007. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 7 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pewzner-Jung Y., S. Ben-Dor, and A. H. Futerman. 2006. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J Biol Chem. 281: 25001–25005. [DOI] [PubMed] [Google Scholar]
- 32.Zheng W., J. Kollmeyer, H. Symolon, A. Momin, E. Munter, E. Wang, S. Kelly, J. C. Allegood, Y. Liu, Q. Peng, et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758 1864–1884. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., B. J. Maurer, Y. Y. Liu, E. Wang, J. C. Allegood, S. Kelly, H. Symolon, Y. Liu, A. H. Merrill, Jr., V. Gouaze-Andersson, et al. 2008. N-(4-Hydroxyphenyl)retinamide increases dihydroceramide and synergizes with dimethylsphingosine to enhance cancer cell killing. Mol. Cancer Ther. 7 2967–2976. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Vasquez F., K. J. Sims, L. A. Cowart, Y. Okamoto, E. O. Voit, and Y. A. Hannun. 2005. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 433 425–430. [DOI] [PubMed] [Google Scholar]
- 35.Cowart L. A., and Y. A. Hannun. 2005. Using genomic and lipidomic strategies to investigate sphingolipid function in the yeast heat-stress response. Biochem. Soc. Trans. 33 1166–1169. [DOI] [PubMed] [Google Scholar]
- 36.Henning P. A., H. J. Merrill, and M. Wang. 2004. Dynamic pathway modeling of sphingolipid metabolism. Conf. Proc. IEEE Eng. Med. Biol. Soc. 4 2913–2916. [DOI] [PubMed] [Google Scholar]
- 37.Degroote S., J. Wolthoorn, and G. van Meer. 2004. The cell biology of glycosphingolipids. Semin. Cell Dev. Biol. 15 375–387. [DOI] [PubMed] [Google Scholar]
- 38.Hakomori S. I. 2008. Structure and function of glycosphingolipids and sphingolipids: recollections and future trends. Biochim. Biophys. Acta. 1780 325–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hannun Y. A., and L. M. Obeid. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9 139–150. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez S. E., S. Milstien, and S. Spiegel. 2007. Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol. Metab. 18 300–307. [DOI] [PubMed] [Google Scholar]
- 41.Urs A. N., E. Dammer, S. Kelly, E. Wang, A. H. Merrill, Jr., and M. B. Sewer. 2007. Steroidogenic factor-1 is a sphingolipid binding protein. Mol. Cell. Endocrinol. 265–266 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han X. 2007. Potential mechanisms contributing to sulfatide depletion at the earliest clinically recognizable stage of Alzheimer's disease: a tale of shotgun lipidomics. J. Neurochem. 103 (Suppl 1): 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koybasi S., C. E. Senkal, K. Sundararaj, S. Spassieva, J. Bielawski, W. Osta, T. A. Day, J. C. Jiang, S. M. Jazwinski, Y. A. Hannun, et al. 2004. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 279 44311–44319. [DOI] [PubMed] [Google Scholar]
- 44.Holland W. L., and S. A. Summers. 2008. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 29 381–402. [DOI] [PMC free article] [PubMed] [Google Scholar]