Abstract
The RD114/simian type D retroviruses, which include the feline endogenous retrovirus RD114, all strains of simian immunosuppressive type D retroviruses, the avian reticuloendotheliosis group including spleen necrosis virus, and baboon endogenous virus, use a common cell-surface receptor for cell entry. We have used a retroviral cDNA library approach, involving transfer and expression of cDNAs from highly infectable HeLa cells to nonpermissive NIH 3T3 mouse cells, to clone and identify this receptor. The cloned cDNA, denoted RDR, is an allele of the previously cloned neutral amino acid transporter ATB0 (SLC1A5). Both RDR and ATB0 serve as retrovirus receptors and both show specific transport of neutral amino acids. We have localized the receptor by radiation hybrid mapping to a region of about 500-kb pairs on the long arm of human chromosome 19 at q13.3. Infection of cells with RD114/type D retroviruses results in impaired amino acid transport, suggesting a mechanism for virus toxicity and immunosuppression. The identification and functional characterization of this retrovirus receptor provide insight into the retrovirus life cycle and pathogenesis and will be an important tool for optimization of gene therapy using vectors derived from RD114/type D retroviruses.
Interaction of the retrovirus surface (SU) glycoprotein with its cognate cell-surface receptor is the major determinant of retrovirus tissue and species tropism (1). In the absence of a specific receptor, virus entry is dramatically attenuated or undetectable. An important technique for the classification of retroviruses by receptor utilization is interference analysis. This approach involves assigning retroviruses to groups based on their ability to block entry by members of the group, but not members of other interference groups. Interference is determined genetically by SU and corresponds to a block at the level of virus entry mediated by binding of SU to its cognate receptor. Retroviruses have evolved to use many different receptors for cell entry that have diverse normal functions in cells, including transport of various solutes, immune recognition, and signaling (2, 3). Identification of virus receptors frequently has provided insights into disease pathogenesis and virus life cycle and has offered fruitful areas for therapeutic intervention (4).
The largest known group of retroviruses that use the same receptor on human cells is the RD114/type D interference group (5, 6). The 10 members of this interference group comprise viruses classified either morphologically as type C or type D or classified by their ability to be transmitted in the host germ line (endogenous) or horizontally (exogenous). When first discovered in the early 1970s, the RD114 retrovirus was considered as one of first candidate human oncogenic viruses (7). It is now known that RD114 is an endogenous feline type C virus that readily infects human, primate, and dog cells, producing a syncytial cytopathic effect (8–10). In view of its ability to transduce human cells, replication-incompetent vectors packaged using the RD114 pseudotype may be important for gene therapy (11). The two other type C viruses in this interference group are spleen necrosis virus (SNV; refs. 6, 12, and 13) and baboon endogenous virus (BaEV; ref. 14). SNV is an avian reticuloendotheliosis virus that causes death or immunodeficiency in ducks, depending on the age at infection, and produces a cytopathic effect in cultured avian, canine, and rat cells (15, 16). Other avian reticuloendotheliosis viruses that share more than 90% nucleic acid homology with SNV (including REV-T and REV-A) all use a common receptor (17).
The majority of type D viruses utilize the same receptor as the type C viruses described above. The type D viruses included in the RD114/type D interference group are the spectacled-langur virus PO-1-Lu, squirrel monkey virus, Mason–Pfizer monkey virus (MPMV, also called SRV-3; ref. 18), and other simian retroviruses subgrouped into separate serotypes (SRV-1, SRV-2, SRV-4, and SRV-5; ref. 19–21). The profound immunodeficiency, retroperitoneal fibrosis, and anemia seen in many species of Asian macaques infected with SRVs is a major cause of morbidity and mortality in captive primates (22, 23). The need to control SRV infection in captive macaques has led to the development of vaccines against several serotypes (24). Although simian retroviruses exhibit a broad cellular tropism, which includes T and B lymphocytes as well as nonlymphoid cells (25), they do not cause a cytopathic effect other than induction of syncytia in Raji human lymphoblastoid cells.
Many laboratories have sought the common cellular receptor used by the RD114/type D virus interference group (5, 26–29). The earliest reports that the determinant of entry was located on human chromosome 19 were published two decades ago (26, 30). Further localization to 19q13.1–13.2 and exclusion of many candidate receptors came from exhaustive analyses performed by Weiss and colleagues (5, 31). In this report we describe cloning of RDR, the receptor for the RD114/type D interference group, using a cDNA expression library. RDR appears to be an allele of the human neutral amino acid transporter hATB0 (32), which also can act as a receptor for the RD114/type D interference group. Both hATB0, which was localized previously by fluorescence in situ hybridization (32, 33), and RDR, which we mapped precisely using radiation hybrid cell lines, are located on the long arm of human chromosome 19 at q13.3. Infection of human cells with replication-competent viruses of this interference group leads to a reduction in transport of amino acids specific for hATB0, which offers a potential explanation for the pathogenicity of these viruses.
MATERIALS AND METHODS
Plasmids, Viruses, and Cell Lines.
Nomenclature for retroviral vectors and pseudotypes conforms to previous usage (34). Retroviral vectors based on murine leukemia viruses (MLV) include: LAPSN (35), which expresses human alkaline phosphatase (AP) and neomycin phosphotransferase (neo); LNCG (36), which expresses neo and the enhanced green fluorescent protein (EGFP; CLONTECH); LXSN, a retroviral expression vector (37); L(RDR)SN, in which a 2-kb EcoRI fragment containing the full coding sequence of RDR was ligated into EcoRI-digested pLXSN; and L(ATB0)SN, in which a 2.1-kb ApoI fragment from plasmid pSPORT-hATB0 [a kind gift from R. Kekuda and V. Ganapathy (32)] was ligated into EcoRI-digested pLXSN. The retrovirus vector plasmid pLXL used for cDNA library expression was made by replacing the simian virus 40 promoter and neo gene of pLXSN with a multiple cloning site.
Helper-free retroviral vectors pseudotyped with gibbon ape leukemia virus (GALV) and RD114 envelope proteins were produced by using PG13 (38) and FLYRD (11) retrovirus packaging cells, respectively. VSV-G pseudotype virus was produced by transient transfection as described (39). Wild-type viruses used include: RD114 (40); SNV [from clone pB101 (41) expressed in D17 cells as described (6)]; MPMV [from clone pSHRM15 (42) expressed in 293 cells]; BaEV [collected from MMΔ cells that are D17 cells expressing a vector containing the lacZ gene, pHM24 (43) and replication-competent BaEV]; and amphotropic MLV (AM-MLV) from clone pAM (44).
Mammalian cells, including HT-1080 human fibrosarcoma cells (ATCC CCL 121), HeLa cervical carcinoma cells (ATCC CCL 2), 293 human kidney cells (ATCC CRL 1573), NIH 3T3 thymidine kinase-deficient mouse embryo fibroblasts (45), BHK-21 hamster cells (ATCC CCL 10), and D17 canine osteosarcoma cells (ATCC CRL 6248) were grown in DMEM supplemented with 10% fetal bovine serum (FBS, HyClone). CHO-K1 Chinese hamster ovary cells (ATCC CCL 61) were grown in α-modified MEM supplemented with 10% fetal bovine serum.
Polyclonal cell lines in which RDR was overexpressed were established by CaPO4-mediated transfection (37). Briefly, NIH 3T3 and BHK cells (5 × 105 cells per 6-cm dish) were transfected separately with pLXSN and pL(RDR)SN and were selected in G418 (750 and 1200 μg/ml active drug for the respective cell lines). D17 cells infected with SNV and MPMV were generated as described previously (46) and were obtained from V. Kewalramani and M. Emerman (6).
Expression Cloning of RDR.
To construct the cDNA expression library, total RNA was isolated from HeLa cells using a modified guanidium thiocyanate method, followed by purification of poly(A)+ RNA using an oligo(dT) cellulose column (47). The first strand of cDNA was synthesized using XhoI-(dT)15 oligo as a primer, and the second strand was random-primed. An EcoRI adapter (5′-AATTCGCGGCCGCGTCGAC-3′ annealed to 5′-GTCGACGCGGCCGCG-3′) was ligated to the cDNA, which subsequently was methylated, digested with XhoI, and size-fractionated. The cDNA library was unidirectionally ligated into EcoRI and SalI sites (SalI and XhoI produce compatible ends) within the extended multiple cloning site of pLXL, which was then introduced into DH10B cells by electroporation.
The retroviral library (complexity of approximately 2.5 × 106 individual clones) was pseudotyped using VSV-G as described (39). NIH 3T3 cells (2 × 106 total) were transduced at an estimated multiplicity of infection of ≤1 virus per cell. Gene expression was induced by exposing cells to sodium butyrate (10 mM) for 16 hr. The NIH 3T3 cell library was exposed to LAPSN(RD114) (titer 106/ml) and selected in G418 (750 μg/ml) for 7 days. Distinct colonies were isolated and their susceptibility to infection by RD114 virus was tested using LNCG(RD114) vector. Expression of GFP from LNCG was assessed either by examination of cells using a Leica MZ-8 stereomicroscope equipped with a 100 W mercury vapor lamp and GFP Plus filters or by flow cytometric quantitation of at least 20,000 propidium iodide (1 μg/ml)-excluding, forward and right-angle light scatter-gated events using a FACSCalibur flow cytometer (Becton Dickinson).
DNA from reinfectable clones was extracted using a modified salt precipitation procedure (Puregene; Gentra Systems). Nested PCRs (eLONGase; Life Technologies, Gaithersburg, MD) were performed in a thermal cycler (GeneAmp PCR System 2400; Perkin–Elmer) for 40 cycles each of 94°C for 30 sec, 58°C for 30 sec, 68°C for 5 min, followed by incubation at 68°C for 10 min. Two primer pairs were used: 5′LXL-C (5′-CCAGCCCTCACTCCTTCTCTAG-3′) and 3′LXL-D (5′-ATGGCGTTACTTAAGCTAGCTTGCCAAACCTAC-3′) followed by 5′LXL-A (5′-CCGGAATTCGCGGCCGCGTCGAC-3′) and 3′LXL-B (5′-GGCCGAGGCGGCCGCTTGTCGAG-3′). PCR products were cloned into pCR2.1-TOPO cloning vector (Invitrogen) according to the manufacturer’s instructions. Dye terminator cycle sequencing was performed at least twice in both directions using an Applied Biosystems DNA Sequencer model 377.
Retrovirus Titer and Vector Rescue.
Retrovirus vector stocks were prepared and vector titer was determined as described (37). All retroviral infections were performed in the presence of Polybrene (4 μg/ml). Staining for AP- and β-galactosidase-positive foci of vector-transduced cells was performed as described (48, 49). Vector rescue was used to demonstrate that NIH 3T3 cell susceptibility to transduction by RD114-pseudotype vectors was conferred by viruses in the retroviral library. NIH 3T3 cells containing the cDNA library were transfected with plasmids pSX2 (encoding the 10A1 MLV envelope) and pLGPS (encoding gag-pol regions of Moloney MLV) to rescue the retroviral cDNA expression vectors as described (50). Conditioned medium harvested from the transfectants was applied to naïve NIH 3T3 cells, which then were tested for susceptibility to infection with LNCG(RD114). Because G418-resistant cells containing receptor-candidate cDNAs should also contain LAPSN, the NIH 3T3 cells were also stained for AP (51) as a positive control for vector rescue.
Production of virus from HT-1080 and HeLa cells after exposure to replication-competent viruses was demonstrated by marker rescue using D17/LAPSN cells (52). Conditioned medium from cells was transferred to D17/LAPSN cells in the presence of Polybrene (4 μg/ml). After incubation for 2 weeks to allow spread of virus, conditioned medium from these cells was applied to naïve D17 cells and transfer of the LAPSN vector was measured by staining cells for AP activity.
Amino Acid Transport.
Cells seeded at 2 × 105/well in 24-well dishes (Costar 3524) were cultured overnight, and amino acid transport was assayed as described previously (53). Briefly, cells were washed twice with transport buffer (32) at 37°C (5-min washes) and were incubated for 2 min at 25°C in transport buffer containing l-[3-3H]-alanine (20 nM, 72.3 Ci/mmol; Sigma), l-[3H(G)]-serine (20 nM, 19.7 Ci/mmol; NEN), l-[2,3-3H]-glutamic acid (20 nM, 24 Ci/mmol; NEN), or l-[3,4-3H(N)]-glutamine (20 nM, 34.6 Ci/mmol; NEN). Transport was terminated by aspiration of the buffer followed by three rinses in ice-cold PBS. Cells were harvested in lysis buffer (0.2% SDS/0.2 M NaOH), and cell-associated radioactivity was determined. All assays were performed in triplicate. Statistical significance was determined by ANOVA using the statview 4.5 software package (Abacus Concepts, Berkeley, CA).
Chromosomal Localization.
The chromosomal location of the human RDR gene was identified using the Stanford Radiation Hybrid Mapping Panel G3 (54). PCR was performed on DNA extracted from each of the 83 G3 hybrid clones using primers R1b15FN (5′-TGGCTGCTGGAGTACATGTG-3′) and R1b15RO (5′-CCCAGTGGGGGCTAGAATTC-3′) to produce a predicted 196-bp product (details are provided at http://www-shgc.stanford.edu/Mapping/rh/procedure/rhassaynew.html). The resulting vector was submitted to the Stanford Human Genome Center Rhserver for chromosomal localization.
RESULTS
Identification and Cloning of the RD114 Receptor.
Human cells are highly susceptible to transduction with RD114-pseudotype retrovirus vectors [titers ≥105 focus-forming units (ffu)/ml on HT-1080 and HeLa cell lines] whereas murine cells are not susceptible (titer <1 ffu/ml on NIH 3T3 cells). To screen for the retrovirus receptor, a HeLa cell cDNA library was constructed by cloning of the cDNA into a retroviral expression vector contained in a bacterial plasmid. Characterization of the library by PCR analysis revealed that 93% of clones contained cDNA inserts with an average size of 2.4 kb (range 0.4–6.0 kb). The presence of cDNAs corresponding to the large transcript of the transferrin receptor (5 kb), the rare transcript from the c-myc gene, and cDNAs for G3PDH and β-actin was demonstrated by PCR using cDNA-specific primers. Colony hybridization using a β-actin probe demonstrated a minimum β-actin frequency of 0.1%.
We introduced the HeLa cDNA retroviral expression library into murine NIH 3T3 cells by infection at a relatively low multiplicity of infection so that the majority of infectants would contain single-copy provirus inserts. The library-containing cells were then screened for susceptibility to RD114-pseudotype vector [LAPSN(RD114)] transduction by selecting them in G418 after exposure to the vector. Of 73 G418-resistant NIH 3T3 clones obtained from the primary screen, 67 (92%) were reinfectable with a different RD114-pseudotype vector, LNCG(RD114), as determined by the acquisition of cellular green fluorescence.
To demonstrate that susceptibility to RD114 transduction had been conferred by a cDNA introduced in a retroviral expression vector, we performed a vector rescue experiment on six independent clones. Transfer of cDNA-containing vectors from all six reinfectable clones rendered naïve NIH 3T3 cells sensitive to LNCG(RD114) transduction. The same vector rescue procedure performed on six NIH 3T3 clones that contained the library, but were not susceptible to infection by RD114, did not confer the phenotype on naïve NIH 3T3. This result confirmed that the RD114 susceptibility exhibited by these clones was conferred by vectors expressing specific cDNAs contained in the retroviral library.
Products between 800 and 2,900 bp were identified after nested PCR was performed on DNA extracted from 18 reinfectable NIH 3T3 clones. Of these, several larger products were selected for subcloning and sequencing. The sequence from clone R1b 15–1 revealed substantial homology to a human “system B” neutral amino acid transporter (hATB0). The cDNA from this clone conferred susceptibility to RD114 infection (see below) and consequently was renamed RDR. The single ORF between bases 22 and 1,644 of the cDNA insert encodes a predicted protein of 541 aa with a molecular mass of 57 kDa. RDR shares 97% amino acid identity with hATB0 (32) and 85% amino acid identity with both the presumed rabbit homologue riATB0 (55) and a possible murine homologue ASCT2 (56). A refined neural network analysis (57) of the RDR sequence predicts 9 transmembrane regions with the 53 amino-terminal residues located intracytoplasmically and the 96 carboxyl-terminal residues located outside the cell. As shown in Fig. 1, two canonical N-glycosylation sites (58) occur in the second of five predicted extracellular domains of RDR. The sequence of RDR also includes 7 casein kinase II sites, 2 protein kinase C phosphorylation sites, 21 N-myristoylation sites, and signatures 1 and 2 of the sodium/dicarboxylate symporter family. The 57-kDa predicted molecular mass for unglycosylated RDR matches the observed molecular mass of 58–60 kDa obtained when SRV-1 was used in immunoprecipitation experiments to identify its cognate receptor (28).
Figure 1.
Amino acid sequence of human RDR. The amino terminus is predicted to be intracytoplasmic and the carboxyl terminus is predicted to be extracellular. Predicted transmembrane regions (underlined) and N-glycosylation sites (bold) are indicated.
The sequence of RDR (GenBank accession no. AF102826) differs from the published sequence of the human neutral amino acid transporter hATB0 (accession no. U53347, ref. 32). In the coding region of RDR, there are four frameshift mutations and seven point mutations compared with the published hATB0 sequence. To further investigate these differences, we resequenced the original DNA clone of hATB0 obtained from the authors and confirmed the presence of the point mutations but not the frame shift mutations (GenBank accession no. AF105230). This result indicated that the putative frameshift mutations were errors in the original sequence of hATB0. The remaining 7-nt differences between RDR and hATB0 result in 5-aa differences from hATB0 at positions 51L → Q, 460I → V, 467D → G, 470V → A, and 515D → G. As such, RDR shares 99% amino acid identity with resequenced hATB0.
A partial sequence of hATB0, named SLC1A5, had been obtained previously by random sequencing of a human pancreatic islet cDNA library (33). The gene for SLC1A5 was localized to 19q13.3 and was shown to be polymorphic in its 3′ untranslated (GT)n region with seven codominant alleles in a Caucasian sample. Sequencing of hATB0 and RDR revealed a (GT)18 repeat in the 3′ untranslated regions. Additional scattered nucleotide differences are present in the coding regions of published partial sequences, e.g., accession no. T11057.
RDR Confers Susceptibility to Members of the RD114/Type D Retrovirus Interference Group.
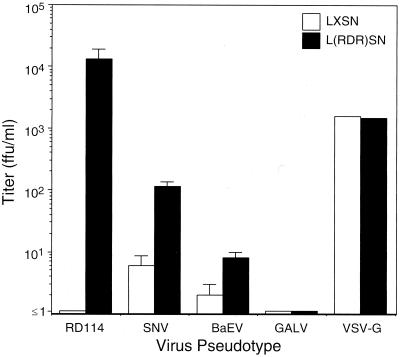
The susceptibility of RDR-expressing NIH 3T3 cells to vectors with various pseudotypes is summarized in Fig. 2. In this and other experiments, the titer of LAPSN(RD114) vector on cells expressing RDR was 104–105 ffu/ml compared with cells lacking RDR on which the titer was ≤1 ffu/ml. To confirm that RDR did not modify cellular susceptibility to retrovirus transduction in a nonspecific manner, we showed that titers of a GALV-pseudotype vector (which does not infect murine cells) and a VSV-G-pseudotype vector (which does infect murine cells) were similar in RDR-expressing and control cells. Additionally, NIH 3T3 cells expressing hATB0 from pL(ATB0)SN were susceptible to infection with LAPSN(RD114) vector (titer 106 ffu/ml), compared with a titer of ≤1 ffu/ml in control cells expressing pLXSN (n = 2 experiments, each performed in triplicate). The difference in RD114-pseudotype vector titer between NIH 3T3 cells expressing hATB0 versus RDR could be a result of differences in the cDNA sequences, expression levels, or other factors. Nevertheless, the fact that the titer of RD114-pseudotype vector on human cells (approximately 5 × 105) is similar to that observed for NIH 3T3 cells expressing RDR or hATB0 shows that susceptibility to RD114 infection is complemented completely by these surface molecules.
Figure 2.
Retrovirus vector titers on polyclonal NIH 3T3 cells transfected with the RDR expression vector pL(RDR)SN or the empty expression vector pLXSN. Both SNV and BaEV pseudotypes represent replication-competent virus, whereas RD114, GALV, and VSV-G were helper-free. The titer of replication-competent viral stocks used in these experiments was 4.1 × 104 AP+ ffu/ml for SNV and 3.2 × 104 lacZ+ ffu/ml for BaEV when assayed on D17 cells. The titer of LAPSN(GALV) was 5.3 × 105 AP+ ffu/ml assayed on human cells. Each bar represents the mean ± SD of triplicate measurements, except for VSV-G, which represents the mean of duplicates. A representative experiment is shown; each determination was performed at least twice.
In a similar experiment in which polyclonal BHK cells containing LXSN or L(RDR)SN were tested for susceptibility to an RD114-pseudotype vector, we observed an increase from ≤2 ffu/ml in the former to 2.0 × 103 ffu/ml in the latter. Again, GALV and VSV-G vector titers did not vary between the two BHK populations.
RDR expression in NIH 3T3 cells also conferred an increase in susceptibility to infection by SNV and BaEV, although the effect was less pronounced than that seen with RD114 (Fig. 2). When tested on human HT-1080 cells, titers of the same stock of SNV and BaEV were >100-fold lower than those determined using D17 cells (respectively, 20 ffu/ml compared with 4 × 104 ffu/ml for SNV, and 1 × 102 ffu/ml compared with 3 × 104 ffu/ml for BaEV).
Chromosomal Localization of RDR.
We designed PCR primers using the 3′ untranslated region of RDR that produce a single product when used to amplify human genomic DNA but that do not amplify sequences in hamster genomic DNA (data not shown). These primers were used in PCR analyses performed on DNA from 83 human/hamster radiation hybrids that comprise the Stanford G3 Mapping Panel. The results of PCRs for localization of RDR produced the vector 0000000000 0000001010 0010000010 1000110000 0001000001 1000000000 0000010000 0010000010 001 in which a hybrid containing (“1”) or not containing (“0”) the human RDR sequence is indicated. This radiation hybrid mapping localized RDR to within 13 centiRay of marker SHGC-57346 (logarithm of odds score 12.10) located on the long arm of chromosome 19 at position q13.3. Consistent with this physical location, on the Genome Database (Johns Hopkins University, Baltimore, MD, 1998) cytogenetic map the SLC1A5 gene is placed between genes for the CD37 antigen and UNR (ubiquitously expressed nuclear receptor).
Infection of Cells with Viruses in the RD114/Type D Interference Group Leads to Specific Reduction in Amino Acid Transport.
To demonstrate that RDR can function as a transporter of neutral amino acids, we examined substrate specificity in polyclonal NIH 3T3 cells containing LXSN or L(RDR)SN. As summarized in Table 1, in contrast to the control amino acid (glutamate), uptake of neutral amino acids (alanine and glutamine) was increased in cells expressing RDR. In four experiments, the percentage increase of specific alanine uptake in NIH 3T3 cells was 29 ± 15%.
Table 1.
RDR increases uptake of neutral amino acids
| [3H]amino acid | Radioactivity uptake by NIH 3T3 cells containing
|
RDR-specific change, % | |
|---|---|---|---|
| LXSN | L(RDR)SN | ||
| Alanine | 5,200 ± 600 | 6,400 ± 200 | 23 |
| Serine | 46,800 ± 2,100 | 49,300 ± 4,600 | 5.3 |
| Glutamine | 22,400 ± 700 | 25,000 ± 1,300 | 12 |
| Glutamate | 6,400 ± 500 | 5,900 ± 300 | −7.0 |
Amino acid transport in NIH 3T3 cells expressing either the empty vector pLXSN or the receptor-containing vector pL(RDR)SN was examined. Uptake of 20 nM [3H]amino acid was measured for 2 min at 25°C in medium containing sodium. The data are presented as mean uptake (cpm/well per min) ±SD of triplicate measurements. Alanine uptake was measured in four independent experiments (mean increase of 29 ± 15%), whereas uptake measurements for the other amino acids were performed once.
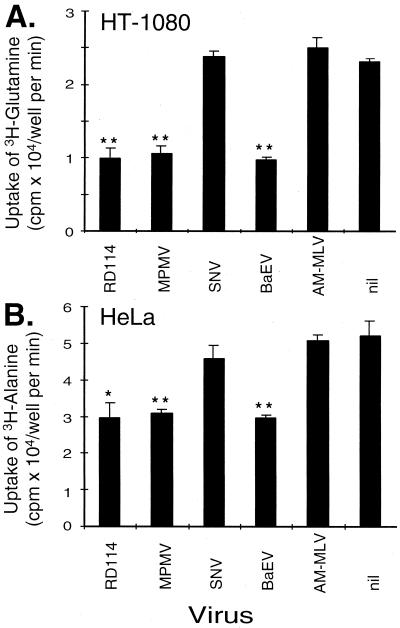
Cellular uptake of alanine and glutamine, both specifically transported by hATB0, was examined in HT-1080 and HeLa cells that had been infected with wild-type viruses. As shown in Fig. 3, RD114, MPMV, and BaEV each reduced specific amino acid transport in both cell lines. A similar pattern of reduced transport of glutamine and alanine was demonstrated in HeLa and HT-1080 cells, respectively (data not shown). Amphotropic virus did not have an effect on neutral amino acid transport (Fig. 3).
Figure 3.
Reduced amino acid transport in cells infected with viruses that use hATB0 for entry. Uptake of glutamine in HT-1080 cells (A) and alanine in HeLa cells (B) was examined in cells cultured after exposure to replication-competent virus indicated on the x axis. The presence of replication competent virus was demonstrated by marker rescue of HT-1080 and HeLa cells exposed to RD114, MPMV, BaEV, and AM-MLV, but was not detected in either cell type exposed to SNV or nil. (Bar represents mean ± SD of triplicate measurements.) Statistical significance of P ≤ 0.01 (∗) and of P ≤ 0.001 (∗∗) was demonstrated by ANOVA in which each result was compared with the result in nil. This experiment was performed twice with similar results.
Productive viral infection (102–106 ffu/ml) was demonstrated by marker rescue assay of HT-1080 and HeLa cells exposed to RD114, MPMV, BaEV, and AM-MLV. Wild-type virus was not detected in conditioned medium assayed from cells exposed to SNV or no virus control. The absence of productive infection by SNV correlates with its inability to reduce neutral amino acid transport in these cells.
DISCUSSION
In this report we describe the functional identification and cloning of the long-sought receptor for members of the RD114/type D retrovirus interference group. We used a retroviral expression library derived from infectable HeLa cells to confer RD114 susceptibility on nonpermissive NIH 3T3 cells. Using vector rescue, we confirmed that cDNA contained within vector sequences present in reinfectable clones was responsible for conferring RD114 susceptibility. We expressed the cloned cDNA in nonpermissive cells and demonstrated its ability to encode a functional receptor for RD114, SNV, and BaEV. The single ORF encodes a protein, RDR, that is closely related to the system B0 amino acid transporter, hATB0, which also acts as a receptor for RD114. Using radiation hybrid mapping, we obtained fine chromosomal localization of RDR within human 19q13.3. This localization concurs with previous independent mapping of hATB0 and SLC1A5 by fluorescence in situ hybridization (32, 33). Finally, we confirmed that RDR functions as a neutral amino acid transporter and showed that infection of human cells with replication-competent viruses of the RD114/type D retrovirus interference group reduces uptake of neutral amino acids. Whether RDR functions as a primary binding receptor for the retroviral Env protein or acts, for example, as a coreceptor awaits further studies on binding between purified components of this system.
The RD114/type D simian retrovirus receptor was localized previously to the long arm of human chromosome 19 at q13.1–13.2 using a panel of seven human/rodent hybrid cells (31, 59). The precise radiation hybrid mapping facilitated by the cloning of RDR localizes the RD114/type D simian retrovirus receptor to 19q13.3. The exclusion in the earlier study of human chromosome 19q13.3 was based on two clones [JDA 16 and 13 (60)], which, in hindsight, could not exclude 19q13.3. Although we cannot rule out the possibility that RDR is a separate gene resulting from genomic duplication at 19q13.3, there are two reasons why RDR most likely represents an allele of hATB0. First, we have observed that the determinant of RD114 entry is localized by phenotypic analysis of the Stanford G3 Radiation Hybrid Mapping Panel to precisely the same genomic locus as described here (J.E.J.R., J.-L.B., and A.D.M., unpublished results). Second, only one or two alleles of SCL1A5 were detected in each of 60 unrelated human samples analyzed for polymorphisms (ref. 33 and Eugenia M. Jones, personal communication), indicating the presence of a single diploid gene encoding hATB0 and RDR.
The cloning of RDR adds yet another solute transport molecule to the list of cell-surface molecules that serve as receptors for retroviruses, including the cationic amino acid transporter mCAT-1 used by ecotropic MLVs and the phosphate transporters Pit1 and Pit2 used by GALV and amphotropic MLVs, respectively. Solute transport is sodium-dependent for hATB0 (32) and presumably for RDR, and for Pit1 and Pit2 (61). Solute transport by mCAT-1 is sodium-dependent for the neutral amino acids cysteine and homoserine, but not for the basic amino acids lysine, arginine, and ornithine (62, 63). Perhaps the portions of these transporters responsible for sodium cotransport play a common role in retrovirus binding or entry. Conservation of RDR recognition among the relatively recently evolved type C viruses and the more ancient type D viruses present in Old World and New World monkeys remains an intriguing observation.
An explanation as to why human RDR may not function as efficiently as a receptor for BaEV and SNV in rodent cells may come from experiments demonstrating differences in the host range of known members of this interference group, possibly because of differential N-linked glycosylation (29, 64). We and others have observed up to a 105-fold increase in apparent titer of RD114-pseudotyped virus on murine, hamster, and feline cells after treatment with the glycosylation inhibitor tunicamycin (refs. 29 and 40; data not shown). Therefore, it may be that the RD114 virus-binding site on RDR is located at one of the two predicted N-linked glycosylation sites on its second extracellular domain. Furthermore, it is known that SNV and other members of the RD114/type D interference group exhibit a posttranscriptional block to replication and possible inefficient preintegration in certain primate cells (65). It is possible that inefficient processing of virus postentry in rodent cells could explain these observations.
A human packaging cell line that uses the RD114 envelope has been constructed for use as an efficient, complement-resistant method of gene delivery in vivo (11). The hATB0 gene is expressed as a single, 2.9-kb transcript in human placenta, lung, skeletal muscle, kidney, and pancreas (32). Therefore, transduction of these tissues should be possible by using pseudotypes from any viruses of the RD114/type D retrovirus interference group. In addition, knowledge of the identity of the receptor used by the RD114 virus suggests interventions that could improve transduction rates in the ex vivo therapeutic setting. For example, up-regulation of hATB0 has been demonstrated in human cells treated with epidermal growth factor or the neuroprotective agent aurintricarboxylic acid (66) and also in bovine cells cultured in amino acid-deprived medium (67). In the context of human gene therapy using repopulating hemopoietic stem cells, expression of RDR on limiting numbers of primitive cells with defined phenotypes now may be quantified using reverse–transcriptase PCR.
We have shown that infection of cells by wild-type viruses of the RD114/type D interference group can impair neutral amino acid transport. Consequently, we suggest that a receptor-mediated mechanism might account for the pathogenesis of diseases such as SRV- and SNV-induced immunodeficiencies. In such a scenario, chronically infected cell types exhibiting the greatest impairment of specific neutral amino acid transport essential for their metabolism would be predicted to lose proliferative capacity and become functionally impaired, such as is seen with B and T lymphocytes in SRV-induced lethal immunodeficiency. Furthermore, if the diseases are a result of a relative deficiency of intracellular neutral amino acid stores, dietary or parenteral supplementation may offer a useful therapeutic intervention. This hypothesis does not preclude a role for the immunosuppressive peptide previously found in the transmembrane region of many mammalian retrovirus env genes of differing interference groups, including the RD114/type D retrovirus interference group (68). Identification of the receptor for the RD114/type D retrovirus interference group may facilitate further studies on the pathogenesis of simian immunodeficiency and other diseases caused by the viruses within this large interference group.
Acknowledgments
We thank Shannon Brady for assistance with the radiation hybrid analysis, Ramesh Kekuda and Vadivel Ganapathy for the hATB0 cDNA, and Galina Filippova and Michael Emerman for advice. These studies were supported by National Institutes of Health Grants HL54881 and HL36444. J.E.J.R. was supported by Fellowship DRG081 of the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation, and J.-L.B. was supported by a fellowship from the Association Française contre les Myopathies.
ABBREVIATIONS
- MLV
murine leukemia virus
- GALV
gibbon ape leukemia virus
- ffu
focus-forming units
- SNV
spleen necrosis virus
- BaEV
baboon endogenous virus
- MPMV
Mason–Pfizer monkey virus
- AP
alkaline phosphatase
Footnotes
References
- 1.Weiss R A. In: The Retroviridae. Levy J A, editor. Vol. 2. New York: Plenum; 1993. pp. 1–108. [Google Scholar]
- 2.Wimer E. In: Cellular Receptors for Animal Retroviruses. Wimer E, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 1–13. [Google Scholar]
- 3.Hunter E. In: Retroviruses. Coffin J M, Hughes S H, Varmus H, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 71–120. [Google Scholar]
- 4.Schnell M J, Johnson J E, Buonocore L, Rose J K. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 5.Sommerfelt M A, Weiss R A. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 6.Kewalramani V N, Panganiban A T, Emerman M. J Virol. 1992;66:3026–3031. doi: 10.1128/jvi.66.5.3026-3031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAllister R M, Nicolson M, Gardner M B, Rongey R W, Rasheed S, Sarma P S, Huebner R J, Hatanaka M, Oroszlan S, Gilden R V, et al. Nat New Biol. 1972;235:3–6. doi: 10.1038/newbio235003a0. [DOI] [PubMed] [Google Scholar]
- 8.Klement V, McAllister R M. Virology. 1972;50:305–308. doi: 10.1016/0042-6822(72)90379-0. [DOI] [PubMed] [Google Scholar]
- 9.Nelson-Rees W A, Klement V, Peterson W D, Jr, Weaver J F. J Natl Cancer Inst. 1973;50:1129–1135. doi: 10.1093/jnci/50.5.1129. [DOI] [PubMed] [Google Scholar]
- 10.Niman H L, Stephenson J R, Gardner M B, Roy-Burman P. Nature (London) 1977;266:357–360. doi: 10.1038/266357a0. [DOI] [PubMed] [Google Scholar]
- 11.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trager W. Proc Soc Exp Biol Med. 1959;101:578–582. doi: 10.3181/00379727-101-25023. [DOI] [PubMed] [Google Scholar]
- 13.Koo H M, Gu J, Varela-Echavarria A, Ron Y, Dougherty J P. J Virol. 1992;66:3448–3454. doi: 10.1128/jvi.66.6.3448-3454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalter S S, Helmke R J, Panigel M, Heberling R L, Felsburg P J, Axelrod L R. Science. 1973;179:1332–1333. doi: 10.1126/science.179.4080.1332. [DOI] [PubMed] [Google Scholar]
- 15.Temin H M, Kassner V K. J Virol. 1974;13:291–297. doi: 10.1128/jvi.13.2.291-297.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshet E, Temin H M. J Virol. 1979;31:376–388. doi: 10.1128/jvi.31.2.376-388.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federspiel M J, Crittenden L B, Hughes S H. Virology. 1989;173:167–177. doi: 10.1016/0042-6822(89)90232-8. [DOI] [PubMed] [Google Scholar]
- 18.Chopra H C, Mason M M. Cancer Res. 1970;30:2081–2086. [PubMed] [Google Scholar]
- 19.Marx P A, Maul D H, Osborn K G, Lerche N W, Moody P, Lowenstine L J, Henrickson R V, Arthur L O, Gilden R V, Gravell M, et al. Science. 1984;223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 20.Daniel M D, King N W, Letvin N L, Hunt R D, Sehgal P K, Desrosiers R C. Science. 1984;223:602–605. doi: 10.1126/science.6695172. [DOI] [PubMed] [Google Scholar]
- 21.Stromberg K, Benveniste R E, Arthur L O, Rabin H, Giddens W E, Jr, Ochs H D, Morton W R, Tsai C C. Science. 1984;224:289–292. doi: 10.1126/science.6200929. [DOI] [PubMed] [Google Scholar]
- 22.Gardner M B, Luciw P, Lerche N, Marx P. Adv Vet Sci Comp Med. 1988;32:171–226. doi: 10.1016/b978-0-12-039232-2.50011-6. [DOI] [PubMed] [Google Scholar]
- 23.Marx P A, Bryant M L, Osborn K G, Maul D H, Lerche N W, Lowenstein L J, Kluge D, Zaiss C P, Henrickson R V, Shiigi S M, et al. J Virol. 1985;56:571–578. doi: 10.1128/jvi.56.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brody B A, Hunter E, Kluge J D, Lasarow R, Gardner M, Marx P A. J Virol. 1992;66:3950–3954. doi: 10.1128/jvi.66.6.3950-3954.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maul D H, Zaiss C P, MacKenzie M R, Shiigi S M, Marx P A, Gardner M B. J Virol. 1988;62:1768–1773. doi: 10.1128/jvi.62.5.1768-1773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown S, Oie H K, Gazdar A F, Minna J D, Francke U. Cell. 1979;18:135–143. doi: 10.1016/0092-8674(79)90362-3. [DOI] [PubMed] [Google Scholar]
- 27.Thiry L, Cogniaux-LeClerc J, Olislager R, Sprecher-Goldberger S, Buekens P. J Virol. 1983;48:697–708. doi: 10.1128/jvi.48.3.697-708.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres J V, Werner L L, Malley A, Benjamin E. J Med Primatol. 1991;20:218–221. [PubMed] [Google Scholar]
- 29.Koo H M, Parthasarathi S, Ron Y, Dougherty J P. Virology. 1994;205:345–351. doi: 10.1006/viro.1994.1651. [DOI] [PubMed] [Google Scholar]
- 30.Brown S, Oie H, Francke U, Gazdar A F, Minna J D. Cytogenet Cell Genet. 1978;22:239–242. doi: 10.1159/000130945. [DOI] [PubMed] [Google Scholar]
- 31.Sommerfelt M A, Williams B P, McKnight A, Goodfellow P N, Weiss R A. J Virol. 1990;64:6214–6220. doi: 10.1128/jvi.64.12.6214-6220.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kekuda R, Prasad P D, Fei Y J, Torres-Zamorano V, Sinha S, Yang-Feng T L, Leibach F H, Ganapathy V. J Biol Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 33.Jones E M C, Menzel S, Espinosa R, III, Le Beau M M, Bell G I, Takeda J. Genomics. 1994;23:490–491. doi: 10.1006/geno.1994.1529. [DOI] [PubMed] [Google Scholar]
- 34.Miller A D, Wolgamot G. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller D G, Edwards R H, Miller A D. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasko J E J, Kiem H-P, Morris J C, Gottschalk R J, Peterson L J, Andrews R G, Miller A D. Blood. 1997;90:118a. (abstr.). [Google Scholar]
- 37.Miller A D, Rosman G J. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 38.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartz S R, Vodicka M A. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 40.Dunn K J, Yuan C C, Blair D G. J Virol. 1993;67:4704–4711. doi: 10.1128/jvi.67.8.4704-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandyopadhyay P K, Temin H M. Mol Cell Biol. 1984;4:743–748. doi: 10.1128/mcb.4.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee S S, Hui H X, Hunter E. J Virol. 1990;64:3844–3852. doi: 10.1128/jvi.64.8.3844-3852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikawa T, Fischman D A, Dougherty J P, Brown A M. Exp Cell Res. 1991;195:516–523. doi: 10.1016/0014-4827(91)90404-i. [DOI] [PubMed] [Google Scholar]
- 44.Miller A D, Law M F, Verma I M. Mol Cell Biol. 1985;5:431–437. doi: 10.1128/mcb.5.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonham L, Wolgamot G, Miller A D. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 48.Fields-Berry S C, Halliday A L, Cepko C L. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halbert C L, Standaert T A, Aitken M L, Alexander I E, Russell D W, Miller A D. J Virol. 1997;71:5932–5941. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller A D, Chen F. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halbert C L, Alexander I E, Wolgamot G M, Miller A D. J Virol. 1995;69:1473–1479. doi: 10.1128/jvi.69.3.1473-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolgamot G, Rasko J E J, Miller A D. J Virol. 1998;72:10242–10245. doi: 10.1128/jvi.72.12.10242-10245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kilberg M S. Methods Enzymol. 1989;173:564–575. doi: 10.1016/s0076-6879(89)73039-1. [DOI] [PubMed] [Google Scholar]
- 54.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 55.Kekuda R, Torres-Zamorano V, Fei Y J, Prasad P D, Li H W, Mader L D, Leibach F H, Ganapathy V. Am J Physiol. 1997;272:G1463–G1472. doi: 10.1152/ajpgi.1997.272.6.G1463. [DOI] [PubMed] [Google Scholar]
- 56.Utsunomiya-Tate N, Endou H, Kanai Y. J Biol Chem. 1996;271:14883–14890. doi: 10.1074/jbc.271.25.14883. [DOI] [PubMed] [Google Scholar]
- 57.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bairoch A, Bucher P, Hofmann K. Nucleic Acids Res. 1996;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneda Y, Hayes H, Uchida T, Yoshida M C, Okada Y. Chromosoma. 1987;95:8–12. doi: 10.1007/BF00293835. [DOI] [PubMed] [Google Scholar]
- 60.Williams B P, Daniels G L, Pym B, Sheer D, Povey S, Okubo Y, Andrews P W, Goodfellow P N. Immunogenetics. 1988;27:322–329. doi: 10.1007/BF00395127. [DOI] [PubMed] [Google Scholar]
- 61.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Kavanaugh M P, North R A, Kabat D. Nature (London) 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 63.Kim J W, Closs E I, Albritton L M, Cunningham J M. Nature (London) 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 64.Schnitzer T J, Weiss R A, Zavada J. J Virol. 1977;23:449–454. doi: 10.1128/jvi.23.3.449-454.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koo H M, Brown A M, Ron Y, Dougherty J P. J Virol. 1991;65:4769–4776. doi: 10.1128/jvi.65.9.4769-4776.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torres-Zamorano V, Kekuda R, Leibach F H, Ganapathy V. Biochim Biophys Acta. 1997;1356:258–270. doi: 10.1016/s0167-4889(96)00178-4. [DOI] [PubMed] [Google Scholar]
- 67.Plakidou-Dymock S, Tanner M J, McGivan J D. Biochem J. 1994;301:399–405. doi: 10.1042/bj3010399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brody B A, Kimball M G, Hunter E. Virology. 1994;202:673–683. doi: 10.1006/viro.1994.1389. [DOI] [PubMed] [Google Scholar]