Abstract
Cells obtain FAs either from LPL-catalyzed hydrolysis of lipoprotein triglyceride or from unesterified FFAs associated with albumin. LPL also influences uptake of esterified lipids such as cholesteryl and retinyl esters that are not hydrolyzed in the plasma. This process might not involve LPL enzymatic activity. LPL is regulated by feeding/fasting, insulin, and exercise. Although a number of molecules may affect cellular uptake of FFAs, the best characterized is CD36. Genetic deletion of this multiligand receptor reduces FFA uptake into skeletal muscle, heart, and adipose tissue, and impairs intestinal chylomicron production and clearance of lipoproteins from the blood. CD36 is regulated by some of the same factors that regulate LPL, including insulin, muscle contraction, and fasting, in part, via ubiquitination. LPL and CD36 actions in various tissues coordinate biodistribution of fat-derived calories.
Keywords: lipase, fatty acids, CD36, insulin actions, triglyceride
There are several pathways that allow the uptake of circulating lipids into cells. Cell surface receptors mediate the uptake of whole lipoproteins. FA uptake is more similar to that of the other major blood nutrient, glucose. Unlike glucose, which is water soluble, FAs are either associated with albumin, so-called free (FFA), or are esterified as a component of triglycerides (TGs), phospholipids, and esterified cholesterol. Esterified FAs within VLDL and chylomicrons comprise >90% of blood FAs. Nonetheless, the rapid plasma turnover of FFA has the potential to supply large amounts of energy to tissues.
LIPID UPTAKE VIA THE LIPOLYSIS PATHWAY
The importance of the lipolysis reaction in the catabolism of TG-rich lipoproteins was shown more than 50 years ago by the discovery that intravenous heparin injection leads to the release of LPL into the plasma and reduces postprandial lipemia. Subsequent studies established that LPL is robustly expressed in tissues like adipose and muscle that require large influxes of FAs for storage or energy and that LPL is the primary enzyme responsible for chylomicron- and VLDL-TG lipolysis. However, several questions on the role of this enzyme in fuel partitioning and tissue lipid uptake, its regulation, and its importance for vascular biology remained to be answered.
Genetic modulation of LPL protein and its sites of expression have partially elucidated how LPL affects physiology and has confirmed prior in vitro studies. Although humans with LPL deficiency develop severe hypertriglyceridemia and are at risk for pancreatitis, mice with spontaneous or genetic loss of LPL die within 24 h of birth. Prior to death, LPL-deficient mice develop hypoglycemia, a process that is partially abrogated by ketone body production via LPL expression in the liver (1). In contrast, tissue-specific overexpression of LPL in skeletal muscle and liver increased cellular stores of TG and probably other lipids and led to insulin resistance (2). Loss of LPL in adipose tissue and its overexpression in skeletal muscle reduced adipose development when these mice were crossed onto the obesity ob/ob background (3). However, in nonobese mice, adipose stores were normal. This suggests compensation by de novo TG synthesis or uptake of plasma lipids through other pathways (4). In pancreatic β cells, both overexpression and deletion of LPL impaired insulin secretion (5). Although tissue-specific effects of LPL are still being explored, the initial hypothesis of Greenwood (6) that LPL serves as a “gatekeeper” to direct calories to specific tissues is clearly correct.
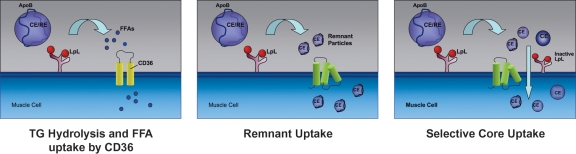
LPL increases uptake of lipids other than TG (Fig. 1), e.g., selective uptake of cholesteryl esters by skeletal muscle is mediated by catalytically inactive LPL (7). LPL might act like a neutral lipid transfer protein, akin to cholesteryl ester transfer protein, or during remodeling of the lipoprotein particle, small lipid particles containing core and surface lipid and protein exclusive of apoB could be shed from the lipoproteins. These particles would be similar or perhaps identical to the lipid/apoprotein complexes that are known to transfer to HDL. LPL also augments LDL uptake either by approximation of LDL to the LDL receptor or by uptake along with recycling of proteoglycans on the cell surface. The recent development of a mouse with a defect of proteoglycan sulfation associated with increased postprandial lipemia (8) is consistent with this type of lipoprotein uptake process.
Fig. 1.
LPL-mediated uptake of lipids. Left panel: LPL hydrolyzes lipoprotein TG releasing FFAs that are internalized by cells. Much of this uptake is via cell surface receptors such as CD36. Middle panel: LPL also creates remnant lipoproteins that interact with cell surface lipoprotein receptors. Right panel: Another option for uptake of core lipids is that lipolysis creates small particles that contain both surface and core lipid as well as apoproteins, exclusive of apoB. Inactive LPL (iLPL) on the cell surface might facilitate cellular internalization of these particles. Such a pathway might allow cholesteryl and retinyl ester uptake into “high lipolysis” organs such as the heart.
Using heart-specific LPL knockout mice, the role of LPL was studied in the setting of normal uptake of FFA. The presence of a marked increase in glucose uptake (9) and the development of heart dysfunction with aging and increased afterload (10) confirmed that FFAs cannot substitute for loss of LPL. As had been reported in vitro (11, 12), reduction in peroxisome proliferator-activated receptor (PPAR) downstream gene expression confirmed the hypothesis that LPL creates ligands for this transcription factor.
Metabolism of fat-soluble vitamins is also affected by LPL actions. Overexpression of LPL in skeletal muscle increased α-tocopherol (13). Dietary retinyl ester is a marker for chylomicron metabolism. Tissue vitamin A circulates in chylomicrons and in association with circulating retinol binding protein. Retinol binding protein knockout mice are visually impaired at birth, a defect that improves as the mice age (14). This is likely due to tissue acquisition of dietary retinyl ester, a process that correlates with LPL expression (15).
Arterial expression of LPL is a marker of atherosclerosis. Zilversmit postulated that LPL converts chylomicrons into atherogenic remnant lipoproteins (16). Lack of LPL in macrophages leads to less atherosclerosis (17). However, mechanisms other than those originally postulated might be operative. LPL increases the association of LDL with matrix; FFAs and lipolysis alter endothelial cell barrier function and cause vascular inflammation (18).
NUTRITIONAL AND HORMONAL REGULATION OF LPL
LPL actions within tissues are modulated at both the transcriptional and posttranscriptional levels. The latter might involve actions of the glycosylphosphatidylinositol HDL binding protein (GPIHBP) protein (19), angiopoietin-like proteins, which reduce LPL dimer formation (20), and the recently described lipase maturation factor (21). LPL regulation is tissue specific. LPL is present in the liver during fetal and early postnatal life but is then suppressed by a putative transcriptional regulatory mechanism, perhaps involving a novel transcription factor, termed RF-1-LPL, which binds to an NF-1-like site in the region of the glucocorticoid response element. This extinction of the hepatic expression of LPL is also under the influence of thyroid hormone and glucocorticoids.
In the mammary gland, synthesis of LPL is induced during late pregnancy and lactation, perhaps via the effects of prolactin (22). Partially dedifferentiated and delipidated adipocytes, rather than epithelial cells, are the source of the lipase. In brown adipose tissue, LPL activity is increased during cold exposure by both a transcriptional and translational/posttranslational mechanism that involves β-adrenergic stimulation. Stress, both chronic and acute, decreases LPL activity in white adipose tissue but increases the LPL activity in cardiac and skeletal muscle and the adrenal glands. These changes may be mediated by catecholamines (23).
The LPL response to feeding and fasting is also tissue specific. In rodents and humans, this regulation in white adipose tissue and muscle exists mostly at the posttranslational level. Adipose tissue LPL activity is high after feeding and low during fasting, whereas in most studies, the opposite is true in the heart and skeletal muscle. Although in human subjects an insulin infusion had a divergent effect on LPL activity in adipose tissue (increased) and skeletal muscle (decreased), 2 weeks of a high-carbohydrate diet or high-fat diet increased the LPL response to feeding in both adipose tissue and skeletal muscle (24). In adipose tissue only, there was a significant difference between the two diets in the LPL meal response (high-carbohydrate diet > high-fat diet). In rats and humans, modulation of adipose tissue LPL is not accompanied by changes in LPL mRNA or LPL mass. One likely possibility is that the rapid downregulation of adipose LPL is due to angiopoietin-like protein 4 inactivation of LPL (20).
A more complex mechanism for LPL regulation exists in the heart. The heparin-releasable LPL activity increases several-fold with fasting. A similar mechanism to that in adipose tissue appears to modulate the LPL activity in the heart by a transition between active and inactive forms of LPL (25). In addition, alterations in the distribution of LPL between the vascular endothelium and other sites within the heart explain some of the differences in the enzyme activity with fasting and feeding (26).
Insulin is a major regulator of LPL activity in adipose tissue. During adipocyte differentiation, insulin increases LPL gene transcription; in mature adipocytes or adipose tissue, insulin stimulates LPL by increasing the level of LPL mRNA and regulating LPL activity through both posttranscriptional and posttranslational mechanisms. Glucose also increases adipose tissue LPL activity. The glucose stimulatory effect seems to work predominantly through the glycosylation of LPL, which is essential for LPL catalytic activity and secretion. In addition, glucose also stimulates LPL synthetic rate and potentiates the stimulatory effect of insulin but does not affect the level of LPL mRNA.
REGULATION OF FFA UPTAKE VIA THE FA TRANSLOCASE CD36
FFAs liberated from circulating lipoproteins and FFAs bound to albumin transfer into the cell via predominantly a saturable protein-facilitated process (27, 28). Several proteins have been implicated in facilitating FFA uptake and more may still await identification, but the scavenger receptor CD36 has been the best characterized as a FA translocase. First identified on platelets as GPIV, an 88-kDa thrombospondin and collagen receptor, CD36 was linked to lipid metabolism in 1993 when it acquired two new functions as a macrophage receptor for oxidized LDL and as an adipocyte receptor/transporter for long-chain FAs. CD36 membrane topology is proposed to include two transmembrane domains with both N and C termini in the cytoplasm (29). The purified protein binds long- but not short-chain FAs in vitro and a putative FA binding site (residues 127 and 279) was postulated based on similarity of the predicted CD36 secondary sequence with that of human muscle FA binding protein (30).
CD36 deficiency in mice significantly impairs FA uptake by heart, skeletal muscle, and adipose tissue (31). Fasting plasma TG and FA levels are increased. The decrease in muscle FA utilization enhances insulin sensitivity of the tissue (32) and is protective against diet-induced obesity (33). Muscle-targeted CD36 overexpression reduces levels of plasma FAs and TG (34) and associates with hyperglycemia and hyperinsulinemia. In the spontaneously hypertensive rat, mutations in the CD36 gene were linked to the hyperlipidemia and insulin resistance observed in this model and the symptoms were improved by transgenic expression of wild-type CD36 (35).
CD36 is expressed throughout the digestive tract including on taste bud cells in the tongue (36), in the stomach, the small intestine, and the colon (37). In the small intestine, CD36 exhibits a proximal to distal decreasing expression gradient (38). It is now clear that the protein plays an important role in chylomicron production (39) in part by facilitating uptake of FA and cholesterol by proximal enterocytes (38) where chylomicron formation is most efficient. CD36 may also facilitate intracellular trafficking of FAs and cholesterol for packaging into lipoproteins, but this remains to be explored. Despite the proximal defects in lipid uptake and chylomicron formation, net intestinal lipid absorption is not altered except for very-long chain FAs (40). However, the slowed absorption is associated with a reduction in fat intake (41). Clearance of blood lipoproteins is also defective in CD36 null mice, which may reflect feedback inhibition of tissue LPL by FA (39, 42).
CD36 binding and uptake of FA at the level of taste bud cells may contribute to fat taste perception and to the regulation of fat preference (36, 41). FA interaction with the protein triggers a signaling cascade that results in neurotransmitter release and relay of signal to the central nervous system (43). In the small intestine, production of the satiety messenger oleoylethanolamide after fat ingestion is dependent on CD36-facilitated oleic acid uptake (44).
In muscle, CD36 levels are chronically upregulated by insulin and muscle contraction (45). CD36 is a gene target of the PPAR in a tissue-specific fashion. In muscle during fasting, activation of PPARδ upregulates CD36 to increase FA utilization (46).
CD36 function is acutely regulated by changes in its subcellular distribution; the protein is present in intracellular vesicles (e.g., endosomes, endoplasmic reticulum, and lysosomes) and on mitochondria (47). The forkhead transcription factor FoxO1 (48) activated with low insulin states, AMP-activated protein kinase activation, and muscular contraction (45) recruits muscle CD36 from intracellular stores to the plasma membrane and enhance FA uptake. The signaling pathways involved in CD36 trafficking are largely unclear, but a role of the small GTPase Rab11a and its effector proteins has been suggested (45). CD36 trafficking may also be regulated by alteration of its ubiquitination, which modifies its protein interactions, subcellular distribution, and turnover (49). CD36 ubiquitination on lysines 469 and 472 in the C-terminal domain is inhibited by insulin and enhanced by FAs (50).
Common polymorphisms in the CD36 gene that significantly affect blood lipid levels and susceptibility to metabolic disease have been identified in humans (51). CD36 deficiency is relatively common (2–7%) in persons of Asian and African descent and is associated with many of the phenotypic alterations described in rodents, such as abnormalities of blood lipids, delayed clearance of blood FAs and TG, and defective FA uptake by the myocardium (52).
CONCLUSIONS
There is a redundancy of the pathways that mediate uptake of FAs and also of fat-soluble vitamins. The uptake pathways involving LPL and CD36 appear to be modulated under the same physiologic and pathological conditions. How these pathways overlap and the molecular details of their regulation remains unclear but can now be studied in genetically modified animals.
Published, JLR Papers in Press, November 24, 2008.
References
- 1.Merkel M., P. H. Weinstock, T. Chajek-Shaul, H. Radner, B. Yin, J. L. Breslow, and I. J. Goldberg. 1998. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J. Clin. Invest. 102 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J. K., J. J. Fillmore, Y. Chen, C. Yu, I. K. Moore, M. Pypaert, E. P. Lutz, Y. Kako, W. Velez-Carrasco, I. J. Goldberg, et al. 2001. Tissue-specific overexpression of lipoprotein lipase causes tissue- specific insulin resistance. Proc. Natl. Acad. Sci. USA. 98 7522–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstock P. H., S. Levak-Frank, L. C. Hudgins, H. Radner, J. M. Friedman, R. Zechner, and J. L. Breslow. 1997. Lipoprotein lipase controls fatty acid entry into adipose tissue, but fat mass is preserved by endogenous synthesis in mice deficient in adipose tissue lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 94 10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kratky D., R. Zimmermann, E. M. Wagner, J. G. Strauss, W. Jin, G. M. Kostner, G. Haemmerle, D. J. Rader, and R. Zechner. 2005. Endothelial lipase provides an alternative pathway for FFA uptake in lipoprotein lipase-deficient mouse adipose tissue. J. Clin. Invest. 115 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappan K. L., Z. Pan, G. Kwon, C. A. Marshall, T. Coleman, I. J. Goldberg, M. L. McDaniel, and C. F. Semenkovich. 2005. Pancreatic beta-cell lipoprotein lipase independently regulates islet glucose metabolism and normal insulin secretion. J. Biol. Chem. 280 9023–9029. [DOI] [PubMed] [Google Scholar]
- 6.Greenwood M. R. 1985. The relationship of enzyme activity to feeding behavior in rats: lipoprotein lipase as the metabolic gatekeeper. Int. J. Obes. 9 (Suppl. 1): 67–70. [PubMed] [Google Scholar]
- 7.Merkel M., J. Heeren, W. Dudeck, F. Rinninger, H. Radner, J. L. Breslow, I. J. Goldberg, R. Zechner, and H. Greten. 2002. Inactive lipoprotein lipase (LPL) alone increases selective cholesterol ester uptake in vivo, whereas in the presence of active LPL it also increases triglyceride hydrolysis and whole particle lipoprotein uptake. J. Biol. Chem. 277 7405–7411. [DOI] [PubMed] [Google Scholar]
- 8.MacArthur J. M., J. R. Bishop, K. I. Stanford, L. Wang, A. Bensadoun, J. L. Witztum, and J. D. Esko. 2007. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J. Clin. Invest. 117 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augustus A., H. Yagyu, G. Haemmerle, A. Bensadoun, R. K. Vikramadithyan, S. Y. Park, J. K. Kim, R. Zechner, and I. J. Goldberg. 2004. Cardiac-specific knock-out of lipoprotein lipase alters plasma lipoprotein triglyceride metabolism and cardiac gene expression. J. Biol. Chem. 279 25050–25057. [DOI] [PubMed] [Google Scholar]
- 10.Augustus A. S., J. Buchanan, T. S. Park, K. Hirata, H. L. Noh, J. Sun, S. Homma, J. D'Armiento, E. D. Abel, and I. J. Goldberg. 2006. Loss of lipoprotein lipase-derived fatty acids leads to increased cardiac glucose metabolism and heart dysfunction. J. Biol. Chem. 281 8716–8723. [DOI] [PubMed] [Google Scholar]
- 11.Ziouzenkova O., S. Perrey, L. Asatryan, J. Hwang, K. L. MacNaul, D. E. Moller, D. J. Rader, A. Sevanian, R. Zechner, G. Hoefler, et al. 2003. Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: evidence for an antiinflammatory role for lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 100 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chawla A., C. H. Lee, Y. Barak, W. He, J. Rosenfeld, D. Liao, J. Han, H. Kang, and R. M. Evans. 2003. PPARdelta is a very low-density lipoprotein sensor in macrophages. Proc. Natl. Acad. Sci. USA. 100 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattler W., S. Levak-Frank, H. Radner, G. M. Kostner, and R. Zechner. 1996. Muscle-specific overexpression of lipoprotein lipase in transgenic mice results in increased alpha-tocopherol levels in skeletal muscle. Biochem. J. 318 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel S., R. Piantedosi, S. M. O'Byrne, Y. Kako, L. Quadro, M. E. Gottesman, I. J. Goldberg, and W. S. Blaner. 2002. Retinol-binding protein-deficient mice: biochemical basis for impaired vision. Biochemistry. 41 15360–15368. [DOI] [PubMed] [Google Scholar]
- 15.van Bennekum A. M., Y. Kako, P. H. Weinstock, E. H. Harrison, R. J. Deckelbaum, I. J. Goldberg, and W. S. Blaner. 1999. Lipoprotein lipase expression level influences tissue clearance of chylomicron retinyl ester. J. Lipid Res. 40 565–574. [PubMed] [Google Scholar]
- 16.Zilversmit D. B. 1973. A proposal linking atherogenesis to the interaction of endothelial lipoprotein lipase with triglyceride-rich lipoproteins. Circ. Res. 33 633–638. [DOI] [PubMed] [Google Scholar]
- 17.Babaev V. R., S. Fazio, L. A. Gleaves, K. J. Carter, C. F. Semenkovich, and M. F. Linton. 1999. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in vivo. J. Clin. Invest. 103 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, L., R. Gill, T. L. Pedersen, L. J. Higgins, J. W. Newman, and J. C. Rutledge. 2008. Triglyceride-rich lipoprotein lipolysis releases neutral and oxidized free fatty acids that induce endothelial cell inflammation. J. Lipid Res. In press. [DOI] [PMC free article] [PubMed]
- 19.Beigneux A. P., B. S. Davies, P. Gin, M. M. Weinstein, E. Farber, X. Qiao, F. Peale, S. Bunting, R. L. Walzem, J. S. Wong, et al. 2007. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukonina V., A. Lookene, T. Olivecrona, and G. Olivecrona. 2006. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc. Natl. Acad. Sci. USA. 103 17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterfy M., O. Ben-Zeev, H. Z. Mao, D. Weissglas-Volkov, B. E. Aouizerat, C. R. Pullinger, P. H. Frost, J. P. Kane, M. J. Malloy, K. Reue, et al. 2007. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat. Genet. 39 1483–1487. [DOI] [PubMed] [Google Scholar]
- 22.Ling C., L. Svensson, B. Oden, B. Weijdegard, B. Eden, S. Eden, and H. Billig. 2003. Identification of functional prolactin (PRL) receptor gene expression: PRL inhibits lipoprotein lipase activity in human white adipose tissue. J. Clin. Endocrinol. Metab. 88 1804–1808. [DOI] [PubMed] [Google Scholar]
- 23.Ricart-Jane D., P. Cejudo-Martin, J. Peinado-Onsurbe, M. D. Lopez-Tejero, and M. Llobera. 2005. Changes in lipoprotein lipase modulate tissue energy supply during stress. J. Appl. Physiol. 99 1343–1351. [DOI] [PubMed] [Google Scholar]
- 24.Yost T. J., D. R. Jensen, B. R. Haugen, and R. H. Eckel. 1998. Effect of dietary macronutrient composition on tissue-specific lipoprotein lipase activity and insulin action in normal-weight subjects. Am. J. Clin. Nutr. 68 296–302. [DOI] [PubMed] [Google Scholar]
- 25.Wu G., L. Zhang, J. Gupta, G. Olivecrona, and T. Olivecrona. 2007. A transcription-dependent mechanism, akin to that in adipose tissue, modulates lipoprotein lipase activity in rat heart. Am. J. Physiol. Endocrinol. Metab. 293 E908–E915. [DOI] [PubMed] [Google Scholar]
- 26.Ruge T., M. Bergo, M. Hultin, G. Olivecrona, and T. Olivecrona. 2000. Nutritional regulation of binding sites for lipoprotein lipase in rat heart. Am. J. Physiol. Endocrinol. Metab. 278 E211–E218. [DOI] [PubMed] [Google Scholar]
- 27.Abumrad N., C. Harmon, and A. Ibrahimi. 1998. Membrane transport of long-chain fatty acids. Evidence for a facilitated process. J. Lipid Res. 39 2309–2318. [PubMed] [Google Scholar]
- 28.Berk P. D., and D. D. Stump. 1999. Mechanisms of cellular uptake of long chain free fatty acids. Mol. Cell. Biochem. 192 17–31. [PubMed] [Google Scholar]
- 29.Febbraio M., D. P. Hajjar, and R. L. Silverstein. 2001. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 108 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baillie A. G. S., C. T. Coburn, and N. A. Abumrad. 1996. Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J. Membr. Biol. 153 75–81. [DOI] [PubMed] [Google Scholar]
- 31.Coburn C. T., F. F. Knapp, Jr., M. Febbraio, A. L. Beets, R. L. Silverstein, and N. A. Abumrad. 2000. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275 32523–32529. [DOI] [PubMed] [Google Scholar]
- 32.Hajri T., X. X. Han, A. Bonen, and N. A. Abumrad. 2002. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J. Clin. Invest. 109 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajri T., A. M. Hall, D. R. Jensen, T. A. Pietka, V. A. Drover, H. Tao, R. Eckel, and N. A. Abumrad. 2007. CD36-facilitated fatty acid uptake inhibits leptin production and signaling in adipose tissue. Diabetes. 56 1872–1880. [DOI] [PubMed] [Google Scholar]
- 34.Ibrahimi A., A. Bonen, W. D. Blinn, T. Hajri, X. Li, K. Zhong, R. Cameron, and N. A. Abumrad. 1999. Muscle-specific overexpression of FAT/CD36 enhances fatty acid oxidation by contracting muscle, reduces plasma triglycerides and fatty acids, and increases plasma glucose and insulin. J. Biol. Chem. 274 26761–26766. [DOI] [PubMed] [Google Scholar]
- 35.Pravenec M., V. Landa, V. Zidek, A. Musilova, V. Kren, L. Kazdova, T. J. Aitman, A. M. Glazier, A. Ibrahimi, N. A. Abumrad, et al. 2001. Transgenic rescue of defective Cd36 ameliorates insulin resistance in spontaneously hypertensive rats. Nat. Genet. 27 156–158. [DOI] [PubMed] [Google Scholar]
- 36.Laugerette F., P. Passilly-Degrace, B. Patris, I. Niot, M. Febbraio, J. P. Montmayeur, and P. Besnard. 2005. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 115 3177–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M., Y. Yang, E. Braunstein, K. E. Georgeson, and C. M. Harmon. 2001. Gut expression and regulation of FAT/CD36: possible role in fatty acid transport in rat enterocytes. Am. J. Physiol. Endocrinol. Metab. 281 E916–E923. [DOI] [PubMed] [Google Scholar]
- 38.Nassir F., B. Wilson, X. Han, R. W. Gross, and N. A. Abumrad. 2007. CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J. Biol. Chem. 282 19493–19501. [DOI] [PubMed] [Google Scholar]
- 39.Drover V. A., M. Ajmal, F. Nassir, N. O. Davidson, A. M. Nauli, D. Sahoo, P. Tso, and N. A. Abumrad. 2005. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J. Clin. Invest. 115 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drover V. A., D. V. Nguyen, C. C. Bastie, Y. F. Darlington, N. A. Abumrad, J. E. Pessin, E. London, D. Sahoo, and M. C. Phillips. 2008. CD36 mediates both cellular uptake of very long chain fatty acids and their intestinal absorption in mice. J. Biol. Chem. 283 13108–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sclafani A., K. Ackroff, and N. A. Abumrad. 2007. CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293 R1823–R1832. [DOI] [PubMed] [Google Scholar]
- 42.Goudriaan J. R., M. A. den Boer, P. C. Rensen, M. Febbraio, F. Kuipers, J. A. Romijn, L. M. Havekes, and P. J. Voshol. 2005. CD36 deficiency in mice impairs lipoprotein lipase-mediated triglyceride clearance. J. Lipid Res. 46 2175–2181. [DOI] [PubMed] [Google Scholar]
- 43.El-Yassimi A., A. Hichami, P. Besnard, and N. A. Khan. 2008. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J. Biol. Chem. 283 12949–12959. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz G. J., J. Fu, G. Astarita, X. Li, S. Gaetani, P. Campolongo, V. Cuomo, and D. Piomelli. 2008. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 8 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwenk R. W., J. J. Luiken, A. Bonen, and J. F. Glatz. 2008. Regulation of sarcolemmal glucose and fatty acid transporters in cardiac disease. Cardiovasc. Res. 79 249–258. [DOI] [PubMed] [Google Scholar]
- 46.Nahle Z., M. Hsieh, T. Pietka, C. T. Coburn, P. A. Grimaldi, M. Q. Zhang, D. Das, and N. A. Abumrad. 2008. CD36-dependent regulation of muscle FoxO1 and PDK4 in the PPAR delta/beta-mediated adaptation to metabolic stress. J. Biol. Chem. 283 14317–14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holloway G. P., J. J. Luiken, J. F. Glatz, L. L. Spriet, and A. Bonen. 2008. Contribution of FAT/CD36 to the regulation of skeletal muscle fatty acid oxidation: an overview. Acta Physiol (Oxf). 194 293–309. [DOI] [PubMed] [Google Scholar]
- 48.Bastie C. C., Z. Nahle, T. McLoughlin, K. Esser, W. Zhang, T. Unterman, and N. A. Abumrad. 2005. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J. Biol. Chem. 280 14222–14229. [DOI] [PubMed] [Google Scholar]
- 49.Miranda M., and A. Sorkin. 2007. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol. Interv. 7 157–167. [DOI] [PubMed] [Google Scholar]
- 50.Smith J., X. Su, R. El-Maghrabi, P. D. Stahl, and N. A. Abumrad. 2008. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J. Biol. Chem. 283 13578–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love-Gregory L., R. Sherva, L. Sun, J. Wasson, T. Schappe, A. Doria, D. C. Rao, S. C. Hunt, S. Klein, R. J. Neuman, et al. 2008. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 17 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita S., K. I. Hirano, T. Kuwasako, M. Janabi, Y. Toyama, M. Ishigami, and N. Sakai. 2007. Physiological and pathological roles of a multi-ligand receptor CD36 in atherogenesis; insights from CD36-deficient patients. Mol. Cell. Biochem. 299 19–22. [DOI] [PubMed] [Google Scholar]