Abstract
Domains are the building blocks of all globular proteins and present one of the most useful levels at which protein function can be understood. Through recombination and duplication of a limited set of domains, proteomes evolved and the collection of protein superfamilies in an organism formed. As such, the presence of a shared domain can be regarded as an indicator of similar function and evolutionary history, but it does not necessarily imply it since convergent evolution may give rise to similar gene functions as well as architectures.
Through the wealth of sequences and annotation data brought about by genomics, evolutionary links can be sought for via homology relationships and comparative genomics, structural modeling and phylogenetics. The goal hereby is not only to predict the function of newly discovered proteins, but also to spell out their pathway of evolution and, possibly, identify their most likely origin. This can ultimately help to understand protein function and functional relationships of protein families. Additionally, through comparison with transcriptional data, evolutionary data can be linked to gene (and genome) activity and thus allow for the identification of common principles behind fast evolving proteins and relatively stable ones.
In this review, we describe the basic principles of studying protein (domain) evolution and illustrate recent developments in molecular evolution and give valuable new insights in the field of comparative genomics. As an example, we include here molecular models of the multiple PDZ domain protein MUPP-1 and present a simple comparative genomic view on its structural course of evolution.
Key Words: Domain, phylogeny, alignment, MUPP, PDZ, molecular evolution, protein folding, MPDZ, molecular modeling, multiple PDZ domain protein.
COMPARATIVE STRUCTURAL AND FUNCTIONAL GENOMICS
The genome projects of the last decade have produced a staggering amount of sequence data, but most of the identified genes lack experimental determination of biological function or even in some instances identification. The advances in bioinformatics have allowed large-scale genome comparisons, and efforts are well under way to make similar use of comparative functional and structural genomic approaches. However, the wealth of comparative genomic data generated has yet to be followed by a comparable gain of structural and functional information.
The annotation of genes, the prediction of new genes and the allocation of regulatory elements to date largely relies on evolutionary relationships for which genome comparison is fundamental [1, 2]. In essence, comparative genomics is based on the assumption that the two (or more) analyzed genomes share a common ancestor and that the bases in the sequence of each organism are the result of evolution acting on the genome of this mutual ancestor.
In general, evolution forms and molds genomes through two processes, namely mutational forces that generate random changes (i.e., point mutations or insertion-deletions [indels]) and selection pressures which can be positive, negative or neutral with regard to the presence of the mutation in the next generation [3, 4]. The combined effect of mutation and selection can subsequently be calculated and presented in a rate matrix, which denotes the probability of a mutation from one amino acid (or nucleotide) into another for a given period of time [5]. In turn, the rate matrix can be used to calculate alignments of two or more functional sequences. These functional sequences are, by definition, functions that are under evolutionary selection and are often a sequence of amino acids. However, they can, for example, also be transcription factor binding sites or RNA structures (e.g. microRNAs or viral RNA genomes). Commonly used rate matrices are BLOSUM and PAM [5, 6], which can readily be found implemented in BLAST and other well known sequence alignment programs [7-10].
As a result, a specific gene or protein of unknown function and biological importance can be compared to the sequence of a set of proteins with characterized functions. From these, the best matching group can be selected based on the number of domains and the nature of these domains. This information can be used to annotate the predicted gene or protein [2, 11-13].
Indeed, comparing genomes provides new insights into the biology of organisms whose hereditary material is under scrutiny. Some recent papers of comparisons between prokaryotes (e.g., γ-proteobacteria) [14, 15], insects (e.g., A. gambiae to D. melanogaster) [16, 17], mammals (e.g., M. musculus to H. sapiens) [18, 19], but also more distant comparisons between yeast and human genomes [20] are good examples of this approach. Furthermore, these studies have shed light upon transcriptional regulation [21-25], horizontal gene transfer [14, 24, 26], conservation of proteome networks [20, 27, 28] and strain-specific adaptations [29]. The combined data in GenBank and other databases now covers sequences for over 200.000 species with at least 50 complete genomes, which makes numerous more genome comparisons feasible [30-32]. But comparative genomics, especially when combined with proteomics, protein folding and microarray data, offers far more than just that; it can be used to explicate the evolution of proteins and the structures that make up proteins: the domains. In this review we describe the approaches currently available to elucidate the evolutionary history of proteins and their domains. We also provide examples, based on the PDZ domains of the Multiple PDZ Domain Protein-1 (MUPP-1; MPDZ) [33] and the single PDZ domain protein Disheveled (Dsh) [34]. MUPP-1 is an important scaffolding protein, which could potentially play important roles in lipid raft assembly [35], in viral entry [36] and in cancer progression [37]. Dsh, with two different additional protein binding domains, a DIX and a DEP domain, plays a central role in development of invertebrates and vertebrates [38].
SEQUENCE ALIGNMENT AND PHYLOGENY
Central biological features like metabolism, transcription and cell cycle progression are conserved from prokaryotes and single cell eukaryotes to humans [39, 40]. This conservation motivated and established the use of model organisms for studying conserved processes that are difficult or expensive to assess in higher organisms. Technological advances over the past two decades have led to the accumulation of genome-wide sequence data for many different species (see e.g., http://www.ensembl.org), but in order to use these sequences they have to be compared to each other in either pair-wise alignments (e.g., used in BLAST) or multiple sequence alignments, in which multiple sequences are compared simultaneously to each other (e.g., employed in ClustalX, Phylip and Muscle (see Table 1)).
Table 1.
List of Resources and Databases Relevant to Comparative Genomics
Alignments can also be subdivided based on the terms global and local. When whole genomes are aligned, bases are lined up by inserting gaps in sequences to account for (hypothetical) insertions or deletions that have taken place since diversification from the common ancestor. Indeed, this can be performed from one end to the other, as global implies, but when working with small genomes of several thousand base pairs or with entire chromosomes of hundred million base pairs it will need processing power and will be time consuming. Therefore, it is mostly applied to relatively short gene or protein sequence data, although web-based alignments can also be browsed (e.g., http://www.dcode.org). For the longer genomic nucleic acid sequences, a focus on regions of (local) high similarity is more feasible; the low sequence similarity regions are then ignored, which makes the procedure altogether much faster.
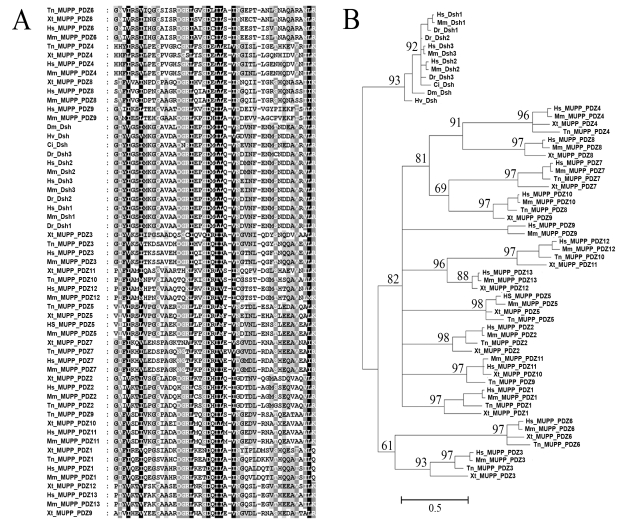
Automated alignments commonly employ a scoring procedure to find the best alignment possible for the input sequences. This scoring takes into account the number of identical residues, the number of different residues, and the size and number of gaps present in the alignment. Each different residue and bigger or extra gap will result in a penalty. Additionally, different penalties are created for the differences between for example transversions and transitions; with the latter being more common and thus favored over transversions [41]. However, the optimized alignment may not be the true one, since parameters can vary from species to species [42]. It is therefore recommended to manually check alignments and improve them (see Table 1 for programs). Fig. (1A) shows an example alignment of a number of PDZ domains with different shadings representing the amount of conservation (100, 75 or 50%) at a particular position in the sequence.
Fig. (1).
Example of sequence alignment and phylogeny. (A) Alignment of PDZ domains of the multiple (thirteen) PDZ domain protein MUPP-1 [33] and Disheveled (Dsh) [34] of several organisms. PDZ domains are modular interaction domains that recognize and bind to 4 C-terminal residues of the target domain, although other binding principles have also been shown. Black shading indicates 100% conservation, while the lighter grays indicates 75% or 50% conservation. Abbreviations used are Hs (Homo sapiens), Mm (Mus musculus), Xt (Xenopus tropicalis), Tn (Tetraodon Nigroviridis), Dr (Danio rerio), Ci (Ciona intestinalis) and Hv (Hydra vulgaris). (B) Evolutionary tree inferred by Bayesian Phylogeny (MrBayes) [46], rooted to the Dsh outgroup sequences. The tree shows clustering of most PDZ domains of MUPP-1 according to their sequence number in the protein. However, after PDZ 7 the numbers mix. Tn PDZ 8 clusters with Xt PDZ 9 and Hs PDZ 10, which suggests a domain duplication event. The clustering of Xt PDZ 8 with Hs PDZ 8 (near PDZ 4) and the separate clustering of Hs PDZ 9 suggest together one insertion (of PDZ 8) and one duplication event (of PDZ 9). To explain this most plausible relationship we have also presented this more visually in our structural model in Fig. (2B). It is important to note that MUPP-1 of Tetraodon nigroviridis contains 10, of Xenopus tropicalis 12 and of Homo sapiens 13 PDZ domains. Numbers indicated represent Bayesian posterior support values and all sequences used were obtained and analyzed as described previously [60].
Evolutionary distances can easily be estimated from small sequence alignments and can subsequently be used to create phylogenies, but also approximate divergence times, rates of evolution and ancestry sequences can be delineated from them. For phylogenetic analysis, multiple software packages are now available that often use one of these approaches: Maximum Likelihood [43, 44], Maximum Parsimony [7, 45], Neighbor Joining [7, 9] or Bayesian Estimation [46, 47] (see also Table 1). To provide an example of such a phylogenetic tree, we used MrBayes to calculate, over 100,000 generations and a mixed rate matrix set, the best tree topology for the alignment given in Fig. (1A).
Since, the MUPP-1 protein of Tetraodon nigroviridis has 10 domains, Xenopus tropicalis 12 and Homo sapiens 13 one hypothesis could be that the last domain of the “ten domain structure” duplicated two to three times to make up for the extra 2 or 3 domains found in the higher vertebrates. If this holds true, the last three PDZ domains should cluster closely together in the phylogenetic tree. However, this appears to be not the case: Tetraodon nigroviridis PDZ 8 clusters with Xenopus tropicalis PDZ 9 and Homo sapiens PDZ 10, which suggests at least one domain duplication event in the middle of the protein. The separate clustering of Xenopus tropicalis PDZ 8 with Homo sapiens PDZ 8 points to an insertion event in their common ancestor, however. Of course, we can not exclude from this small analysis that the domain was already present in the very early vertebrates and only lost in Tetraodon. We will try to shine more light on this with a structural model of these events in Fig. (2B).
Fig. (2).
Structural modeling of MUPP-1 PDZ domains and hypothetical model for internal domain duplications. (A) Molecular modeling of the thirteen human PDZ domains of MUPP-1 with Swiss-Model Workspace and Swiss-PBD Viewer 3.7 [83]. (B) In figure 1B, we compared the MUPP-1 PDZ domains of 4 different species. Of these four species, Tetraodon nigroviridis MUPP-1 consists of 10 PDZ domains, Xenopus tropicalis of 12 and Homo sapiens of 13 PDZ domains. Phylogenetic analyses implied that PDZ 8 of the Tetraodon MUPP-1 structure duplicated before at least twice to form the extra 2/3 PDZ domains present in the Xenopus and Homo sapiens structures. We therefore applied molecular modeling to these PDZ domains to visually support these findings. We modeled PDZ domains 7-9 of Tetraodon nigroviridis, domains 7-10 of Xenopus tropicalis and domains 7-11 of Homo sapiens. Indeed, PDZ 8 of Tetraodon, seems structurally related to 8 and 9 of Xenopus and 8-10 of Homo sapiens. However, within this group of six Xenopus PDZ 8 and Homo sapiens PDZ 8 appear to form a separate group. The most parsimonious explanation (and taking into account both the structural and phylogenetic data) therefore suggests one insertion event and one duplication event.
All phylogenetic information is extremely dependent on a proper alignment and not so much on the programs used to infer phylogeny [48]. Recently, software has been developed to combine the alignment procedure and phylogenetic analysis in one single program [47]. Current versions of this software can, however, only handle a limited set of sequences.
PROTEIN DOMAIN CLASSIFICATION AND SUPERFAMILIES
By definition, a domain is a structural, functional, but also an evolutionary component of a protein. Domain duplication and reorganization play important roles in evolution. It has been estimated that at least 70% of the domains duplicated in prokaryotes. In eukaryotes this number is presumed to be even higher, ranging to up to 90% [49]. Not surprisingly, many proteins comprise of more than one domain [1, 50, 51].
Domains are essential and versatile evolutionary elements that have been used to create from a relatively limited set an enormous and diverse assembly of proteins. Many protein family resources (e.g., Prosite and Pfam (see Table 1)) present a hierarchical classification that is almost fully dependent on sequence similarity and motif identification. Close relatives, sharing for example >50% sequence identity and often also functional properties, are grouped into families and subfamilies (e.g. PRINTS (see Table 1)). In turn, these families are grouped with other families into superfamilies [49, 52], with which they share for example ~25% sequence similarity. For a recent review on the function of these databases see reference [13].
PROTEIN DOMAIN FOLDING
After sequence analysis, the question arises whether sequence divergence is correlated with structural divergence and ultimately functional divergence. In the 1970s technologies (NMR and X-ray crystallography) for determining the 3D structure of domains and proteins became established. It was found that protein structures are primarily composed of α-helical and β-strand secondary structures (see Fig. 2 for a PDZ domain model structure) and there usually is a clear way to achieve optimal packing of the hydrophobic residues in the core of the protein (or sometimes outside, in case of a transmembrane protein).
As the number of solved structures increased it quickly became evident that protein (domain) structures are much more conserved (~50%) than the protein (amino acid) sequence (~5%) [53]. For this reason, it is possible that protein structures and their models can be used to find close as well as very distant relatives. Indeed, sometimes it is difficult to recognize divergent relatives solely through sequence comparison and often for these cases, there are no features present indicative of mutual functional properties [54]. There are two possible explanations: both domains or proteins have evolved from two different ancestral proteins; or they are two extremely distant relatives that started out from the same evolutionary ancestor [50, 54]. To distinguish between these possibilities, it is important to look at the current understanding of domain evolution. It is believed that the small set of protein domains known to date, descended from an even smaller group of ancestral domains. Unlike the raw protein sequence, the core of the protein domain is largely stable as it must be functionally conserved (i.e., selection is on function) and relies on inter-residue dependence. It is likely that protein evolution took place – or rather started – at the periphery of the relatively constant core. Indeed, it was shown that in pair-wise alignments, the amount of indels correlates with the evolutionary distance of proteins [4, 55, 56]. The structures most susceptible to point mutations, insertions or deletions are typically surface loops [57]. Unless mutations in these areas are neutralized, the number of changes will accumulate and eventually generate new polypeptide folds. Subsequently, positive selection will favor some of these newly arisen substructures when they become implemented in the biological process.
It should be clear from the above that the process of structural evolution is of a completely different order than that of sequence evolution, which is much faster. The tertiary sequence of a protein contains therefore much more phylogenetic signal and makes it far more likely to find linkages beyond the timeframe of standard sequence alignments [54]. Indeed, it may not be surprising that, like recognizing distinct sequence similarities, distinct folds and structures can be identified and classified as well. Examples are SCOP and CATH (see Table 1), which are linked to the Protein Data Bank (PDB) that stores protein structural data. Moreover, structural information can be used to verify and support phylogenetic data. As an example we modeled the differently clustering PDZ domains of MUPP-1 (the phylogenetic analyses shown in Fig. (1B) implied one insertion and one duplication event to form the extra 2/3 PDZ domains present in the Xenopus [PDZs 8 and 9] and Homo sapiens structures [PDZs 8-10]). Indeed, PDZ 8 of Tetraodon nigroviridis seems structurally highly related to domain 9 of Xenopus and domain 9 and 10 of Homo sapiens. In other words, either of these structures appears more structurally related to the others in this small group than to any of the other (flanking) PDZ domains, which suggests duplication. The PDZs 8 of Xenopus and Homo sapiens form, however, a separate structural group as the phylogenetic analysis predicted. We therefore propose that the Homo sapiens PDZ 9 originates from a duplication event of the Xenopus PDZ 9 and that Homo sapiens PDZ 8 is a result of an insertion in the common ancestor of Xenopus and Homo sapiens. Our evolutionary model shown in Fig. (2B) can thus be used to confirm the phylogenetic tree shown in Fig. (1B).
Even though domains are recognized by prediction programs, like Pfam and SMART, the actual fold may be different due to intermolecular interactions. Proteins usually contain more than one domain (i.e., multidomain proteins) and have evolved through a process of duplication and recombination of the limited set of protein domains available [51]. This principle not only brought together different enzymatic functions into single protein units (e.g., a catalytic domain and an ATP binding domain resulting in a helicase or kinase), but also combined domains that could co-evolve into one larger superdomain. An example of the latter can be found in the MAGUK family of proteins in which the Src homology 3 (SH3) domain and the Guanylate Kinase (GUK) domain interact intramolecularly to form a superdomain involved in protein-protein interactions [58, 59]. Not surprisingly, the GUK domain in these proteins is often only partially active or lacks activity completely and it was recently found that this loss of GUK activity corresponds with a position further away from the origin in the phylogenetic tree of the MAGUK proteins [60, 61].
GENES AND DOMAIN EVOLUTION BEYOND THE SEQUENCES
Important elements in a gene’s function are its spatial and temporal expression patterns. In recent years, microarray technology has made an extraordinary number of experiments possible that were aimed to map genome-wide expression levels under a variety of conditions [62-65]. For example, transcriptional comparisons have been made to look at for instance aging [66], pathogenicity [67] and non-coding RNAs [68]. Equivalent data is now, in addition to the sequence data, becoming available for dozens of different species and they provide a rich resource for comparative studies.
Unfortunately, the comparison of distantly related organisms can only be done under strictly defined expression conditions since gene expressions are not static. Indeed, by thoroughly controlling research conditions, comparisons between different (sub)species were made for conditions like embryogenesis, metamorphosis, sex-dependency and mutation rates [65, 69-72]. Other studies including diverse organisms such as yeasts, plants and primates, have revealed valuable information on promoter types and whether or not genes had previously undergone a duplication event [64, 65, 73, 74].
However, more evolutionary distant organisms may react differently to the same stimulus, which undermines comparison of gene expression data. To overcome this limitation, the association of co-expression data of genes and of expression signatures has been developed in addition to a direct comparison of individual gene expression changes [62]. Firstly, the co-expression between gene pairs is determined for each individual organism (within-species comparison) and this is then compared to the co-expression entities of other organisms. This approach focuses on the similarity and differences of the orthologous genes within their expression networks and this can be compared when species differences do not allow direct comparison at a specific condition. This system already has been applied for several species and it has revealed that both species-specific parts of the expression networks are combinations of conserved and newly evolved modules [62, 75, 76].
Another benefit of comparing co-expression of genes is that often functional entities can be discovered and, subsequently, new leads can be gained for functional interpretation. The approach can be combined with the search for common cis-regulatory elements at the promoter regions or applied to other similarity measures between genes, such as protein-protein interactions, phosphorylation networks or ligand-binding specificities [77-79].
CONCLUDING REMARKS
Finding evolutionary relationships for genes, proteins or protein domains is mostly based on orthology and thus on best sequence matches. Identifying these and categorizing them depends largely on multiple sequence alignments and this will in most cases give good indications for function and fold. However, this approach usually discards apparent ambiguities that arise from species-specific duplications or losses and may therefore introduce extensive biases [80]. Biases may also derive from the method of alignment, the phylogenetic analysis and the sample size used [47, 48, 81]. Therefore, care should be taken to not regard orthology as a pure one-to-one relationship, but as a family of homologous relations [64] and to select for the appropriate method of analysis [48, 81].
Genome and proteome comparisons can be performed by looking at expression data and, preferably, co-expression patterns or protein-protein and phosphorylation interactions. In the end, it will be the ultimate challenge to combine all comparative data (sequence, structure, expression, interaction and function) into one biological network. Indeed, only through putting together data obtained from protein-protein interactions and co-expression networks, conserved functional cell cycle complexes shared among yeast, plants, worms and humans have been revealed [82]. Expectantly, with these approaches we will be able to clearly distinguish how different biological mechanisms integrate, mold and flow along the forces of evolution. This is certainly an exciting and stimulatory time for interdisciplinary genomic research.
REFERENCES
- 1.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- 4.Bin Qian RAG. Distribution of indel lengths. Proteins Struct. Funct. Genet. 2001;45:102–104. doi: 10.1002/prot.1129. [DOI] [PubMed] [Google Scholar]
- 5.Dayhoff MO, Schwarz RM, Orcut BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Washington DC: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 6.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP version 3.63. Seattle: Dept of Genetics, Univ of Washington; 2004. [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucle. Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson WR. Comparison of methods for searching protein sequence databases. Protein Sci. 1995;4:1145–1160. doi: 10.1002/pro.5560040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotech. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 13.Attwood TK. The role of pattern databases in sequence analysis. Brief. Bioinformatics. 2000;1:45–49. doi: 10.1093/bib/1.1.45. [DOI] [PubMed] [Google Scholar]
- 14.Lerat E, Daubin V, Moran NA. From Gene Trees to Organismal Phylogeny in Prokaryotes:The Case of the gamma-Proteobacteria. PLoS Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comas I, ntilde aki Moya A, eacute Gonz aacute lez-Candelas F. From Phylogenetics to Phylogenomics: The Evolutionary Relationships of Insect Endosymbiotic &b.gamma;-Proteobacteria as a Test Case. Syst. Biol. 2007;56:1–16. doi: 10.1080/10635150601109759. [DOI] [PubMed] [Google Scholar]
- 16.Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM. The Interactive Fly: Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149 – 159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 17.Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 19.Consortium MGS. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 20.Gavin A, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A, Cruciat C, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier M, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 21.Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucl. Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, McCarter JP, Blaxter ML. A transcriptomic analysis of the phylum Nematoda. Nat. Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Prabhakar S, Chanan S, Cheng J, Rubin E, Boffelli D. Detection of weakly conserved ancestral mammalian regulatory sequences by primate comparisons. Genome Biol. 2007;8:R1. doi: 10.1186/gb-2007-8-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price M, Dehal P, Arkin A. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lercher MJ, Pal C. Integration of Horizontally Transferred Genes into Regulatory Interaction Networks Takes Many Million Years. Mol. Biol. Evol. 2007;msm283 doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- 27.Wyder S, Kriventseva E, Schroder R, Kadowaki T, Zdobnov E. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 2007;8:R242. doi: 10.1186/gb-2007-8-11-r242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Grus WE, Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SL, Hung C-S, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, Maskeri B, Hansen NF, Schwartz MS, Weber RJ, Kent WJ, Karolchik D, Bruen TC, Bevan R, Cutler DJ, Schwartz S, Elnitski L, Idol JR, Prasad AB, Lee-Lin SQ, Maduro VVB, Summers TJ, Portnoy ME, Dietrich NL, Akhter N, Ayele K, Benjamin B, Cariaga K, Brinkley CP, Brooks SY, Granite S, Guan X, Gupta J, Haghighi P, Ho SL, Huang MC, Karlins E, Laric PL, Legaspi R, Lim MJ, Maduro QL, Masiello CA, Mastrian SD, McCloskey JC, Pearson R, Stantripop S, Tiongson EE, Tran JT, Tsurgeon C, Vogt JL, Walker MA, Wetherby KD, Wiggins LS, Young AC, Zhang LH, Osoegawa K, Zhu B, Zhao B, Shu CL, De Jong PJ, Lawrence CE, Smit AF, Chakravarti A, Haussler D, Green P, Miller W, Green ED. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 31.Premzl M, Gready JE, Jermiin LS, Simonic T, Marshall Graves JA. Evolution of Vertebrate Genes Related to Prion and Shadoo Proteins--Clues from Comparative Genomic Analysis. Mol. Biol. Evol. 2004;21:2210–2231. doi: 10.1093/molbev/msh245. [DOI] [PubMed] [Google Scholar]
- 32.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullmer C, Schmuck K, Figge A, Lübbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- 34.Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- 35.Ackermann F, Zitranski N, Heydecke D, Wilhelm B, Gudermann T, Boekhoff I. The Multi-PDZ domain protein MUPP1 as a lipid raft-associated scaffolding protein controlling the acrosome reaction in mammalian spermatozoa. J. Cell. Physiol. 2008;214:757–768. doi: 10.1002/jcp.21272. [DOI] [PubMed] [Google Scholar]
- 36.Coyne CB, Voelker T, Pichla SL, Bergelson JM. The coxsackievirus and adenovirus receptor interacts with the multi-PDZ domain protein-1 (MUPP-1) within the tight junction. J. Biol. Chem. 2004;279:48079–48084. doi: 10.1074/jbc.M409061200. [DOI] [PubMed] [Google Scholar]
- 37.Martin TA, Watkins G, Mansel RE, Jiang WG. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer. 2004;40:2717–2725. doi: 10.1016/j.ejca.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Wharton Jr KA. Runnin' with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Development. Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 39.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 40.Miller W, Makova KD, Nekrutenko A, Hardison RC. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg MS, Kumar S. Heterogeneity of nucleotide frequencies among evolutionary lineages and phylogenetic inference. Mol. Biol. Evol. 2003;20:610–621. doi: 10.1093/molbev/msg067. [DOI] [PubMed] [Google Scholar]
- 42.Vingron M, Waterman MS. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J. Mol. Biol. 1994;235:1–12. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368 – 376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 44.Guindon Sp, Gascuel O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 45.Swofford D. PAUP* 4.0. Sinauer Associates; 2001. [Google Scholar]
- 46.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 47.Lunter G, Miklos I, Drummond A, Jensen J, Hein J. Bayesian coestimation of phylogeny and sequence alignment. BMC Bioinformatics. 2005;6:83. doi: 10.1186/1471-2105-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Filipski A. Multiple sequence alignment: In pursuit of homologous DNA positions. Genome Res. 2007;17:127–135. doi: 10.1101/gr.5232407. [DOI] [PubMed] [Google Scholar]
- 49.Apic G, Gough J, Teichmann SA. An insight into domain combinations. Bioinformatics. 2001;17:S83–89. doi: 10.1093/bioinformatics/17.suppl_1.s83. [DOI] [PubMed] [Google Scholar]
- 50.Han J, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 51.Wolf YI, Grishin NV, Koonin EV. Estimating the number of protein folds and families from complete genome data. J. Mol. Biol. 2000;299:897–904. doi: 10.1006/jmbi.2000.3786. [DOI] [PubMed] [Google Scholar]
- 52.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucl. Acids Res. 2007;35:D308–313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orengo CA, Thornton JM. Protein families and their evolution: a structural perspective. Ann. Rev. Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 55.Benner SA, Cohen MA, Gonnet GH. Empirical and structural models for insertions and deletions in the divergent evolution of proteins. J. Mol. Biol. 1993;20:1065–1082. doi: 10.1006/jmbi.1993.1105. [DOI] [PubMed] [Google Scholar]
- 56.Pascarella S, Argos P. Analysis of insertions/deletions in protein structures. J. Mol. Biol. 1992;224:461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- 57.Panchenko A, Madej T. Structural similarity of loops in protein families: toward the understanding of protein evolution. BMC Evol. Biol. 2005;5:10. doi: 10.1186/1471-2148-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavares GA, Panepucci EH, Brunger AT. Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. Mol. Cell. 2001;8:1313–1325. doi: 10.1016/s1097-2765(01)00416-6. [DOI] [PubMed] [Google Scholar]
- 59.McGee AW, Bredt DS. Identification of an Intramolecular Interaction between the SH3 and Guanylate Kinase Domains of PSD-95. J. Biol. Chem. 1999;274:17431–17436. doi: 10.1074/jbc.274.25.17431. [DOI] [PubMed] [Google Scholar]
- 60.te Velthuis A, Admiraal J, Bagowski C. Molecular evolution of the MAGUK family in metazoan genomes. BMC Evol. Biol. 2007;7:129. doi: 10.1186/1471-2148-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olsen O, Bredt DS. Functional Analysis of the Nucleotide Binding Domain of Membrane-associated Guanylate Kinases. J. Biol. Chem. 2003;278:6873–6878. doi: 10.1074/jbc.M210165200. [DOI] [PubMed] [Google Scholar]
- 62.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:294–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 63.Bergmann S, Ihmels J, Barkai N. Similarities and Differences in Genome-Wide Expression Data of Six Organisms. PLoS Biol. 2004;2:e9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tirosh I, Bilu Y, Barkai N. Comparative biology: beyond sequence analysis. Curr. Opin. Biotechnol. 2007;18:371–377. doi: 10.1016/j.copbio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Hooper SD, Boue S, Krause R, Jensen LJ, Mason CE, Ghanim M, White KP, Furlong EEM, Bork P. Identification of tightly regulated groups of genes during Drosophila melanogaster embryogenesis. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin C-S, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 67.Jeon J, Park S, Chi M, Choi J, Park J, Rho H, Kim S, Goh J, Yoo S, Choi J, Park J, Yi M, Yang S, Kwon M, Han S, Kim BR, Khang CH, Park B, Lim S, Jung K, Kong S, Karunakaran M, Oh H, Kim H, Kim S, Park J, Kang S, Choi W, Kang S, Lee Y. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 68.Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome Res. 2008;18:242–251. doi: 10.1101/gr.6887408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 70.Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 71.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 72.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 73.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 74.Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 75.Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Conservation and Coevolution in the Scale-Free Human Gene Coexpression Network. Mol. Biol. Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- 76.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB TP. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucl. Acids Res. 2008;36:D684–688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-Species Sequence Comparisons: A Review of Methods and Available Resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blouin C, Butt D, Roger AJ. Impact of Taxon Sampling on the Estimation of Rates of Evolution at Sites. Mol. Biol. Evol. 2005;22:784–791. doi: 10.1093/molbev/msi065. [DOI] [PubMed] [Google Scholar]
- 82.Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 83.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]