Abstract
Ribosomal RNA (rRNA) is transcribed from the ribosomal DNA (rDNA) genes by RNA polymerase I (Pol I). Despite being responsible for the majority of transcription in growing cells, Pol I regulation is poorly understood compared to Pol II. To gain new insights into rDNA transcriptional regulation, we developed a genetic assay in Saccharomyces cerevisiae that detects alterations in transcription from the centromere-proximal rDNA gene of the tandem array. Changes in Pol I transcription at this gene alter the expression of an adjacent, modified URA3 reporter cassette (mURA3) such that reductions in Pol I transcription induce growth on synthetic media lacking uracil. Increases in Pol I transcription induce growth on media containing 5-FOA. A transposon mutagenesis screen was performed with the reporter strain to identify genes that play a role in modulating rDNA transcription. Mutations in 68 different genes were identified, several of which were already known to function in chromatin modification and the regulation of Pol II transcription. Among the other classes of genes were those encoding proteasome subunits and multiple kinases and phosphatases that function in nutrient and stress signaling pathways. Fourteen genes were previously uncharacterized and have been named as regulators of rDNA transcription (RRT).
RIBOSOME biogenesis is a complex, tightly regulated process consisting of ribosomal DNA (rDNA) transcription, ribosomal RNA (rRNA) processing, ribosomal protein synthesis, and ribosome assembly. Its regulation is important for the control of cellular growth because it dictates the availability of the number of ribosomes required for efficient protein synthesis. Of particular importance is the synthesis of rRNA by RNA polymerase I (Pol I), which comprises the majority of all transcription in growing yeast (Saccharomyces cerevisiae) (Warner 1999). Ribosomal RNA synthesis rates are very high in these cells, but as nutrients in the growth medium are depleted and cells stop growing in stationary phase, the rate of rDNA transcription is greatly diminished (Ju and Warner 1994). One of the common hallmarks shared by rapidly proliferating cancer cells, or by hypertrophic cardiomyocytes in response to hypertension, is an increase in rDNA transcription (Hannan and Rothblum 1995; White 2005). Determining how cells activate and repress rDNA transcription in response to external nutrient and stress signals is therefore imperative for understanding the mechanisms of uncontrolled cell growth in such diseases. Yeast is an excellent model system to address such a problem.
Yeast rDNA consists of a single tandem array on the right arm of chromosome XII containing ∼150–200 head-to-tail repeats of the 9.1-kb rDNA gene. Each rDNA gene is transcribed by Pol I toward the centromere-proximal end of the array to produce a large 35S rRNA precursor, which is then co- and post-transcriptionally processed into the 18S, 5.8S, and 25S rRNAs (for review see Nomura 2001). Although only ∼50% of the rDNA genes are transcribed in a proliferating yeast cell (Dammann et al. 1993), each active gene is highly transcribed by ∼50 Pol I enzymes (French et al. 2003). Upon entry into stationary phase, the reduction in rate of rDNA transcription is accompanied by a lower percentage of rDNA genes that are actively transcribed and maintained in an open, psoralen-accessible chromatin state (Dammann et al. 1993). The histone deacetylase Rpd3 is required for closing the rDNA genes during stationary phase, although the mechanism by which this occurs remains uncertain (Sandmeier et al. 2002).
The rDNA in yeast can be transcribed by RNA polymerase II (Pol II) under certain conditions, although this is normally prevented by the Pol I transcription factor UAF through an unknown mechanism (Vu et al. 1999). Pol II-transcribed reporter genes inserted into the rDNA are silenced in a Pol I- and UAF-dependent manner (Buck et al. 2002; Cioci et al. 2003). This type of silencing (known as rDNA silencing) also requires the NAD+-dependent histone deacetylase Sir2 (Bryk et al. 1997; Fritze et al. 1997; Sandmeier et al. 2002). In contrast, mutating SIR2 has no measurable effect on Pol I transcription (Sandmeier et al. 2002). The mechanism of how UAF and Pol I function in silencing Pol II transcription remains unknown, but the involvement of Sir2 implies that environmental changes that alter rRNA synthesis rates could have significant effects on the rDNA chromatin structure.
In yeast, formation of the Pol I preinitiation complex requires four transcription factors: upstream activation factor (UAF), core factor (CF), TATA-binding protein, and Rrn3 (Nomura 2001). Rrn3 directly associates with Pol I, and the differential phosphorylation of Rrn3 regulates Pol I function (Milkereit and Tschochner 1998; Fath et al. 2001). Additional phosphorylation sites on Pol I subunits have been proposed to regulate Pol I function (Gerber et al. 2008). Rrn3 is equivalent to TIF-IA in mammals (Bodem et al. 2000), and CF is analogous to the mammalian SL1 complex (Lalo et al. 1996). A mammalian equivalent of UAF has not been identified, but the HMG-box-containing protein, UBF, functions in the initiation of mammalian Pol I transcription and is important for determining the number of active rDNA genes (Sanij et al. 2008), a function shared by UAF in yeast (Hontz et al. 2008). UBF has also been implicated in Pol I elongation (Stefanovsky et al. 2006) and is a target of regulation by phosphorylation and acetylation mechanisms, as is SL1 (for review see Moss et al. 2007).
The complex nature by which rDNA transcription, rRNA processing, and ribosomal protein gene transcription are coordinated suggests that researchers have only begun to scratch the surface of understanding the mechanisms involved in regulating these processes. In yeast, several signaling pathways are implicated in the regulation of rDNA transcription. Mutants defective in the secretory pathway respond by repressing the transcription of both rDNA and ribosomal protein genes through a protein-kinase-C-dependent regulatory circuit (Thevelein and de Winde 1999). Ribosomal DNA and ribosomal protein gene transcription also respond to signaling by the TOR (for review see Martin et al. 2006 and Tsang and Zheng 2007) and cAMP/PKA (Klein and Struhl 1994; Thevelein and de Winde 1999) pathways upon changes in nutrient availability. The details of how these pathways mediate changes in rDNA transcription, especially at the chromatin level, remain largely unknown. In this study, we developed a genetic assay that detects changes in Pol I transcription and employed it in a genetic screen for mutations that either improve or weaken rDNA transcription. Of the genes identified, several have previously been implicated in the regulation of Pol II transcription via chromatin modification, while others have no prior ties to transcriptional regulation. Importantly, the data provide new insights into the cellular processes that impact on Pol I transcription.
MATERIALS AND METHODS
Yeast strains and growth media:
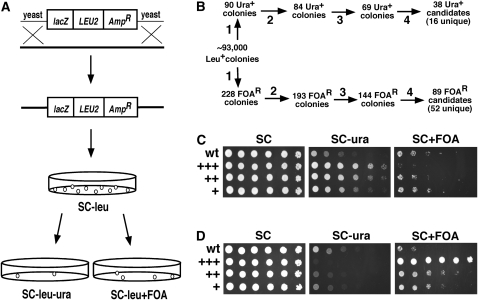
Yeast strains were derived from JB740 (Smith and Boeke 1997). YRH4 and YRH269 contain the mURA3-HIS3 reporter cassette adjacent to the centromere-proximal rDNA gene, where the NTS1 sequence was replaced with a 361-bp sequence derived from pJSS51-9 (Buck et al. 2002). The genetic identifier for this reporter cassette is nts1Δ∷mURA3-HIS3, where HIS3 is used for transformant selection. Strains used in this study are listed in the supporting information, Table S1. YSB519 is similar to YRH4, except that the mURA3-HIS3 reporter replaced the TRP1 gene (Buck et al. 2002). Genes were tagged with 13 copies of the myc epitope (13xMyc) in YRH269 as previously described (Longtine et al. 1998), using primers listed in Table S2. Spot-test growth assays were performed by growing strains as patches on YPD plates. Cells were scraped from the patches, resuspended in 1 ml of sterile H2O, normalized to an OD600 of 1.0, and serially diluted fivefold in 96-well plates. Five microliter aliquots of the dilutions were then spotted onto SC, SC−ura, and SC media containing 0.1% 5-fluoro-orotic acid (5-FOA). We use FOA to represent this compound throughout the text. Recipes for SC media are from Burke et al. (2000). Plates were incubated for 3 days before photographs were taken using an Alpha Innotech imager system.
Transposon mutagenesis:
YRH4 was transformed with NotI-digested plasmid pools that contained fragments of yeast genomic DNA with random mTn3 transposon insertions (Burns et al. 1994), as previously described (Smith et al. 1999). Transformants were selected on SC−leu plates, grown for 3 days, and then replica plated to SC−leu, SC−leu−ura, and SC−leu+FOA (0.2%) to identify mutants with altered rDNA transcription (Figure 2A). To cut down on background, 0.2% FOA was used. Locations of the mTn3 insertions were determined using either a plasmid rescue technique with pRS404, as previously described (Smith et al. 1999), or inverse PCR from genomic DNA (Burke et al. 2000). In both cases, sequencing across the transposon–yeast DNA junction was carried out with primer JS702. Sequences adjacent to the transposon were compared to the Saccharomyces Genome Database using BLAST to identify the exact nucleotide location of the insertion.
Figure 2.—
Experimental design for the transposon mutagenesis screen. (A) A schematic of the transposon mutagenesis and screening procedure. Yeast genomic DNA fragments containing random mTn3 transposon insertions were transformed into YRH4, and transformants were selected on SC−leu plates. Colonies were then replica plated to SC−leu−ura and SC−leu+0.2% FOA plates to identify relevant mutants. (B) Numbers of mutant candidates remaining after multiple steps of screening. Step 1: initial screening of transformants. Step 2: first single-colony isolation and replica plating. Step 3: second single-colony isolation and replica plating. Step 4: backcross test. (C) Examples of phenotype strength from Ura+ mutants isolated from the screen after 1 day (+++), 2 days (++), or 3 days (+) incubation after replica plating. The mutants shown are uaf30-Tn (PS1-174; +++), rrn5-Tn (PS1-212; ++), and nts1-Tn (PS1-170; +). (D) Examples of FOAR mutants isolated after day 1, 2, or 3 of the screen. The mutants shown are rpn8-Tn (PS1-7; +++), bre1-Tn (PS1-26; ++), and ira2-Tn (PS1-117; +).
Reverse transcriptase PCR:
Yeast were grown in 50-ml YPD cultures to an OD600 of ∼1.0. Total RNA was isolated using the hot-acid phenol method (Ausubel et al. 2000). cDNA was synthesized using the Invitrogen SuperScript reverse transcriptase II kit following the manufacturer's instructions. Five micrograms of total RNA was used for each cDNA reaction using either primer 1 (JS635) or the ACT1 primer JS84 at final concentrations of 1 μm. The following oligonucleotides were used for PCR reactions: primer 1 (JS635), primer 2 (JS636), primer 3 (JS639), ACT1 forward (JS83), and ACT1 reverse (JS84). Primers for this study are provided in Table S2. The PCR conditions were as follows: 95° for hot start, 94° for 2 min, 35 cycles of 94° for 1 min, 50° (for ACT1 cDNA) or 55° (for rRNA readthrough cDNA) for 1 min, 72° for 1 min, and 72° for 10 min. The products were run on a 1% agarose/TBE gel.
For the real-time RT–PCR assays, cDNA was produced from 5 μg total RNA using 1 μm primer 4 (JS766) for the rRNA product or 1 μm JS770 for the ACT1 product. The primer set for the rRNA readthrough product was JS765 and JS766, and the primer set for ACT1 was JS769 and JS770. Each 20-μl PCR reaction contained 200 nm of each primer, 10 μl 2× SYBR Green PCR Master Mix (Applied Biosystems), and 1 μl of cDNA (from a 1:625 dilution of the cDNA reaction). The manufacturer's instructions were followed. PCR reactions utilized a four-stage profile: stage 1, 50° for 2 min; stage 2, 95° for 10 min; stage 3, 95° for 15 sec, 60° for 1 min (40×); and stage 4 (dissociation), 95° for 15 sec, 60° for 30 sec, and 95° for 15 sec. Triplicates of each unknown cDNA were run simultaneously on a single 96-well plate, with standard deviation error bars calculated on the basis of the average of three to four independent cultures. Reactions were performed on an Applied Biosystems 7300 real-time PCR machine.
Pulsed-field gel electrophoresis:
The procedure was performed as previously described (Gerring et al. 1991). Yeast spheroplasts in low-melting-point agarose plugs were incubated in 1 ml LET solution (0.5 m EDTA, pH 8; 0.01 m Tris–HCl, pH 7.5) overnight at 37°, then equilibrated at 4° for 1 hr. The LET solution was replaced with 2 mg/ml proteinase K in NDS solution (0.5 m EDTA, pH 8; 0.01 m Tris–HCl, pH 7.5; 0.5 g Sarkosyl) and incubated for 2 days at 50°. The proteinase K/NDS solution was removed and replaced with 1 ml EDTA/Tris (0.05 m EDTA, pH 8; 0.01 m Tris–HCl, pH 7.5). For BamHI digests, the plugs were preequilibrated in 500 μl 1× BamHI buffer (NEB) + 0.1 mg/ml BSA on ice for 1 hr. The buffer was replaced two times. Following the final incubation, 2 μl BamHI enzyme (NEB; 20,000 units/ml) was added and incubated on ice for 2 hr and then shifted to 37° overnight. Plugs were placed into the wells of a 1% agarose/0.5× TBE gel and run for 68 hr on a Bio-Rad CHEF Mapper electrophoresis system. The running conditions were as follows: 120° angle; linear ramp (default); 3.0 V/cm; initial switch time, 300 sec; final switch time, 900 sec with 0.5 TBE× circulated at 14° as the running buffer. Following Southern blotting, the rDNA array released by BamHI was detected by hybridization with a 32P-labeled probe specific for the transcribed 35S region, followed by autoradiography.
Western blotting:
Log-phase yeast cultures (5 ml) were pelleted and then extracted in 0.5 ml 20% trichloroacetic acid (TCA) by vortexing with glass beads as previously described (Tanny et al. 1999). TCA precipitates were resuspended in 200 μl sample buffer (50 mm Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue), 10 μl β-mercaptoethanol, and 50 μl 2M Tris base. Ten-microliter aliquots of the samples were separated on 10% SDS polyacrylamide gels and transferred to an Immobilon-P membrane (Millipore). The membrane was blocked with 5% milk in PBS/0.1% Tween-20 and then probed with a 1:5000 dilution of anti-myc monoclonal antibody 9E10 (Millipore) or with a 1:5000 dilution of antitubulin monoclonal antibody (B-5-1-2, Sigma) in 25 ml PBS/0.1% Tween for 1 hr and then with a 1:5000 dilution of goat anti-rabbit or anti-mouse secondary antibody conjugated to horseradish peroxidase. Proteins were detected by chemiluminescence using ECL (GE Healthcare).
Chromatin immunoprecipitation assays:
The chromatin immunoprecipitation (ChIP) procedure was performed as previously described (Dasgupta et al. 2005). YPD cultures (100 ml) were grown into log phase and crosslinked with 1% formaldehyde for 6 min at 30°. The washed cell pellets were resuspended in 0.6 ml FA-lysis 140 buffer + protease inhibitors [(50 mm HEPES, 140 mm NaCl, 1.0% Triton X-100, 1.0 mm EDTA, 0.1% sodium deoxycholate), 25 ml 100× protease inhibitor cocktail (Sigma), 24 μl 100 mm benzamidine, 6 μl fresh 500 mm PMSF] and disrupted using a mini-bead beater 8 (Biospec Products) at 4°. Cell extracts were sonicated with eight 10-sec pulses (30% output, 90% duty cycle) on ice, then centrifuged at 4°, and the supernatants were transferred to a new microfuge tube. For immunoprecipitations, 1.5 mg of protein for each extract was incubated in a 0.6-ml microfuge tube with FA-lysis 140 solution in a total volume of 0.4 ml. Prior to antibody addition, an RNase A treatment was performed as previously described (Jones et al. 2007). A 1:50 dilution of anti-myc antibody (8 μl) (Millipore; clone 9E10) was added and rotated overnight at 4°. One-tenth of the chromatin extract volume used for the immunoprecipitation (IP) was used as the input control. The tubes were centrifuged at 14,000 rpm for 2 min at 4°, and the supernatant was added to a new 0.6-ml microfuge tube containing 60 μl protein A sepharose beads (50% slurry in FA lysis 140 buffer) and rotated at 4° for 4 hr. Beads were spun at 5000 rpm for 30 sec at room temperature and then washed 4 times with 0.5 ml FA-lysis 140 buffer, 4 times with 0.5 ml FA lysis 500 buffer (50 mm HEPES, 500 mm NaCl, 1.0% Triton X-100, 1 mm EDTA, 0.1% sodium deoxycholate), and 4 times with 0.5 ml LiCl detergent wash buffer (10 mm Tris–HCl, pH 8, 250 mm LiCl, 0.5% NP40, 0.5% sodium deoxycholate, 1.0 mm EDTA). DNA and protein was washed 2 times with 75 μl (5× TE + 1% SDS) and incubated 15 min at 65°. The 150 μl total volume was incubated overnight at 65° to reverse crosslinks. DNA was purified using an Invitrogen PCR purification kit. ChIP DNA was analyzed by real-time PCR. Input DNA was diluted 1:125, and the ± antibody samples were diluted 1:5. Each sample was run in triplicate per plate, with n = 3–4. The fivefold standard curve for each sample used YRH269 input DNA, starting with a 1:25 primary dilution. Values are reported as the ratio of IP sample to the input DNA. P-values were calculated using the two-tailed Student's t-test.
RESULTS
A reporter gene assay that is sensitive to changes in RNA polymerase I transcription:
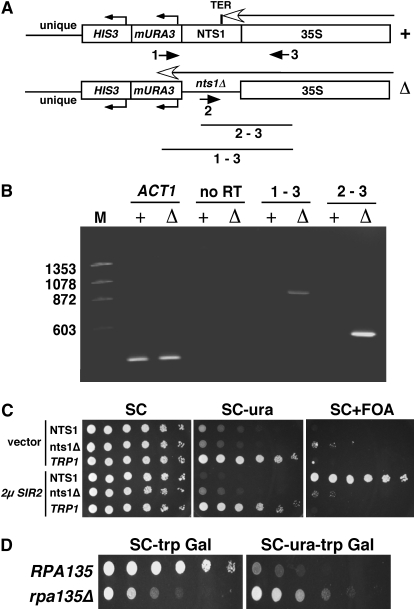
Transcription of yeast rDNA genes terminates within the nontranscribed spacer 1 (NTS1) region at the Reb1-binding site. The 10% of transcription that leaks through this primary terminator is halted by a secondary “fail-safe” termination sequence located downstream of the Reb1 site, also within NTS1 (Reeder et al. 1999). At the centromere-proximal rDNA repeat, the termination sequences in NTS1 prevent Pol I from transcribing into the unique chromosome XII sequence flanking the tandem array (Figure 1A, top). The introduction of a modified URA3 reporter gene driven by a TRP1 promoter (mURA3) into the flanking unique sequence resulted in SIR2-dependent silencing of the reporter (Buck et al. 2002). Repression of the mURA3 reporter gene was retained when the NTS1 sequence was deleted (Figure 1A, bottom), but the repression was no longer dependent on SIR2 (Buck et al. 2002). We hypothesized that deleting the Pol I termination sequences in NTS1 allowed Pol I transcription to proceed into the adjacent mURA3 promoter, thus interfering with its expression.
Figure 1.—
Pol I readthrough transcription inhibits mURA3 expression. (A) Schematics of the centromere-proximal rDNA repeat when the NTS1 sequence is present (YSB425, +) or replaced with a stuffer fragment (YRH4, Δ). Primer 1 was used for cDNA synthesis. PCR products obtained with primer pairs 1–3 and 2–3 are indicated. (B) Ethidium-bromide-stained agarose gel showing RT–PCR products obtained from YSB425 (+) and YRH4 (Δ). The size markers (M) are HaeIII-digested ØX174 DNA (New England Biolabs). ACT1 primers were JS83 and JS84. Without reverse transcription, products were not observed for ACT1 or the rDNA (no RT). (C) The nts1Δ∷mURA3-HIS3 reporter in YRH4 is insensitive to increased SIR2 dosage. YSB425 (NTS1), YRH4 (nts1Δ), and YSB519 (TRP1) were transformed with pRS425 (empty vector) or pSB766 (2μ SIR2 plasmid). Fivefold serial dilutions were spotted onto SC, SC−ura, and SC containing 0.1% 5-FOA (SC+FOA). SC plates were incubated for 2 days, while SC−ura and SC+FOA plates were incubated for 3 days. (D) Pol I transcription is required for mURA3 repression.
To demonstrate that Pol I was transcribing into the adjacent mURA3 reporter gene when NTS1 was deleted, we implemented RT–PCR to detect primary rRNA transcripts from the centromere-proximal rDNA gene that do not properly terminate (Figure 1A, bottom). Total RNA was isolated from a reporter strain that lacks the NTS1 sequence (YRH4; Δ) and a related strain (YSB425; +) that has an intact NTS1 sequence. An antisense mURA3 promoter primer was used to generate cDNA (Figure 1A, primer 1). When the cDNA was PCR amplified using primer 1 and a 35S-specific return primer (primer 3), a product of the expected 886-bp size was observed only when NTS1 was deleted (Figure 1B). Similarly, a 556-bp fragment was observed in the nts1Δ strain when the cDNA was PCR amplified with primer 3 and a nested primer (primer 2) specific for the stuffer fragment that replaced NTS1 (Figure 1B). Equal amounts of RNA were loaded into the cDNA reactions as measured by RT–PCR for the ACT1 transcript, and no product was observed when the RNA was not reverse transcribed. Pol I is therefore able to read into the mURA3 reporter when the terminators are removed, leading to poor expression of mURA3. This result suggested that the expression level of mURA3 in the nts1Δ reporter strain (YRH4) could potentially be used as a surrogate marker for the level of Pol I transcription.
To confirm that the expression of mURA3 in the nts1Δ reporter strain was unaffected by Sir2p-dependent silencing, we overexpressed SIR2 to test whether mURA3 repression was improved (Figure 1C). With typical rDNA silencing, repression of the mURA3 reporter is stronger when SIR2 is overexpressed, leading to even less growth on SC−ura plates (Smith et al. 1998). While repression of mURA3 in the nts1Δ∷mURA3 reporter strain (YRH4) causes poor growth on SC−ura media, it is not strong enough to trigger robust growth on SC medium containing 5-FOA (Figure 1C), which is toxic to cells sufficiently expressing URA3 (Boeke et al. 1984). Improved mURA3 repression would be expected to weaken growth on SC−ura and improve growth on FOA. As predicted, transformation of the NTS1 strain with a high-copy SIR2 plasmid induced stronger silencing of mURA3, as indicated by a lack of growth on SC−ura and by stronger growth on SC+FOA when compared to the empty vector control (Figure 1C). However, the SIR2 plasmid had no effect when NTS1 was deleted or when mURA3 was positioned outside the rDNA at the nonsilenced TRP1 locus (Figure 1C). Promoter interference of mURA3 by Pol I readthrough transcription is therefore unrelated to SIR2-mediated silencing.
To show that mURA3 repression required Pol I readthrough interference, we deleted the RPA135 gene, which encodes the second-largest subunit of Pol I. Viability of the rpa135Δ mutant was maintained on galactose-containing SC media by the presence of a TRP1 plasmid (pNOY199) that expressed the 35S rRNA from a galactose-inducible promoter (Vu et al. 1999). As shown in Figure 1D, repression of mURA3 was eliminated in the rpa135Δ mutant. Together, these results in Figure 1 demonstrate that the nts1Δ∷mURA3-HIS3 reporter strain (YRH4) acts as an indicator of active Pol I transcription from the centromere-proximal rDNA repeat.
A genetic screen for modulators of Pol I transcription:
Because deleting RPA135 eliminated mURA3 repression in the nts1Δ strain, we hypothesized that smaller effects on Pol I transcription that do not cause lethality would also be detectable. For example, mutations in genes that promote Pol I transcription would cause less readthrough interference of mURA3 and an improved Ura+ phenotype, whereas mutations in Pol I repressors would cause more transcriptional readthrough and improved growth on FOA. On the basis of these predictions we carried out a screen in which YRH4 (the nts1Δ∷mURA3-HIS3 reporter strain) was mutagenized by transformation with a library of yeast genomic DNA fragments that harbored random artificial transposon insertions (Burns et al. 1994; Smith et al. 1999) (Figure 2A). Insertion mutations were selected by growth on SC media lacking leucine (SC−leu). We obtained ∼93,000 Leu+ colonies that were replica plated to SC−leu−ura and SC−leu+0.2% FOA plates (Figure 2A). Initially, 90 Ura+ and 228 FOAR colonies were chosen for analysis (Figure 2B, step 1). Following two successive rounds of restreaking for single colonies and replica plating to verify the phenotypes, 69 Ura+ and 144 FOAR candidates remained (Figure 2B, steps 2 and 3). The phenotypic strength for each mutant was classified on the basis of the number of days following replica plating that they were identified: day 1 (+++), day 2 (++), and day 3 (+). Examples for the Ura+ mutants are shown in Figure 2C and the FOAR mutants in Figure 2D. As expected, increased growth on SC−ura correlated with decreased growth on FOA, and vice versa.
From 213 candidates, we were able to pinpoint the site of transposon insertion for 181 (85.0%). The transposon insertions in several mutants occurred at loci such as Ty elements or the rDNA locus that were not expected to cause a phenotype, suggesting that some mutants had secondary mutations or genomic rearrangements unrelated to the transposon insertion. To determine if the mURA3 growth phenotype was linked to the transposon insertion, we crossed each MATα haploid mutant to a MATa strain that did not contain the mURA3 reporter (JS579). Through tetrad dissection of the resulting diploids and replica plating of the spores, we were able to reduce the final number of sequenced candidates with phenotype–transposon (LEU2) linkage down to 38 Ura+ (16 unique) and 89 FOAR (52 unique) (Figure 2B, step 4). The unique genes are listed in Table 1 (Ura+) and in Table 2 (FOAR). All uncharacterized ORFs were renamed regulators of rDNA transcription (RRT). An expanded list describing the transposon insertion site for each mutant and whether the mutation is recessive or dominant is shown in Table S3. Not surprisingly, some of the FOAR mutants contained a transposon insertion within a gene involved in uracil biosynthesis and metabolism, and these likely affect growth on FOA independently of any changes in Pol I transcription. The effects of the mutations on mURA3 expression were predicted to rely on the NTS1 sequence being absent, thus allowing Pol I readthrough. We therefore picked 10 Ura+ and 10 FOAR mutants and crossed them to a reporter strain that retained the NTS1 sequence, followed by tetrad dissection and retesting. As shown in Table S4, the presence of NTS1 usually prevented any effect of the mutations on the mURA3 expression phenotype when compared to the nts1Δ counterparts. Sometimes there were even opposite effects.
TABLE 1.
Genes from Ura+ screen
| Gene | ORF | Function |
|---|---|---|
| Pol I transcription | ||
| RRN5 | YLR141W | UAF subunit |
| UAF30 | YOR295W | UAF subunit |
| Proteasome | ||
| RPN7 | YPR108W | Regulatory subunit |
| Miscellaneous | ||
| ATG2 | YNL242W | Autophagy |
| BNI4 | YNL233W | Targeting subunit for Glc7 phosphatase |
| GAP1 | YKR039W | Amino acid permease |
| HMS1 | YOR032C | myc family txn factor |
| MNN1 | YER001W | Mannosyltransferase |
| MSS4 | YDR208W | Phosphatidylinositol kinase |
| SEY1 | YOR165W | Vesicular trafficking |
| Unknown | ||
| RRT11 | YBR147W | Uncharacterized |
| RRT12 | YCR045C | Uncharacterized |
| RRT13 | YER066W | Uncharacterized |
| RRT14 | YIL127C | Uncharacterized, nucleolar |
| RRT15 | YLR162W-A | Uncharacterized |
| RRT16 | YNL105W | Dubious ORF, overlaps with INP52 |
| Intergenic | ||
| VTS1/PDE2 | YOR359W/YOR360C | |
TABLE 2.
Genes from FOAR screen
| Gene | ORF | Function |
|---|---|---|
| Chromatin-related | ||
| ADA1 | YPL254W | SAGA subunit |
| ADA2 | YDR448W | SAGA subunit |
| GCN5 | YGR252W | SAGA subunit |
| SPT20 | YOL148C | SAGA subunit |
| PHO23 | YNL097C | RPD3 subunit |
| RXT3 | YDL076C | RPD3 subunit |
| SAP30 | YMR263W | RPD3 subunit |
| SIN3 | YOL004W | RPD3 subunit |
| UME1 | YPL139C | RPD3 subunit |
| BRE1 | YDL074C | H2B ubiquitin ligase |
| HOS2 | YGL194C | Histone deacetylase |
| SIR1 | YKR101W | HM silencing factor |
| DPB11 | YJL090C | DNA Pol ɛ subunit |
| Proteasome | ||
| ECM29 | YHL030W | Bridging subunit |
| PRE9 | YGR135W | 20S subunit |
| RPN8 | YOR261C | Regulatory subunit |
| Miscellaneous | ||
| CHS6 | YJL099W | Exomer subunit |
| CIN1 | YOR349W | Tubulin folding factor D |
| IMP2′ | YIL154C | Transcription factor |
| MLC2 | YPR188C | Myosin regulatory light chain |
| MLP1 | YKR095W | Myosin-like protein |
| PAU7 | YAR020C | Seripauperin gene family |
| NTE1 | YML059C | Serine esterase |
| REV3 | YPL167C | DNA polymerase ζ |
| SBA1 | YKL117W | Hsp90-binding chaperone |
| Intragenic | ||
| BMH2/TVP15 | YDR099W/YDR100W | |
| MRP13/RPL11B | YGR084C/YGR085C | |
| Signaling | ||
| CMP2 | YML057W | Calcineurin A isoform |
| GPB1 | YOR371C | cAMP/PKA signaling |
| GPG1 | YGL121C | cAMP/PKA signaling |
| INM1 | YHR046C | Inositol monophosphatase |
| IRA1 | YBR140C | RAS/cAMP signaling |
| IRA2 | YOL081W | RAS/cAMP signaling |
| PPH22 | YDL188C | PP2A catalytic subunit |
| PRR1 | YKL116C | ser/thr protein kinase |
| PSK2 | YOL045W | PAS domain, ser/thr kinase |
| PXL1 | YKR090W | Cdc42 and Rho1 signaling |
| SAC7 | YDR389W | GAP for Rho1 signaling |
| YPK1 | YKL126W | ser/thr kinase, Sphingolipids |
| Metabolism | ||
| AAP1′ | YHR047C | Arg/Ala aminopeptidase |
| AGC1 | YPR021C | Nitrogen metabolism |
| GPM3 | YOL056W | Phosphoglycerate mutase |
| GUT2 | YIL155C | Glycerol utilization |
| PIG2 | YIL045W | Glycogen synthesis |
| Unknown | ||
| RRT1 | YBL048W | Dubious ORF/next to MOH1 |
| RRT2 | YBR246W | Two-hybrid with Rrt4 |
| RRT3 | YDR020C | Recently named DAS2 |
| RRT4 | YDR520C | Two-hybrid with Rrt2 |
| RRT5 | YFR032C | Uncharacterized |
| RRT6 | YGL146C | Uncharacterized |
| RRT7 | YLL030C | Dubious ORF/next to GPI13 |
| RRT8 | YOL048C | Uncharacterized |
| Pyrimidine synthesis | ||
| FUR4 | YBR021W | Uracil permease |
| URA5 | YML106W | De novo pyrimidine synthesis |
| URA6 | YKL024C | De novo pyrimidine synthesis |
Mutants isolated from the genetic screen alter transcription of the centromere-proximal rDNA gene:
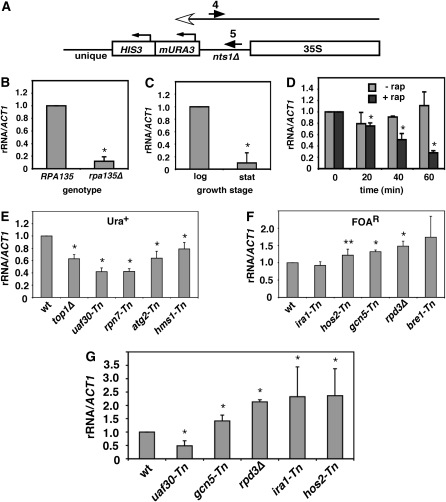
Two Ura+ mutants had transposon insertions in UAF30 and RRN5, both of which encode subunits of the Pol I transcription factor complex UAF (Keys et al. 1996; Siddiqi et al. 2001). Isolating these genes suggested that many of the Ura+ mutants would have Pol I transcription defects. Because severe Pol I defects could cause general slow growth or lethal phenotypes, the changes in rDNA expression were predicted to be mild. We utilized a quantitative real-time RT–PCR (qRT–PCR) assay to measure changes in rRNA production from the centromere-proximal rDNA gene. A cDNA product was synthesized from total RNA using primer 4 (JS765), which is specific for the “stuffer” fragment that replaced NTS1. PCR was then performed on the cDNA using primers 4 (JS765) and 5 (JS766), which amplify a segment of the stuffer transcribed by Pol I (Figure 3A). This rRNA product was then compared to an ACT1 mRNA loading control to indicate the relative changes in rRNA. As expected, the rRNA signal was greatly reduced in mutants or growth conditions that were known to reduce Pol I transcription, including the rpa135Δ mutant (Figure 3B), a wild-type strain grown to stationary phase (Figure 3C), and a wild-type strain treated with rapamycin (Figure 3D). We next tested the effects of various mutants from the screen. As shown in Figure 3E, each Ura+ mutant that we initially tested, including the top1Δ positive control, showed a reduction in rRNA compared to the wild-type strain (YRH4), indicating that the Ura+ phenotype is a good indicator of decreased rRNA transcription from the centromere-proximal repeat. Among these genes was HMS1, which encodes a helix-loop-helix protein with similarity to Myc-family transcription factors that has previously been implicated in regulation of pseudohyphal filamentous growth (Lorenz and Heitman 1998). Hms1 also physically interacts with Cdc14 (Ho et al. 2002), a phosphatase that is a subunit of the nucleolar RENT complex (Shou et al. 1999). Moreover, deletion of HMS1 is synthetically lethal with deletion of topoisomerase I (TOP1) (Tong et al. 2004), which functions in Pol I transcriptional elongation (Schultz et al. 1992; Hontz et al. 2008).
Figure 3.—
qRT–PCR assay for measuring changes in Pol I transcription from the centromere-proximal rDNA gene. (A) Schematic of the mURA3 reporter gene showing two PCR primers (4 and 5) used to specifically detect transcription of the nts1Δ stuffer fragment sequence. (B) Deletion of RPA135 causes a decrease in Pol I transcription. The RPA135 strain was YRH92 and the rpa135Δ strain was YRH95. (C) qRT–PCR assay of YRH4 grown into log phase or early stationary phase (stat). (D) YRH4 was treated with 0.2 mg/ml rapamycin during log-phase growth and analyzed by qRT–PCR at 20-min intervals. (E) Examples of four Ura+ mutants from the screen: uaf30-Tn (PS1-174), rpn7-Tn (PS1-152), atg2-Tn (PS1-184), and hms1-Tn (PS1-186). A deletion mutant for TOP1 was used as a positive control for reduction in Pol I transcription. (F) Examples of four FOAR mutants from the screen: ira1-Tn (PS1-56), hos2-Tn (PS1-120), gcn5-Tn (PS1-72), and bre1-Tn (PS1-26). A deletion mutant for RPD3 (YRH5) was also tested. (G) Examples of five mutants tested by qRT–PCR in stationary phase (4 days at 30°). In A–G, the ratio of rRNA qRT–PCR product to ACT1 RNA qRT–PCR product was calculated and normalized to 1.0 for wild type at time 0. All experiments were performed in triplicate with standard deviation error bars. A single asterisk indicates a P-value of < 0.05, and a double asterisk indicates P < 0.1.
We next tested the effects of several FOAR mutants using the qRT–PCR assay. These mutants were predicted to have increased levels of rDNA transcription at the left repeat. As shown in Figure 3F, the hos2-Tn, gcn5-Tn, and bre1-Tn mutants each resulted in a modest increase in rRNA signal compared to wild type. Several FOAR mutants affected subunits of the large RPD3 histone deacetylase complex Rpd3L (Table 1). Similarly, deleting RPD3 from YRH4 caused a modest increase in rRNA product. One of the exceptions that did not cause an increase in rRNA signal in the qRT–PCR assay was ira1-Tn. Both IRA1 and IRA2, which were isolated from the screen multiple times, are required for proper entry into stationary phase (Russell et al. 1993). Since the qRT–PCR assays were performed with cells growing in YPD media where rDNA transcription is active, it was possible that the ira1-Tn mutant and perhaps other FOAR mutants would have greater effects in the qRT–PCR assay in stationary phase where the rDNA is repressed. We retested several mutants from Figure 3F after growth for 24 hr into late diauxic shift/early stationary phase. The ira1-Tn mutant now showed higher rRNA signal than wild type, and the effects with rpd3Δ and hos2-Tn mutants were more severe than in log phase (compare Figure 3G with Figure 3F). The FOAR phenotype with the nts1Δ∷mURA3-HIS3 reporter is, therefore, an effective indicator of increases in rDNA synthesis from the centromere-proximal rDNA repeat.
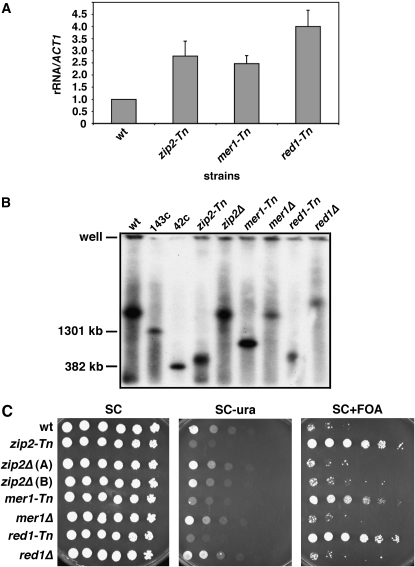
Altering rDNA transcription by changing the tandem array size:
Prior to the backcrossing that eliminated a large number of the mutant candidates, three of the FOAR class had a transposon insertion in ZIP2, RED1, or MER1, genes that specifically function in the formation of the meiotic synaptonemal complex. Each of these mutants produced large increases in centromere-proximal rDNA gene expression in the qRT–PCR assay (Figure 4A). However, the size of their rDNA tandem array was clearly reduced compared to wild type, which had an array size estimated as ∼175 copies (Figure 4B). The reduced rDNA copy number in these mutants could explain the increase in expression of the centromere-proximal rDNA repeat, because a previous study showed that rDNA genes in a 42-copy strain were more highly transcribed than rDNA genes in a 143-copy strain due to compensation (French et al. 2003). To test if that was the case here, we deleted ZIP2, RED1, or MER1 from YRH4 and retested their array size (Figure 4B) and the effects on FOAR growth (Figure 4C). The deletion mutants no longer had short arrays and their FOA and SC−ura phenotypes were the same as wild type, indicating that the effects on rDNA expression were due to the change in array size. As expected, the FOAR phenotype also did not cosegregate with the transposon insertions after tetrad dissections (data not shown), suggesting that the mutants that passed the backcross test (Table 2) are unlikely to have a significant change in array size.
Figure 4.—
Some primary mutants from the genetic screen affect Pol I transcription levels by changing the rDNA tandem array size. (A) qRT–PCR analysis of transposon mutants of ZIP2 (zip2-Tn; PS1-13), MER1 (mer1-Tn; PS1-14), and RED1 (red1-Tn; PS1-115). Values were calculated relative to wild type (YRH4) normalized to 1 (n = 3). (B) Pulsed-field gel electrophoresis on transposon mutants and direct knockouts of ZIP2 (YRH244), MER1 (YRH253), and RED1 (YRH254). Controls for rDNA array size are wild type (YRH4), a 42-copy strain (NOY886), and a 143-copy strain (NOY1051). (C) The same strains from B were tested for effects on the mURA3 reporter using the spot test assay. SC plates were grown for 2 days, and SC−ura and SC+FOA plates for 3 days. P-values for changes were all < 0.01.
Multiple chromatin-modifying factors function in Pol I transcriptional regulation:
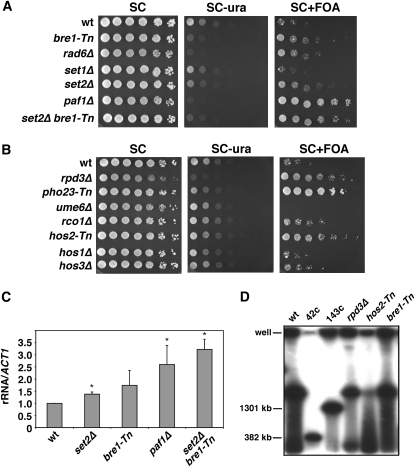
A large number of FOAR candidate genes were previously characterized as chromatin related and/or associated with Pol II transcription. These include Bre1, the E3 ubiquitin ligase for histone H2B (Hwang et al. 2003; Wood et al. 2003); Hos2, an H3/H4 histone deacetylase that is part of the SET3 complex (Pijnappel et al. 2001; Wang et al. 2002); and several subunits each from the SAGA/SLK/ADA histone acetyltransferase complexes and the Rpd3L/RpdS histone deacetylase complexes. Each of these has been previously linked to Pol II transcriptional elongation. We therefore deleted other genes with connections to these factors in elongation, including RAD6, the E2 ubiquitin-conjugating enzyme for H2B; SET1, the H3-K4 methyltransferase; SET2, the H3-K36 methyltransferase; and PAF1, which is a subunit of the conserved PAF complex that functions in Pol II elongation (Squazzo et al. 2002; Krogan et al. 2003; Chu et al. 2007). As shown in Figure 5A, the rad6Δ mutant had an FOAR phenotype similar to the bre1-Tn mutant, as one might have expected. Deletion of SET1 had no effect (Figure 5A), which was interesting because H3-K4 methylation was previously shown to function in rDNA silencing of Pol II-transcribed reporter genes (Briggs et al. 2001). In contrast, deletion of SET2 did cause an increase in growth on FOA (Figure 5A) and caused a higher rRNA signal in the qRT–PCR assay (Figure 5C). Deleting PAF1 resulted in strong growth on FOA (Figure 5A) and in a 2.5-fold increase in the rRNA qRT–PCR assay (Figure 5C). Because loss of PAF1 causes both H2B ubiquitination and H3-K36 methylation defects (Wood et al. 2003; Chu et al. 2007), we tested a set2Δ bre1-Tn double mutant and found similar results to those of the paf1Δ mutant by spot growth assay and qRT–PCR (Figure 5, A and C), consistent with the paf1Δ mutation causing a defect in both pathways.
Figure 5.—
Effects of several chromatin-related genes on transcription of the centromere-proximal rDNA gene. (A) Reporter gene assay with wild type (YRH4), bre1-Tn (PS1-26), rad6Δ (YRH698), set1Δ (YRH132), set2Δ (YRH134), paf1Δ (YRH164), and a set2Δ bre1-Tn double mutant (YRH265). (B) Reporter gene assay with wild type (YRH4), rpd3Δ (YRH5), pho23-Tn (PS1-76), ume6Δ (YRH51), rco1Δ (YRH152), hos2-Tn (PS1-120), hos1Δ (YRH142), and hos3Δ (YRH143). (C) qRT–PCR results of several candidates highlighted in A. Wild type was normalized to 1.0. (D) Pulsed-field gel electrophoresis analysis of the rDNA array size in wild type (YRH4), 42-copy rDNA control (NOY886), 143-copy rDNA control (NOY1051), rpd3Δ (YRH5), hos2-Tn (PS1-120), and bre1-Tn (PS1-26) strains. An asterisk indicates a P-value > 0.05.
Genetic dissection of histone deacetylase complexes that function in Pol I regulation:
Each of the Rpd3-associated genes isolated (SIN3, PHO23, SAP30, RXT3, and UME1) encode subunits of the Rpd3L complex, which represses Pol II transcription at target genes by locally deacetylating histones at the promoter (Kadosh and Struhl 1997). The Rpd3S (small) complex is targeted to the transcribed regions of genes by interactions with histone H3 methylated at K36, a modification catalyzed by Set2 (Carrozza et al. 2005; Keogh et al. 2005). Rpd3S shares some core subunits with the Rpd3L complex such as Sin3 and Ume1, but also contains the unique subunits Eaf3 and Rco1 (Carrozza et al. 2005). To test whether the Rpd3S complex functioned in Pol I transcription repression, we deleted RCO1. As shown in Figure 5B, the rco1Δ mutant had a moderate FOAR phenotype that was not as strong as the Rpd3L-specific mutant, pho23-Tn. Neither reduced growth on SC−ura as well as the rpd3Δ mutant, suggesting that both complexes contribute to Pol I regulation, with most of the activity coming from Rpd3L. The Ume6 subunit of Rpd3L specifically targets deacetylation to certain meiosis-related genes (Kadosh and Struhl 1997). This mutant did not induce FOA resistance (Figure 5B), indicating that the repressive effects are independent of Ume6-regulated genes.
Other histone deacetylases were tested to see if there were general deacetylase effects on rDNA transcription. While the hos2-Tn mutant produced a strong phenotype, deleting genes for other related deacetylases, HOS1 or HOS3, had little effect (Figure 5B), despite their known roles in deacetylating histones in the rDNA locus (Robyr et al. 2002). This result suggested that the Hos1- and Hos3-mediated histone deacetylation likely has another function. To confirm that the chromatin-related mutants do not affect rDNA expression by changing the array size, we measured the rDNA in the rpd3Δ, hos2-Tn, and bre1-Tn mutants by pulsed-field gel electrophoresis and found no difference from the wild-type YRH4 strain (Figure 5D). This is consistent with mutants having altered array sizes being excluded by the backcrossing procedure. It is not clear why the rDNA became degraded in the hos2-Tn mutant. Additional genes deleted from the YRH4 reporter strain during the course of this study, along with their mURA3 growth phenotypes, are listed in Table S5. Among these mutants was rsc1Δ, which produced an FOAR phenotype but also affected mURA3 expression when located at the non-rDNA location TRP1 (Figure S1). Therefore, RSC1 directly affects expression of the Pol II-driven reporter gene. Importantly, rpd3Δ, hos2Δ, and set2Δ mutations had no effect on Ura+ growth when mURA3 was positioned at the TRP1 locus (Figure S1), which is consistent with their effects on this reporter being specific to changes in Pol I transcription.
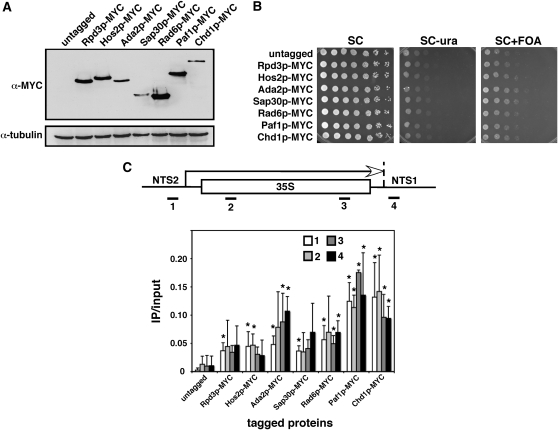
Chromatin-related factors isolated from the screen physically associate with the rDNA:
Changes in rDNA transcription could occur through direct or indirect mechanisms, so it became important to ask whether several of our candidate proteins physically associated with the rDNA locus. Rpd3, Hos2, Ada2, Sap30, Rad6, Paf1, and Chd1 were C-terminally tagged with 13 copies of the Myc epitope at the endogenous chromosomal locus. Each tagged protein was expressed at the predicted size in a Western blot (Figure 6A) and had growth properties similar to the untagged parental strain (YRH269; genotypically identical to YRH4) (Figure 6B). Quantitative ChIP assays were then performed to determine if the tagged proteins associated with four different locations on the rDNA genes (Figure 6C). To increase the chances of detecting a signal at the rDNA locus, we treated each crosslinked extract with RNaseA prior to immunoprecipitation to reduce the massive amount of rRNA associated with the active genes. This technique was previously used to demonstrate that Chd1 is enriched on the transcribed regions of the rDNA genes (Jones et al. 2007). On the basis of this previous study, we used the Myc-tagged Chd1 as a positive control for rDNA association (Figure 6C). CHD1 mutations cause an FOAR phenotype in YRH4. All of the tagged proteins were detected on the rDNA to varying degrees, each with a signal greater than that for the untagged negative control. While some of the increases for the transcribed region and NTS1 were not significant (P > 0.05), all of the promoter interactions at NTS2 were significant (Figure 6C, primer pair 1). All rDNA interactions were significant for Chd1 and Paf1. These results raise the possibility that some factors isolated from the screen, and not previously linked to chromatin, may also directly associate with the rDNA to modulate its transcription. This will be a focus of future studies.
Figure 6.—
Chromatin immunoprecipitations by real-time PCR reveal that several chromatin-related candidate proteins associate with the rDNA. (A) Western blot of steady-state protein levels for each 13xMYC epitope-tagged protein. Tubulin is the loading control. The strains used were as follows: untagged/wild type (YRH269), Rpd3p-MYC (YRH902), Hos2p-MYC (YRH901), Ada2p-MYC (YRH912), Sap30p-MYC (YRH915), Rad6p-MYC (YRH905), Paf1p-MYC (YRH914), and Chd1p-MYC (YRH911). (B) The Myc-tagged proteins do not alter expression of the nts1Δ∷mURA3-HIS3 reporter gene using the spot test assay. (C) Real-time PCR assay showing varying levels of rDNA association for each of the tagged proteins. (Top) Locations of the primer pairs are indicated as 1–4. The arrow represents the direction of Pol I transcription. The ratio of the PCR signal from the immunoprecipitated samples (IP) compared to the input chromatin sample (input) are graphed and compared to the untagged control strain. Ratios with a P-value >0.05 are indicated with an asterisk.
DISCUSSION
Possible mechanisms for mutants to affect Pol I transcription:
The genetic screen performed in this study takes advantage of an inhibitory effect of Pol I when it transcribes into an adjacent Pol II-transcribed reporter gene (mURA3). While we have yet to decipher the exact mechanism of this repression, it is likely that the large number of engaged Pol I molecules can displace Pol II-specific transcription factors. This is similar to the phenomenon of promoter interference (occlusion) that occurs for Pol II-transcribed genes when termination of the upstream gene is defective (Eggermont and Proudfoot 1993). It is clear that an increase or decrease in rDNA transcription from the centromere-proximal repeat affects mURA3 expression, as shown, respectively, by strains with a small array size (Figure 4) or with an rpa135 deletion (Figure 1). In theory, the transcription rate of this single repeat could be affected by changes in Pol I initiation or elongation, the rDNA array size, or the percentage of active genes, as we have recently shown for uaf30 mutants (Hontz et al. 2008). It is also possible that the mutations affect Pol I termination, because there is evidence that some rRNA transcripts manage to terminate without NTS1, as exhibited by a reduction in RT–PCR product when the cDNA is primed from the mURA3 promoter instead of the stuffer fragment (Figure 1B). If some termination does take place in the nts1Δ reporter strain, then some of the effects caused by the chromatin-related mutants could be through an effect on termination. For example, Chd1 has been implicated in Pol I termination via its chromatin-remodeling activity (Jones et al. 2007). Another possibility is that due to the coordination between rRNA processing and transcription, a defect in processing could in turn affect transcription. In any of these scenarios, the screen is still able to identify the mutant as being involved in rRNA synthesis at some level. We did not expect to identify many known positive regulators of Pol I transcription since they are typically essential, although, interestingly, the only nonessential transcription factor (Uaf30) and a viable mutant allele of an essential factor (Rrn5) from the UAF complex were found. Recovering such large numbers of mutants suggests that Pol I transcription is a finely tuned process.
The relationship between Pol II elongation and Pol I transcriptional repression:
The chromatin-related genes that we isolated as FOAR mutants were previously linked physically or genetically to the Pol II elongation process. For example, SAGA-dependent histone acetylation in the transcribed regions of genes helps to promote elongation by evicting nucleosomes (Govind et al. 2007). SAGA also interacts with the Pol II elongation factor TFIIS (Wery et al. 2004). Paf1 has recently been reported to destabilize nucleosomes (Marton and Desiderio 2008), suggesting that Paf1-mediated histone modifications may also be involved in this process. When nucleosomes are reassembled behind the transcribing RNA Pol II, they are methylated on H3-K36 by Set2 (Workman 2006). This mark provides a binding site for the Rpd3S complex via the combined affinity of the Eaf3 chromodomain and Rco1 PHD motif (Li et al. 2007). Rpd3S-mediated histone deacetylation resets the chromatin to a repressive state that prevents spurious transcription from within the open reading frame (Carrozza et al. 2005), a process that also involves Paf1 (Chu et al. 2007). However, Set2 has not been reported to interact with Pol I or with the rDNA, and H3-K36 methylation does not appear to occur in the rDNA (Xiao et al. 2003), suggesting that the effects of Set2 on Pol I transcription may be indirect. SET2 deletion could alter the expression of the Pol II-transcribed ribosomal protein genes, which would then affect the coordination between ribosomal protein gene expression and Pol I transcription/rRNA processing. Another possibility is that chromatin-level defects in Pol I transcription could result in an increase in the number of open rDNA repeats, such as occurs with mutations in Rpd3 (Sandmeier et al. 2002), resulting in an FOAR phenotype as shown here.
Paf1 association with the rDNA was relatively strong in our ChIP assay (Figure 6), and it had a large effect in the qRT–PCR and mURA3 growth assays. A previous study using immunofluorescence microscopy found that when PAF1 was deleted, other subunits of the PAF complex localized to the nucleolus (Porter et al. 2005). The wild-type complex was not detected, but our data indicate that the complex does normally associate with the rDNA. In the same study, it was reported that deleting PAF1 caused an rRNA-processing defect that resulted in the accumulation of unprocessed rRNA precursors. More recent investigations of yeast Paf1 found that it was required for silencing of Pol II genes in the rDNA (Mueller et al. 2006). While our article was being prepared, the PAF complex was also implicated in Pol I elongation (Zhang et al. 2009). Interestingly, deleting the Ctr9 subunit of the PAF complex resulted in an increase in the percentage of rDNA genes in the array that were transcribed (Zhang et al. 2009). This would increase the frequency at which the centromere-proximal rDNA gene was transcribed, perhaps explaining why our assays detected greater transcription of the centromere-proximal rDNA gene. Alternatively, Paf1 could be required for limiting the number of rDNA genes that are transcribed, thus acting in a repressive capacity. It will be interesting to further dissect the role of Paf1 in Pol I transcriptional regulation.
Interestingly, there are several Pol II-transcribed genes within the yeast rDNA, including TAR1 (Coelho et al. 2002). Pol II also produces cryptic unstable transcripts from the nontranscribed spacer that may have roles in regulating rDNA silencing and recombination (Kobayashi and Ganley 2005; Li et al. 2006; Houseley et al. 2007; Vasiljeva et al. 2008). Additionally, the rDNA genes have the capacity to be transcribed by Pol II in mutant strains defective for the Pol I transcription factor UAF (Vu et al. 1999; Oakes et al. 2006). Mutations in certain chromatin-modifying enzymes could alter Pol II transcription within the rDNA, which could in turn influence rRNA synthesis by Pol I. The Nomura lab previously demonstrated that there is a reciprocal relationship between Pol I and Pol II transcription within the rDNA (Cioci et al. 2003). Of course it is also possible that some of the chromatin factors from the screen directly repress Pol I transcription through an uncharacterized mechanism, which would be consistent with their rDNA association.
Signaling pathways impact rDNA transcription:
Multiple signaling factors were identified from the FOAR portion of the screen (Table 2). IRA1, IRA2, GPB1, and GPG1 all function in the negative regulation of the cAMP/PKA signaling pathway, which works in parallel with TOR to coordinate the expression of genes required for cell growth (Zurita-Martinez and Cardenas 2005). IRA1 and IRA2 encode GTPase-activating proteins (GAPs) that negatively regulate RAS by converting it from the GTP- to the GDP-bound inactive form, and mutations in these genes result in constitutive activation of RAS and, therefore, of the cAMP/PKA pathway (Broach 1991). GPB1 encodes a protein that inhibits the cAMP/PKA pathway by binding and stabilizing Ira1 and Ira2 (Harashima et al. 2006; Peeters et al. 2006), and GPG1 encodes the gamma subunit required for Gpb1 or Gpb2 to interact with Gpa2 as a heterotrimeric G-protein complex (Harashima and Heitman 2002). Constitutive activation of RAS and the cAMP/PKA pathway prevents the rapamycin-induced repression of Pol I and Pol III transcription (Schmelzle et al. 2004) and the starvation-induced repression of ribosomal protein genes (Klein and Struhl 1994). Furthermore, yeast with constitutively high PKA activity have been shown to upregulate ribosomal protein gene expression by approximately twofold (Klein and Struhl 1994). The identification of these four genes as repressors of rDNA transcription is fully consistent with these findings.
Another intriguing signaling gene from the FOAR mutants is YPK1, which encodes a serine/threonine protein kinase that is part of the sphingolipid long-chain base (LCB)-mediated signaling pathway. LCBs are intracellular signaling molecules in yeast (see Dickson et al. 2006 for review). Their concentrations rapidly, but transiently, increase in response to cellular stress. Ypk1 can be activated by LCBs, but upstream kinases Pkh1 and -2 are also activated by LCBs and can then activate Ypk1. Ypk1 and its paralog, Ypk2, then regulate cell-wall integrity, actin dynamics, translation, endocytosis, and cell growth. It is also important to note that Ypk1 and Ypk2 are downstream of the TOR-signaling kinase TORC2. Taken together, the data from our screen implicate multiple, diverse signaling pathways in the regulation of Pol I transcription, many of which require further investigation.
Novel genes that influence rDNA transcription:
Fourteen previously uncharacterized genes, designated as RRTs, were identified from our screen. Transposon insertions in RRT1–RRT8 caused an FOAR phenotype, whereas insertions in RRT11–RRT16 caused a Ura+ phenotype. Little is known about most of the Ura+ rrt mutants, although RRT14 (YIL127C) encodes a nucleolar protein that interacts with Utp5, a subunit of the nucleolar tUTP complex that functions in linking rRNA processing with transcription of the rDNA (Gallagher et al. 2004). Among the FOAR mutants, the Rrt2 and Rrt4 proteins have been shown to interact in a two-hybrid assay (Ito et al. 2001), and deletion of RRT3 (YDR020C, also known as DAS2) suppresses the 6-azauracil and mycophenolic acid sensitivity of a TFIIS mutant (S. Chavez, personal communication). TFIIS is a Pol II elongation factor (Wind and Reines 2000). Other genes isolated from the DAS2 screen were connected with the regulation of ribosomal protein genes (S. Chavez, personal communication). Finally, the transposon insertions for three of the rrt mutants occurred in dubious open reading frames that may not encode functional genes. In these cases, it is possible that the transposon insertions instead affected the expression of an adjoining gene. In conclusion, this genetic screen has identified many characterized and uncharacterized genes that affect the level of rDNA transcription from the centromere-proximal rDNA gene. Future analysis will focus on determining the mechanisms of action.
Acknowledgments
We thank Ashley Blair and Marisol Santisteban for assistance with qRT–PCR, Dan Burke for advice on inverse PCR, Stephen Buck for assistance and advice with yeast strains, Masayasu Nomura for providing strains, Sebastian Chavez for critically reading the manuscript and providing results prior to publication, David Schneider for communicating results prior to publication, and Ann Beyer for critical review of the manuscript. We also thank Patrick Grant, David Auble, and members of the Smith and Beyer labs for helpful discussions. Funding for this study was provided by grants from the National Institutes of Health (GM61692) and the American Heart Association (grant-in-aid 0555490 and 0755633) to J.S.S.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.100313/DC1.
References
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 2000. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf et al., 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie et al., 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach, J. R., 1991. Ras-regulated signaling processes in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 1 370–377. [DOI] [PubMed] [Google Scholar]
- Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel et al., 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11 255–269. [DOI] [PubMed] [Google Scholar]
- Buck, S. W., J. J. Sandmeier and J. S. Smith, 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111 1003–1014. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8 1087–1105. [DOI] [PubMed] [Google Scholar]
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson et al., 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581–592. [DOI] [PubMed] [Google Scholar]
- Chu, Y., R. Simic, M. H. Warner, K. M. Arndt and G. Prelich, 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26 4646–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci, F., L. Vu, K. Eliason, M. Oakes, I. N. Siddiqi et al., 2003. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12 135–145. [DOI] [PubMed] [Google Scholar]
- Coelho, P. S., A. C. Bryan, A. Kumar, G. S. Shadel and M. Snyder, 2002. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 16 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller and J. M. Sogo, 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, A., S. A. Juedes, R. O. Sprouse and D. T. Auble, 2005. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 24 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, R. C., C. Sumanasekera and R. L. Lester, 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 45 447–465. [DOI] [PubMed] [Google Scholar]
- Eggermont, J., and N. J. Proudfoot, 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 12 2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath, S., P. Milkereit, G. Peyroche, M. Riva, C. Carles et al., 2001. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. USA 98 14334–14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, S. L., Y. N. Osheim, F. Cioci, M. Nomura and A. L. Beyer, 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze, C. E., K. Verschueren, R. Strich and R. E. Esposito, 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim et al., 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18 2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, J., A. Reiter, R. Steinbauer, S. Jakob, C. D. Kuhn et al., 2008. Site specific phosphorylation of yeast RNA polymerase I. Nucleic Acids Res. 36 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring, S. L., C. Connelly and P. Hieter, 1991. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 194 57–77. [DOI] [PubMed] [Google Scholar]
- Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer and A. G. Hinnebusch, 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25 31–42. [DOI] [PubMed] [Google Scholar]
- Hannan, R. D., and L. I. Rothblum, 1995. Regulation of ribosomal DNA transcription during neonatal cardiomyocyte hypertrophy. Cardiovasc. Res. 30 501–510. [PubMed] [Google Scholar]
- Harashima, T., and J. Heitman, 2002. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol. Cell 10 163–173. [DOI] [PubMed] [Google Scholar]
- Harashima, T., S. Anderson, J. R. Yates, III and J. Heitman, 2006. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 22 819–830. [DOI] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415 180–183. [DOI] [PubMed] [Google Scholar]
- Hontz, R. D., S. L. French, M. L. Oakes, P. Tongaonkar, M. Nomura et al., 2008. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol. Cell. Biol. 28 6709–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley, J., K. Kotovic, A. El Hage and D. Tollervey, 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26 4996–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone et al., 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11 261–266. [DOI] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, H. S., J. Kawauchi, P. Braglia, C. M. Alen, N. A. Kent et al., 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, Q., and J. R. Warner, 1994. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10 151–157. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89 365–371. [DOI] [PubMed] [Google Scholar]
- Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny et al., 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 593–605. [DOI] [PubMed] [Google Scholar]
- Keys, D. A., B.-S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu et al., 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10 887–903. [DOI] [PubMed] [Google Scholar]
- Klein, C., and K. Struhl, 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., and A. R. Ganley, 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309 1581–1584. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt et al., 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11 721–729. [DOI] [PubMed] [Google Scholar]
- Lalo, D., J. S. Steffan, J. A. Dodd and M. Nomura, 1996. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem. 271 21062–21067. [DOI] [PubMed] [Google Scholar]
- Li, B., M. Gogol, M. Carey, D. Lee, C. Seidel et al., 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316 1050–1054. [DOI] [PubMed] [Google Scholar]
- Li, C., J. E. Mueller and M. Bryk, 2006. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol. Biol. Cell 17 3848–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., and J. Heitman, 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150 1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. E., T. Powers and M. N. Hall, 2006. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 4 259–260. [DOI] [PubMed] [Google Scholar]
- Marton, H. A., and S. Desiderio, 2008. The Paf1 complex promotes displacement of histones upon rapid induction of transcription by RNA polymerase II. BMC Mol. Biol. 9 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., and H. Tschochner, 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, T., F. Langlois, T. Gagnon-Kugler and V. Stefanovsky, 2007. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol. Life Sci. 64 29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. E., M. Canze and M. Bryk, 2006. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 173 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, M., 2001. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 66 555–565. [DOI] [PubMed] [Google Scholar]
- Oakes, M. L., I. Siddiqi, S. L. French, L. Vu, M. Sato et al., 2006. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol. Cell. Biol. 26 3889–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, T., W. Louwet, R. Gelade, D. Nauwelaers, J. M. Thevelein et al., 2006. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 103 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte et al., 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, S. E., K. L. Penheiter and J. A. Jaehning, 2005. Separation of the Saccharomyces cerevisiae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryot. Cell 4 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder, R. H., P. Guevara and J. G. Roan, 1999. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 19 7369–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang et al., 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109 437–446. [DOI] [PubMed] [Google Scholar]
- Russell, M., J. Bradshaw-Rouse, D. Markwardt and W. Heideman, 1993. Changes in gene expression in the Ras/adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol. Biol. Cell 4 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo et al., 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij, E., G. Poortinga, K. Sharkey, S. Hung, T. P. Holloway et al., 2008. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183 1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T., T. Beck, D. E. Martin and M. N. Hall, 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, M. C., S. J. Brill, Q. Ju, R. Sternglanz and R. H. Reeder, 1992. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 6 1332–1341. [DOI] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Siddiqi, I. N., J. A. Dodd, L. Vu, K. Eliason, M. L. Oakes et al., 2001. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 20 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., and J. D. Boeke, 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11 241–254. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, L. Pillus and J. D. Boeke, 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., E. Caputo and J. D. Boeke, 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic et al., 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L. I. Rothblum and T. Moss, 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21 629–639. [DOI] [PubMed] [Google Scholar]
- Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz and D. Moazed, 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99 735–745. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., and J. H. de Winde, 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33 904–918. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Tsang, C. K., and X. F. Zheng, 2007. TOR-in(g) the nucleus. Cell Cycle 6 25–29. [DOI] [PubMed] [Google Scholar]
- Vasiljeva, L., M. Kim, N. Terzi, L. M. Soares and S. Buratowski, 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29 313–323. [DOI] [PubMed] [Google Scholar]
- Vu, L., I. Siddiqi, B.-S. Lee, C. A. Josaitis and M. Nomura, 1999. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase I. Proc. Natl. Acad. Sci. USA 96 4390–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., S. K. Kurdistani and M. Grunstein, 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298 1412–1414. [DOI] [PubMed] [Google Scholar]
- Warner, J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24 437–440. [DOI] [PubMed] [Google Scholar]
- Wery, M., E. Shematorova, B. Van Driessche, J. Vandenhaute, P. Thuriaux et al., 2004. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 23 4232–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. J., 2005. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6 69–78. [DOI] [PubMed] [Google Scholar]
- Wind, M., and D. Reines, 2000. Transcription elongation factor SII. BioEssays 22 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt et al., 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11 267–274. [DOI] [PubMed] [Google Scholar]
- Workman, J. L., 2006. Nucleosome displacement in transcription. Genes Dev. 20 2009–2017. [DOI] [PubMed] [Google Scholar]
- Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall et al., 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., M. L. Sikes, A. L. Beyer and D. A. Schneider, 2009. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc. Natl. Acad. Sci. USA 106 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez, S. A., and M. E. Cardenas, 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]