Abstract
The completion of genome-sequencing projects for a number of fungi set the stage for detailed investigations of proteins. We report the generation of versatile expression vectors for detection and isolation of proteins and protein complexes in the filamentous fungus Neurospora crassa. The vectors, which can be adapted for other fungi, contain C- or N-terminal FLAG, HA, Myc, GFP, or HAT–FLAG epitope tags with a flexible poly-glycine linker and include sequences for targeting to the his-3 locus in Neurospora. To introduce mutations at native loci, we also made a series of knock-in vectors containing epitope tags followed by the selectable marker hph (resulting in hygromycin resistance) flanked by two loxP sites. We adapted the Cre/loxP system for Neurospora, allowing the selectable marker hph to be excised by introduction of Cre recombinase into a strain containing a knock-in cassette. Additionally, a protein purification method was developed on the basis of the HAT–FLAG tandem affinity tag system, which was used to purify HETEROCHROMATIN PROTEIN 1 (HP1) and associated proteins from Neurospora. As expected on the basis of yeast two-hybrid and co-immunoprecipitation (Co-IP) experiments, the Neurospora DNA methyltransferase DIM-2 was found in a complex with HP1. Features of the new vectors allowed for verification of an interaction between HP1 and DIM-2 in vivo by Co-IP assays on proteins expressed either from their native loci or from the his-3 locus.
THE filamentous ascomycete Neurospora crassa has served as a model eukaryote for more than 70 years (Davis 2000; Davis and Perkins 2002). Fifty years ago Beadle and Tatum were awarded a Nobel Prize for their “one-gene, one-enzyme” research project with Neurospora and numerous subsequent studies with the organism have provided important insights into metabolism, gene regulation, chromosome behavior, DNA repair, DNA methylation, genome defense, photobiology, circadian rhythms, differentiation, development, and other biological phenomena of relevance to higher eukaryotes (Dunlap et al. 2007). Simultaneously, Neurospora researchers developed a variety of biochemical and genetic tools. Recently, sequencing of the N. crassa genome (Galagan et al. 2003) greatly stimulated the field, in part by immediately revealing ∼10,000 predicted genes (Borkovich et al. 2004). Bioinformatics data are accessible via public databases, and microarrays covering all the predicted genes are available from the Fungal Genetics Stock Center and used for transcriptional profiling (Kasuga et al. 2005; Tian et al. 2007; Kasuga and Glass 2008) and for microarray-based genetic mapping (Lewis et al. 2007). Moreover, a systematic gene knockout project to disrupt all the predicted genes is well underway (Colot et al. 2006). Although genetic resources for Neurospora are plentiful and invaluable, proteomics technology for this and other filamentous fungi are less developed. We have sought to improve this situation.
Proteins obviously play critical roles in diverse biological processes through interactions with other proteins, DNA, RNA, and small molecules both inside and outside cells. Antibodies are extremely valuable for the detection and isolation of proteins in vivo, but generation of antibodies specific for a protein of interest is not always successful and can be costly. An alternative approach is to tag proteins of interest with epitopes that can be detected with commercially available antibodies (Jarvik and Telmer 1998; Brizzard 2008). Although this obviates the need to raise antibodies, epitope tags fused to either the C terminus or the N terminus may influence protein folding, mask neighboring domains or signal sequences, inhibit protein processing and modification, and interfere with proper protein function (Booher and Kaiser 2008; Sung et al. 2008). Similarly, the use of multiple epitopes can increase sensitivity, but bulky epitopes can also interfere with protein function. A flexible polypeptide linker, such as a 10-glycine chain (flexible due to its absence of side groups) placed between an epitope and a tagged protein, can minimize such problems and improve the accessibility of an epitope to antibodies (Borjigin and Nathans 1994; Sabourin et al. 2007). We and others previously constructed first-generation expression vectors for Neurospora with epitope tags such as FLAG, hemagglutinin (HA), Myc, green fluorescent protein (GFP), and red fluorescent protein, but their cloning sites and other features were not always consistent (Freitag et al. 2004b; Freitag and Selker 2005; He et al. 2005; Dementhon et al. 2006; Kawabata and Inoue 2007). Thus it was laborious to generate different tagged constructs, as is generally necessary to obtain fully functional and detectable epitope-tagged proteins because of the peculiarities of specific epitopes placed at C or N termini (Booher and Kaiser 2008; Sung et al. 2008). Here we report standardized, advanced multi-purpose vectors that simplify simultaneous construction of a family of epitope-tagged expression constructs.
We also developed materials and methods to identify protein complexes. Identification of proteins interacting with a given target protein is an excellent initial strategy for characterizing protein complexes (Pandey and Mann 2000; Gavin et al. 2002; Cravatt et al. 2007). Yeast two-hybrid assays have been widely used, including in Neurospora (Li and Borkovich 2006; Honda and Selker 2008), for identification of interacting proteins. The utility of this approach is limited, however, because it is time-consuming, is normally limited to detection of pairwise interactions, and does not generally allow for post-translational modifications that may create binding sites for partner proteins. In the past decade, mass spectrometry (MS) has become the method of choice for characterization of protein complexes (Aebersold and Mann 2003). Typically, epitope-tagged proteins are affinity-purified from cellular extracts, and associated proteins are then identified by MS (Chang 2006). A feature of this method is that it can identify all the components associated, directly or indirectly, with epitope-tagged proteins of interest under natural conditions. To reduce background from nonspecific binding proteins, it is common to employ a two-step purification using a tandem affinity purification (TAP) tag, which consists of two different high-affinity tags (Rigaut et al. 1999). Each TAP tag has advantages and disadvantages (Chang 2006). A Myc-6xHis TAP tag purification system has been successfully used in Neurospora (He et al. 2002, 2005), but the FLAG-6xHis system is more economical than any other available affinity purification systems and has been demonstrated to work well in mammals and Drosophila (Yang et al. 2006; Ooi et al. 2007). Unfortunately, the high charge of the 6xHis tag can influence folding of 6xHis-tagged proteins. A natural 19-amino-acid poly-histidine affinity tag (HAT) derived from chicken lactate dehydrogenase is preferable to the 6xHis epitope because the overall charge of the HAT epitope is lower (Chaga et al. 1999).
After identifying potential interacting proteins, researchers can use different tagged epitopes to verify their interaction by co-immunoprecipitation (Co-IP) from extracts. In Neurospora, it is common to target epitope-tagged genes to the his-3 locus by replacing a his-3 mutation with wild-type sequences (Margolin et al. 1997; Lee et al. 2003), while in yeast the high efficiency of homologous recombination makes it easy to express epitope-tagged proteins from their native loci (Longtine et al. 1998), facilitating coexpression of multiple constructs. Most eukaryotes, including Neurospora, have a strong nonhomologous end-joining (NHEJ) system that results in ectopic insertions of exogenous DNA, and this complicates strategies based on homologous recombination. This problem has recently been overcome in Neurospora, however, by utilization of mutants (e.g., mus-51, mus-52, or mus-53) that are defective in NHEJ and therefore are only capable of homologous recombination (Ninomiya et al. 2004; Ishibashi et al. 2006). The current Neurospora gene knock-out project uses Saccharomyces cerevisiae for assembly of constructs, taking advantage of the fact that yeast needs only 30–40 nucleotides of sequence homology for recombination (Oldenburg et al. 1997). Following their example (Colot et al. 2006), we used this approach to generate knock-in constructs for Neurospora, incorporating features of the Cre/loxP site-specific recombination system (Yu and Bradley 2001). The bacteriophage Cre recombinase specifically recognizes two loxP sites and leads to reciprocal recombination between them, resulting in excision, inversion, and translocation (Van Duyne 2001). This system functions in various organisms, including some fungi (Krappmann et al. 2005; Forment et al. 2006), and has been exploited for various purposes, including excision of selectable markers that are no longer required or desirable.
Here we report the development of convenient entry and expression vectors to build N- or C-terminal fusion proteins with FLAG, HA, Myc, GFP, or HAT–FLAG TAP tags and poly-glycine flexible linkers for expression at native loci or at his-3. To demonstrate the utility of these tools, we purified HETEROCHROMATIN PROTEIN 1 (HP1)-associated proteins using the HAT–FLAG purification system and identified DIM-2 among the interacting proteins by MS analysis. The DIM-2–HP1 interaction was then verified in vivo by Co-IP using the epitope-tagging system. We also demonstrate that the Cre/loxP system works efficiently in Neurospora and allows for “recycling” of the popular hygromycin-resistance selectable marker, hph. Key plasmids constructed in this study will be made available through the Fungal Genetics Stock Center.
MATERIALS AND METHODS
Neurospora strains and isolation of genomic DNA:
N. crassa strains used in this study are listed in supporting information, Table S1 and were grown, maintained, and crossed according to standard published procedures (Davis 2000). For isolation of genomic DNA, Neurospora strains were grown with shaking in Vogel's minimal medium with 1.5% sucrose and required supplements at 32° for 2 days; genomic DNA was isolated and used for PCR and Southern hybridizations as described previously (Freitag et al. 2004a).
Construction of yeast shuttle vector containing a 10xGly linker:
All primers and synthetic oligonucleotides used in this study are listed in Table S2. Synthetic oligonucleotides #1790 and #1791, which contain a 10xGly linker sequence, an EcoRI site on one end, and a PacI site followed by a HindIII site on the other end, were incubated at room temperature for 30 min after boiling for 5 min. The annealed oligonucleotides were digested with EcoRI and HindIII and inserted into EcoRI + HindIII-digested pRS416 yeast shuttle vector (NEB), yielding pRS416-10xGly.
Construction of his-3-targeting vectors containing C-terminal 3xFLAG or HAT–FLAG tags:
We amplified a fragment containing the 3xFLAG sequence by PCR using the p3xFLAG CMV14 vector (Sigma) as the template with forward primer #2014 containing a PacI site and reverse primer #2015 containing an EcoRI site. The PCR products were digested with PacI and EcoRI and inserted into PacI + EcoRI-digested pMF270, which is a his-3 targeting vector containing a C-terminal 3xHA-epitope tag. In this way, we replaced the 3xHA tag with the 3xFLAG tag, yielding pCCG∷C-3xFLAG. In some cases, fewer than 10 glycines were included in the poly-glycine linker to render constructions practical. To construct a vector containing a HAT∷5xGly∷FLAG tag, a fragment containing a HAT tag was amplified by PCR from the vector pHAT10 (Clontech) with forward primer #2036, which contains a PvuI site, and reverse primer #2037, which contains a 5xGly linker and a PacI site followed by a XhoI site. The PCR fragments were digested with XhoI and PvuI and inserted into pRS416-10xGly digested with XhoI and PacI, which produce ends compatible with PvuI, yielding pRS416-10xGly-HAT-5xGly. A fragment containing a HAT∷5xGly∷FLAG tag was generated by PCR using pRS416-10xGly-HAT-5xGly as template with forward primer #1939, which contains a 1xFLAG sequence followed by a PacI site, and reverse primer #1940, which contains an AscI site. The PCR products were digested with PacI and AscI and inserted into PacI + AscI-digested pMF270, yielding pCCG∷C-HAT∷FLAG.
Construction of his-3-targeting vectors containing a 10xGly linker followed by C-terminal 3xFLAG, GFP, or HAT–FLAG tags:
We amplified a fragment containing the 3xFLAG tag by PCR using the p3xFLAG CMV14 vector (Sigma) as the template with forward primer #2084, which contains a 10xGly linker plus a PacI site, and reverse primer #2015, which contains an EcoRI site. The PCR products were digested with PacI and EcoRI and inserted into PacI + EcoRI-digested pMF270 to replace the 3xHA tag with the 10xGly∷3xFLAG tag, yielding pCCG∷C-Gly∷3xFLAG. pCCG∷C-Gly∷GFP, which is a his-3 targeting vector containing a 10xGly linker followed by a GFP tag at its C terminus, was similarly generated using forward primer #2085, reverse primer #2086, and pMF272 (Freitag et al. 2004b) as template. A fragment containing a 10xGly∷HAT∷5xGly∷FLAG tag was amplified by PCR with forward primer #1938 containing a PacI site, reverse primer #1940 containing an AscI site, and pRS416-10xGly-HAT-5xGly as the template. The PCR products were digested with PacI and AscI and inserted into PacI + AscI-digested pMF270, yielding pCCG∷C-Gly∷HAT∷FLAG.
Construction of his-3-targeting vectors containing N-terminal 3xFLAG, GFP, 3xMyc, 3xHA, or FLAG–HAT tags followed by a poly-glycine linker:
We amplified a fragment containing the Neurospora ccg-1 promoter (Pccg-1) (McNally and Free 1988) by PCR from pMF270 with forward primer #1954, which contains an EcoRI site, and reverse primer #1953, which contains a BamHI site. The PCR products were digested with EcoRI and BamHI and inserted into pBM61 digested with the corresponding restriction enzymes (Margolin et al. 1997), yielding pCCG-N. To obtain a fragment containing the FLAG tag followed by a HAT tag, we amplified the HAT tag sequence by PCR from the pCCG∷C-HAT∷FLAG vector with forward primer #1967, which contains a BamHI site followed by a start codon (ATG), a single FLAG sequence, and a 3xGly linker, and reverse primer #1946, which contains a 5xGly tail sequence followed by AscI, PacI, and SpeI sites. The PCR products were digested with BamHI and SpeI and inserted into BamHI + SpeI-digested pCCG-N, yielding pCCG∷N-FLAG∷HAT. A fragment containing the 3xFLAG tag was amplified by PCR from pCCG∷C-FLAG with forward primer #2341, which contains a BamHI site followed by a start codon (ATG), and reverse primer #2342, which contains an 8xGly linker followed by an AscI site. The PCR products were digested with BamHI and AscI and inserted into pCCG∷N-FLAG∷HAT digested with the corresponding restriction enzymes to replace its FLAG∷HAT tag with the 3xFLAG tag, yielding pCCG∷N-3xFLAG. pCCG∷N-GFP and pCCG∷N-3xMyc, which are his-3 targeting vectors with N-terminal GFP or 3xMyc tags followed by 8xGly linkers, were similarly generated using forward primer #2343, reverse primer #2344, and pMF272 as template or forward primer #2345, reverse primer #2346, and pMF276 as template. To generate pN-3xHA, a fragment containing a 3xHA tag was amplified by PCR from pMF270 with forward primer #2347, which contains a BglII site, and reverse primer #2348 and then introduced into SmaI + SpeI-digested pBM61.
Construction of vectors to target dim-2-3xFLAG or hpo-gfp to the his-3 locus:
A 500-bp segment of dim-2, including the promoter region, was amplified by PCR with forward primer #1970, which contains a NotI site and the reverse primer #2087, which contains an XbaI site. The PCR product and the vector pCCG∷C-3xFLAG were digested with NotI and XbaI and ligated to replace the Pccg-1 with the dim-2 promoter. The plasmid containing the dim-2 promoter was then digested with XbaI and PacI and ligated with a fragment containing the coding region of the dim-2 gene, which had been amplified by PCR from Neurospora genomic DNA with forward primer #1517, which contains an XbaI site, reverse primer #1518, which contains a PacI site, and digested with XbaI and PacI. For the HP1 construct, a 500-bp promoter region of hpo was amplified by PCR with primers #2066 and #2067. The coding region of hpo was cut out from pMF308 (Freitag et al. 2004a) by digestion with BamHI and XbaI. The two fragments were inserted into pCCG∷C-GFP. The constructs for GFP-tagged HP1 and FLAG-tagged DIM-2 were linearized and inserted at the his-3 locus of the hpo strain N3395 and the dim-2 null mutant N3396, respectively, by the gene replacement method previously described (Margolin et al. 1997).
Construction of knock-in vectors containing the 10xGly linker followed by a 3xFLAG, 3xHA, GFP, or 13xMyc tag at the C terminus and the hph gene flanked by loxP sequences:
We amplified a fragment containing the bacterial hph gene, which confers hygromycin resistance (hygR) in Neurospora, from pCSN44 (Staben et al. 1989) by PCR with forward primer #1977, which contains a SpeI site, and reverse primer #1976, which contains a loxP sequence followed by a BamHI site. The PCR products were digested with SpeI and BamHI and inserted into SpeI + BamHI-digested vector pZErO-2 (Clontech), yielding pZErO-hph∷loxP. To insert epitope tags and a 10xGly linker for construction of knock-in modules, we first generated constructs containing the hpo gene with a 10xGly linker followed by epitope tags and then used them as templates for PCR. A fragment of the hpo coding region with its promoter was amplified by PCR from genomic DNA of Neurospora N623 with forward primer #2040, which contains a BamHI site, and reverse primer #2041, which contains a MfeI site. The PCR products were digested with BamHI and MfeI and inserted into pRS416-10xGly between the BamHI and EcoRI sites (ends compatible with those generated by MfeI), yielding pRS416-HP1∷10xGly. A fragment containing the hpo gene with a 10xGly tail was cut out from pRS416-HP1∷10xGly by digestion of NotI and PacI and inserted into pCCG∷C-3xFLAG, pMF270, pMF272, or pMF276 between the NotI and PacI sites, yielding pHP1∷10xGly∷3xFLAG, pHP1∷10xGly∷3xHA, pHP1∷10xGly∷GFP, or pHP1∷10xGly∷13xMyc, respectively. Finally, we used forward primer #1979, containing a XhoI site, and reverse primer #1978, which contains the loxP sequence followed by an XbaI site, to amplify the 10xGly and epitope tags from pHP1∷10xGly∷3xFLAG, pHP1∷10xGly∷3xHA, pHP1∷10xGly∷GFP, or pHP1∷10xGly∷13xMyc. The PCR products then were digested with XhoI and XbaI and inserted into pZErO-hph∷loxP between the XhoI and SpeI sites (compatible overhang with XbaI), yielding p3xFLAG∷hph∷loxP, p3xHA∷hph∷loxP, pGFP∷hph∷loxP, or p13xMyc∷hph∷loxP, respectively.
Engineering of epitope-tagged proteins for expression of genes at their native loci:
We modified a gene knock-out procedure (Colot et al. 2006) to serve as a convenient knock-in strategy for Neurospora genes. We used recombination-mediated plasmid construction in S. cerevisiae (Oldenburg et al. 1997) to make knock-in cassettes. The system worked well: 58 of 66 transformants (87.9%) from 12 different constructs were found to have generated the intended cassettes. Yeast strains were grown and maintained according to the Yeast Protocol Handbook (Clontech), and the S. cerevisiae strain PJ69-4A was used as the host strain (James et al. 1996). For the DIM-2-FLAG knock-in cassette, pRS416 was linearized by digestion with BamHI and EcoRI, and a 3xFLAG knock-in module (3xFLAG∷loxP∷hph∷loxP) was isolated from p3xFLAG∷hph∷loxP by digestion with KpnI and XhoI. A 1-kb fragment including the 3′-end of the dim-2 coding region, without the stop codon, was amplified by PCR with forward primer #1988, which contains 29 nt of homology at one end of the linearized pRS416, and reverse primer #2013, which contains 29 nt of homology at the 5′-end of the 3xFLAG knock-in module. A 500-bp fragment of the 3′ dim-2 flanking region was amplified with forward primer #1989, which contains 29 nt of homology with the 3′-end of the 3xFLAG knock-in module, and reverse primer #1990, which contains 29 nt of homology at the other end of the linearized pRS416. The yeast strain PJ64-4A was cotransformed with linearized pRS416, the 3xFLAG knock-in module, and the two PCR products for assembly in yeast by using its endogenous homologous recombination system (Oldenburg et al. 1997). Plasmids were extracted from pooled transformants and introduced into Escherichia coli DH5α cells by electroporation. The correct cassette was cut out by digestion with XhoI and XbaI and transformed into the Δmus-52 Neurospora strain (N2930) by electroporation. The subsequent steps were performed as described (Colot et al. 2006). The HP1–GFP knock-in cassette was similarly generated using primers: #1996, #1997, #1998, and #1999.
Construction of a strain with cre recombinase driven by the Pccg-1 at the his-3 locus:
The cre recombinase gene of bacteriophage P1 was amplified by PCR from plasmid HZ-73 (gift of H. Zong) with forward primer #2459, which contains an XbaI site, and reverse primer #2460, which contains an EcoRI site. The product was then digested with XbaI and EcoRI and inserted into pCCG∷C-3xFLAG between the XbaI and EcoRI sites, yielding pCCG∷Cre. This plasmid was then linearized by digestion with DraI and inserted at the his-3 locus of a strain (N3322) containing an hpo-gfp∷loxP∷hph+∷loxP cassette by our gene replacement method (Margolin et al. 1997). Ten transformants were confirmed to have correct integration events by Southern blotting and were used for further analysis.
Visualization of HP1–GFP by fluorescence microscopy:
A suspension of conidia was spotted on a slide glass and immediately covered by a glass coverslip. Pictures were taken using an Axioplan2 fluorescence microscope and an AxioCam HRm digital camera (Carl Zeiss, Thornwood, NY).
Construction of a strain containing hpo-HAT-FLAG at the his-3 locus:
A fragment containing the hpo gene with a 10xGly tail was cut out from pRS416-HP1∷10xGly by digestion with NotI and PacI and inserted into pCCG∷C-HAT∷FLAG between the NotI and PacI sites, yielding pHP1∷HAT∷FLAG. The HP1–HAT–FLAG construct was linearized by digestion with SapI and NheI and inserted at the his-3 locus of hpo strain N2556 by gene replacement. Correct integration and proper DNA methylation were confirmed for the transformants by Southern blotting, and a homokaryotic strain (N3278) was selected for subsequent study.
Purification of HP1 complex(es) using a HAT–FLAG tag:
Neurospora strain N3278 was grown at 32° for 7 days in two 250-ml flasks, each containing 50 ml of Vogel's medium with 1.5% sucrose and 1.5% agar. The conidia were suspended in 100 ml of fresh medium and filtered through cheesecloth. The suspended conidia were inoculated into two 2.8-liter flasks, each containing 1000 ml of medium and grown for 20 hr at 32° with shaking at 150 rpm. Cells were harvested by filtration and washed twice with 500 ml water. The collected tissue (∼50 g) was pressed between paper towels to remove extra water and then quickly frozen in liquid nitrogen. The frozen tissue was ground with a mortar and pestle to a fine powder and suspended in 500 ml of ice-cold nuclear extraction buffer without EDTA [15 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 20 μm ZnCl2, 5% glycerol, 1 mm PMSF (Sigma), 1 μg/ml leupeptin (Roche), 1 μg/ml pepstatin (Roche), and 1 μg/ml E-64 (Roche)] and centrifuged at 10,000 × g for 20 min at 4°. One milliliter of 10% NP 40 (final concentration 0.02%) and 15 ml of elution buffer (20 mm Tris–HCl, pH 7.4, 150 mm NaCl, 0.01% NP40, and 150 mm imidazole) was added to the supernatant to give a final concentration of 4.5 mm of imidazole, and it was then centrifuged again at 10,000 × g for 20 min at 4°. The supernatant was transferred carefully into two 250-ml centrifuge bottles and incubated with a 5-ml volume TALON Metal Affinity Resin (Clonetech) with rotation at 4°. The resin was loaded into a column and washed in 200 ml of wash buffer (20 mm Tris–HCl, pH 7.4, 150 mm NaCl, 0.01% NP40, 6 mm imidazole; ∼2 ml/min). The HAT-tagged HP1 protein and associated proteins were eluted in 9 ml elution buffer five times and collected in a 50-ml tube containing 100 μl of 0.5 m EDTA. Proteinase inhibitors (final concentrations: 1 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 μg/ml E-64) and 0.2 ml of 10% NP40 (final concentration: 0.05% NP40) were then added to the eluate. For the second purification, based on the FLAG tag, the eluate was incubated overnight at 4° with 500 μl anti-FLAG M2 affinity gel (A2220, Sigma) with rotation on a 3D-rotator. Immune complexes were collected by centrifugation and washed three times, with rotation at 10-min intervals, in 50 ml of ice-cold TBSE (20 mm Tris–HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 0.05% NP40) containing 1 mm PMSF. The affinity gel was transferred to a 1.5-ml centrifuge tube and incubated in 500 μl of ice-cold TBSE containing 0.5 mg/ml FLAG peptide (H2N-DYKDDDDK-OH, custom order, New England Peptide), 1 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 μg/ml E-64 for 6 hr with rotation at 4°. The gel was sedimented by centrifugation, and the supernatant was transferred into a fresh 1.5-ml centrifuge tube. The gel was briefly washed in 500 μl of ice-cold TBSE and spun down, and the supernatant was combined with the first supernatant. The purified complexes were concentrated with a Microcon YM-10 (Millipore), separated by SDS–PAGE and visualized by Coomassie Blue. Each stained band was cut out and subjected to mass spectrometry.
Co-IP assay:
N. crassa strains were grown at 32° with shaking in 20 × 150-mm glass test tubes containing 7 ml Vogel's minimal medium with 1.5% sucrose and histidine. After 16 hr, tissue was harvested by filtration, washed in PBS [10 mm phosphate, pH 7.4, 137 mm NaCl, and 2.7 mm KCl] and suspended in 1 ml of ice-cold lysis buffer [50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 0.02% NP40, 1 mm EDTA, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 mm PMSF]. Extracts were sonicated (Branson Sonifier 450) three times at 10-min intervals for 20 sec with a duty cycle 80 and output set to 2. After centrifugation at 12,000 rpm for 10 min in a microfuge, aliquots of the supernatants were incubated with 10 μl of anti-FLAG M2 affinity gel. Immune complexes were washed twice in lysis buffer and suspended in SDS-sample buffer. Samples were separated by SDS–PAGE and transferred to PVDF membranes in 48 mm Tris, 39 mm glycine (pH 9.2) containing 20% methanol at 150 mA for 2 hr. Membranes were blocked in 10 mm Tris, pH 7.5, 150 mm NaCl, and 0.05% Tween 20 (TBS-Tween) containing 3% skim milk powder for 30 min and incubated with anti-FLAG M2 antibody in TBS-Tween containing 3% skim milk and anti-GFP antibody (ab290, Abcam) in TBS-Tween containing 3% BSA for 2 hr at room temperature. The tagged proteins were detected using horseradish-peroxidase-conjugated secondary antibodies and SuperSignal West Femto chemiluminescent substrate (Pierce) as instructed by the manufacturer.
RESULTS
Construction of an entry vector with a flexible linker and new vectors for C-terminal epitope tagging:
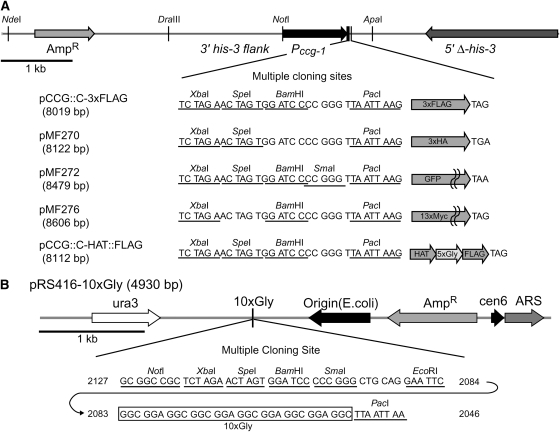
We previously reported the construction of a GFP expression vector (pMF272; Figure 1A) and demonstrated its utility in Neurospora (Freitag et al. 2004a,b). This his-3 targeting vector was built to express GFP under the control of the Neurospora ccg-1 promoter (Pccg-1), which is intensely induced by glucose deprivation or stress (McNally and Free 1988). Similar vectors were generated for different epitopes: 3xHA tag (pMF270), 13xMyc tag (pMF276), 3xFLAG tag (pCCG∷C-3xFLAG), and a HAT–FLAG tandem tag with a 5xGly linker (pCCG∷C-HAT∷FLAG), as illustrated in Figure 1A. All vectors contain the same multiple cloning sites (MCS) followed by epitope tag sequences and an in-frame stop codon; thus, a gene of interest amplified with a primer pair can be introduced simultaneously into several of these expression vectors. The Pccg-1 is particularly useful for visualization of GFP fusion protein in Neurospora because of its strength (Freitag et al. 2004b), and this promoter is helpful in detecting proteins that are nondetectable when expressed under the control of their endogenous promoters. Overexpression can cause artifactual effects, however (Rigaut et al. 1999). We therefore constructed the plasmids so that Pccg-1 can be replaced with a gene of interest with its endogenous promoter using a NotI site and the MCS, which flank the Pccg-1.
Figure 1.—
Entry and first-generation expression vectors. (A) Partial maps of the expression vectors pCCG∷C-3xFLAG (GenBank accession no. FJ456996), pMF270 (3xHA) (FJ456997), pMF272 (GFP) (AY598428) (Freitag et al. 2004b), pMF276 (13xMyc) (FJ456998), and pCCG∷C-HAT∷FLAG (FJ456999) and sequences of their MCSs. Unique restriction sites are underlined and indicated by vertical bars. This set of vectors contain the N. crassa ccg-1 (Pccg-1) promoter (McNally and Free 1988) and a MCS followed by 3xFLAG, 3xHA, GFP, 13xMyc, or 1xFLAG∷5xGly∷HAT. A gene of interest can be inserted into MCS or inserted with its endogenous promoter between the NotI site and the MCS, replacing Pccg-1. The flanking his-3 sequences allow the construct to be targeted to the N. crassa his-3 locus (Margolin et al. 1997). (B) Map and sequence of the MCS of entry vector pRS416-10xGly (FJ457000). The box marks the 10xGly flexible linker. Genes of interest can be inserted into the MCS using restriction enzymes or by homologous recombination in yeast (Oldenburg et al. 1997). The 8-base cutter PacI site was included to transfer the gene with the poly-glycine linker to epitope tag expression vectors.
We previously noted that C-terminal GFP fusions with Neurospora genes are not always successful. It is not uncommon to find undetectable expression of the GFP fusion proteins even though they were overexpressed by the Pccg-1 (Freitag et al. 2004b). One of a number of reasons for this is that a GFP tag can influence folding of the tagged protein, resulting in instability of the GFP fusion proteins. Flexible protein linkers can be used to improve the folding and function of epitope-tagged proteins (Sabourin et al. 2007). We therefore constructed an entry vector containing a flexible poly-glycine linker (10xGly). We inserted a flexible poly-glycine linker into the yeast shuttle vector pRS416 to produce pRS416-10xGly (Figure 1B). After subcloning into the entry vector, a gene of interest with the poly-glycine linker can be easily transferred into a series of our constructed epitope-tagging expression vectors, which contain the same MCS and PacI site. Genes can be introduced into pRS416-10xGly by a conventional subcloning system by using restriction enzyme sites in the MCS or by using a yeast homologous recombination system (Oldenburg et al. 1997).
New vectors for N- and C-terminal epitope tagging with flexible linkers:
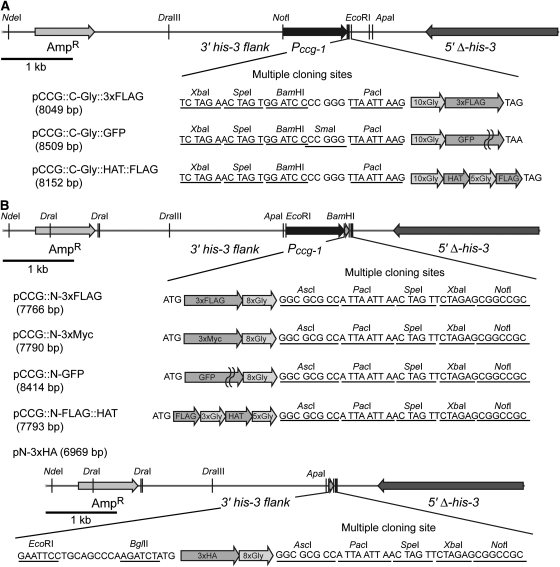
The entry vector system described above allows one to take advantage of previously generated vectors such as pMF272 but requires two cloning steps. To speed building constructs for expressing epitope-tagged proteins with a flexible poly-glycine linker, we generated new vectors containing a poly-glycine linker and the alternative epitopes 3xFLAG (pCCG∷C-Gly∷3xFLAG) and GFP (pCCG∷C-Gly∷GFP) or a HAT–FLAG combination separated by a flexible linker (pCCG∷C-Gly∷HAT∷FLAG) (Figure 2A). These flexible linker vectors sport the features of the first-generation vectors (Figures 1A and 2A), allowing the genes inserted into the previously generated vectors to be directly transferred into the new expression vectors.
Figure 2.—
Advanced expression vectors with N- or C-terminal epitope tags and a poly-glycine flexible linker. (A) Partial maps of C-terminal epitope tag expression vectors pCCG∷C-Gly∷3xFLAG (FJ457001), pCCG∷C-Gly∷GFP (FJ457002), and pCCG∷C-Gly∷HAT∷FLAG (FJ457003) and sequences of their MCSs. Unique restriction sites are underlined and indicated by vertical bars. All vectors contain the N. crassa ccg-1 promoter (Pccg-1) (McNally and Free 1988), a MCS, and a poly-glycine linker in the same frame and direction. (B) Partial maps of the N-terminal epitope-tag expression vectors pCCG∷N-3xFLAG (FJ457004), pCCG∷N-3xMyc (FJ457005), pCCG∷N-GFP (FJ457006), pCCG∷N-FLAG∷HAT (FJ457007), and pN-3xHA (FJ457008) and sequences of their MCS. Unique restriction sites are underlined and indicated by vertical bars. All vectors except pN-3xHA contain Pccg-1, and pN-3xHA has ApaI, EcoRI, and BglII (ends compatible with those generated by BamHI) sites for introduction of the Pccg-1 or the promoter of a gene of interest. All vectors contain a poly-glycine linker and a MCS in the same frame and direction.
In some cases, C-terminal epitope tags interfere with protein function, e.g., by interfering with targeting sites or post-translational modification adjacent to the C terminus, even when a flexible linker is included (Booher and Kaiser 2008). To deal with such situations, we generated a series of new expression vectors for N-terminal epitope tagging with 3xFLAG (pCCG∷N-3xFLAG), 3xMyc (pCCG∷N-3xMyc), GFP (pCCG∷N-GFP), and 3xHA (pN-3xHA) followed by an eight-Gly chain. We also built an N-terminal TAP tag vector: 1xFLAG∷3xGly∷HAT∷5xGly (pCCG∷N-FLAG∷HAT) (Figure 2B). These plasmids are all suitable for targeting to his-3 and all except pN-3xHA contain Pccg-1. This promoter can be replaced with the native promoter of a gene of interest by using an EcoRI or ApaI site and the BamHI site. In the case of pN-3xHA, Pccg-1 or a native promoter can be inserted between the EcoRI or ApaI site and the BglII site (which generates ends compatible with those of BamHI; the 3xHA sequence in pN-3xHA contains a BamHI site). All the vectors contain an identical MCS in the same frame so that a gene of interest can be easily inserted into several of these expression vectors at same time.
Expression of epitope-tagged proteins from genes at their native chromosomal location:
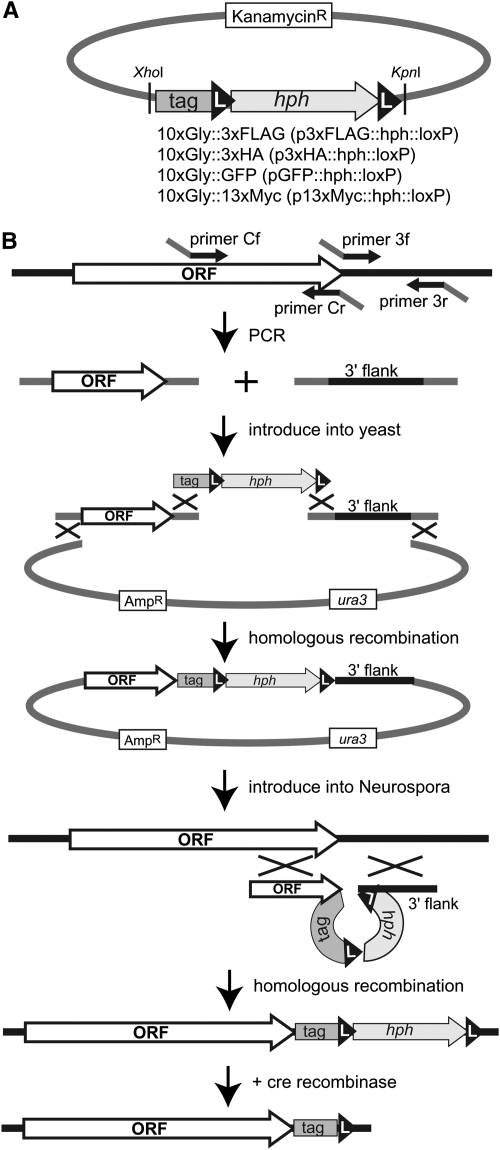
The described vectors possess a common backbone suitable for targeting to Neurospora his-3, similar to the commonly used vector pBM61 and its derivatives (Margolin et al. 1997). We also developed expression vectors for gene replacement by taking advantage of mutants deficient for NHEJ (Δmus-51 or Δmus-52) (Ninomiya et al. 2004). Utilizing the yeast homologous recombination system (Oldenburg et al. 1997), we first generated a knock-in module containing an epitope tag with a flexible poly-glycine linker followed by the selectable marker hph, flanked by loxP sites (Figure 3A). The idea was to utilize the Cre/loxP system to remove the hph gene after it was no longer needed, allowing the valuable marker to be “recycled.” Vectors with four epitope tags (3xFLAG, 3xHA, GFP, and 13xMyc) were made to allow the modules to be transferred from any of them (p3xFLAG∷hph∷loxP, p3xHA∷hph∷loxP, pGFP∷hph∷loxP, or p13xMyc∷hph∷loxP) by digestion with XhoI and KpnI. All the modules contain identical sequences of the 5′- and 3′-ends, allowing constructions of four epitope tag knock-in cassettes with common primers designed to work with the yeast homologous recombination system. For expression of bacteriophage Cre recombinase in Neurospora, we generated the his-3-targeting plasmid pCCG∷Cre, which allows cre to be expressed under the control of Pccg-1 at the his-3 locus. The scheme is illustrated in Figure 3 and described in detail in materials and methods. In brief, fragments immediately before and after the stop codon of a target gene are amplified with four primers containing sequences allowing for assembly in the yeast shuttle vector pRS416. The two PCR products, the linearized pRS416, and the isolated knock-in module are assembled in yeast using the endogenous recombination system. The circular knock-in cassettes are isolated from yeast, amplified, linearized, and introduced into Neurospora, selecting for growth in the presence of hygromycin. The hph marker, conferring hygR, can then be excised by the Cre recombinase, which is expressed from a construct targeted to the his-3 locus.
Figure 3.—
Strategy of creating knock-in constructs containing C-terminal epitope tags and allowing for excision of the selectable marker hph by use of the Cre/loxP system in Neurospora. The knock-in procedure was modified from a gene knock-out procedure described by Colot et al. (2006). (A) Partial map of p3xFLAG∷hph∷loxP (FJ457009), p3xHA∷ hph∷loxP (FJ457010), pGFP∷ hph∷loxP (FJ457011), and p13xMyc∷ hph∷loxP (FJ457012) plasmids. The epitope tag∷ hph∷loxP modules, which contain a 10xGly linker and an epitope tag followed by hph (conferring hygR) flanked by loxP (L) sites, is isolated by digestion with XhoI and KpnI. (B) To build custom knock-in constructs, the C-terminal end of the target gene, without the stop codon, and its 3′ flanking region are amplified by PCR from genomic DNA with primers Cf + Cr and 3f + 3r, respectively. Primers Cf and 3r contain 29 nt of identity to the yeast shuttle vector pRS416, and primers Cr and 3f contain 29 nt in common with each end of the knock-in module. The same sequences serve in all the modules so that the same primers (Cf, Cr, 3f, and 3r) work to make knock-in cassettes with any of the epitope tags. The two PCR products, the module, and the linearized pRS416 are cotransformed into yeast to assemble them by homologous recombination (Oldenburg et al. 1997). Circular plasmids containing the knock-in cassette are extracted from the transformants and amplified in E. coli. The knock-in cassette is then isolated and introduced into the Δmus-52 Neurospora strain. Knock-in strains are selected by growth on medium containing hygromycin. Correct integration of the knock-in cassette and expression of the tagged protein in the transformants are confirmed by Southern blotting and Western blotting, respectively. After removing the Δmus-52 marker by crossing, the hph gene flanked by loxP sequences is excised by introduction of linearized pCCG∷Cre (FJ457013), which contains the cre recombinase.
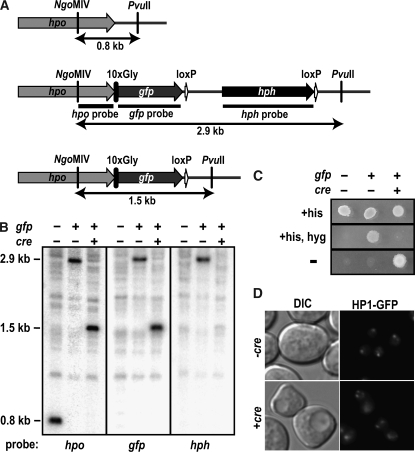
To demonstrate the described system, we prepared two knock-in cassettes, an hpo-gfp∷loxP∷hph∷loxP cassette and a dim-2-3xFLAG∷loxP∷hph∷loxP cassette, to generate GFP-tagged HP1 (HP1–GFP) and 3xFLAG-tagged DIM-2 (DIM-2-FLAG) at their endogenous loci. The two cassettes were separately introduced into Δmus-52 recipient strains and then crossed with mus-52+ strains to recover progeny with the wild-type allele. We confirmed correct integrations of the knock-in sequences by Southern blotting (Figure 4, A and B) and confirmed proper expression by Western blotting with antibodies specific for the respective epitope tags (see Figure 6). In addition, we confirmed that DNA methylation appeared normal in several methylated regions (data not shown), indicating that the tagged proteins are functional since both DIM-2 and HP1 are essential for DNA methylation in Neurospora (Kouzminova and Selker 2001; Freitag et al. 2004a). To excise hph, the linearized pCCG∷Cre was introduced into strains containing the hpo-gfp∷loxP∷hph∷loxP cassette. Ten transformants were isolated and showed correct integrations of cre by Southern blotting (data not shown). Two of these displayed complete and precise excision of the hph (Figure 4, A and B); three displayed a mixture of nuclei with and without precise excision; the remaining five retained the gene (data not shown). Consistent with these data, we found that the two strains showing complete excision restored sensitivity to hygromycin (Figure 4C); the others did not (data not shown). One of the two transformants sensitive to hygromycin appeared to be homokaryotic with respect to the insertion of cre; the other appeared to be heterokaryotic. The strains that were insensitive to hygromycin showed various integrations of cre and did not reveal an obvious relationship between the nature of the integration of cre and the successful excision of hph. Curiously, strains homokaryotic for insertion of cre displayed growth and conidiation defects regardless of whether they showed excision whereas the strain that appeared heterokaryotic for cre, but successfully excised hph, showed normal growth and development. Normal growth and conidiation were restored by removal of cre by crossing with cre− strains. Microscopic visualization of HP1–GFP in strains that excised hph shows normal localization of HP1–GFP (Figure 4D). These data indicate that the Cre/loxP system can work in Neurospora.
Figure 4.—
Application of the Cre/loxP system to excise the hph marker in Neurospora. (A) Schematic of the wild-type hpo locus and the hpo locus after integration of the knock-in cassette containing gfp followed by the hph marker flanked by loxP sites (HP1–GFP knock-in cassette) and after excision of the hph marker by introduction of the cre recombinase (see materials and methods). DNA fragments generated by digestion with NgoMIV and PvuII are indicated by arrows, and the regions used as probes for Southern blotting are indicated with horizontal bars. (B) Southern analysis showing integration of the HP1–GFP knock-in cassette at the hpo locus and excision of the hph marker with the Cre/loxP system. Genomic DNA isolated from strains with (+) or without (−) the gfp-tagged hpo and/or in the presence (+) or absence (−) of cre was digested with NgoMIV and PvuII and used for Southern hybridizations with the indicated probes. (C) Sensitivity to hygromycin in strains containing the hpo-gfp knock-in cassette is restored by excision of hph using the Cre/loxP system. Conidia suspensions of strains with (+) or without (−) the gfp-tag construct at hpo and/or in the presence (+) or absence (−) of cre were plated on minimal medium (−) or medium supplemented with histidine (+his) or hygromycin (+hyg) and grown at 32° overnight. (D) Localization of HP1–GFP is not affected by excision of hph using the Cre/loxP system. Conidia of an hpo-gfp∷loxP∷hph∷loxP; his-3− strain (−cre, N3322) and an hpo-gfp∷loxP; his-3+∷Pccg-1∷cre (+cre, N3684) strain were examined by differential interference contrast (DIC) and fluorescence (HP1–GFP) microscopy.
Figure 6.—
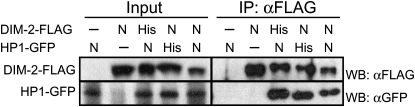
Co-IP assays of epitope-tagged DIM-2 and HP1 expressed at his-3 or at native loci confirm their interaction. Extracts from strains with or without (−) the FLAG-tagged dim-2 gene and/or the GFP-tagged hpo gene at the his-3 (His) locus or its native loci (N) were immunoprecipitated with anti-FLAG antibodies. Input and immunoprecipitation (IP) samples were fractionated, transferred to PVDF membranes, and immunoblotted with anti-FLAG antibodies or anti-GFP antibodies, as indicated. The tested strains, in order, were N3322, N3323, N3415, N3418, and N3436.
Isolation of HP1-associated proteins using the HAT–FLAG purification system:
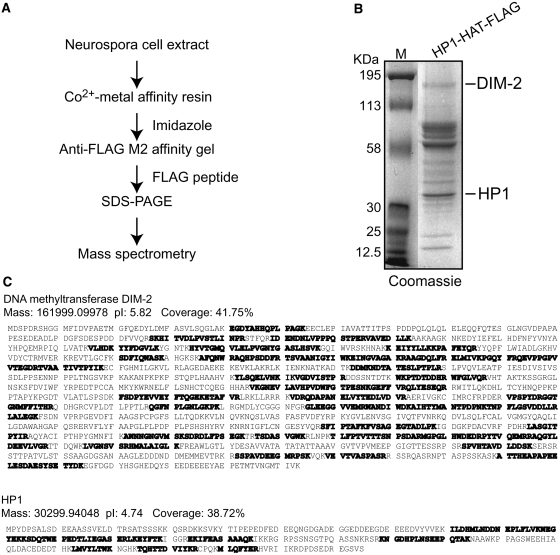
We wanted to develop an efficient purification method to identify proteins associated with a protein of interest in Neurospora. Tandem affinity tags have been demonstrated to work efficiently in various organisms (Chang 2006). Among various possible TAP tags, the FLAG-6xHis system is attractive due to its low cost and high efficiency. Purification using the natural HAT tag was shown to be superior to that using the simple 6xHis tag, however (Chaga et al. 1999), prompting us to build our HAT–FLAG, his-3-targeting vector system (Figures 1A and 2). To test the utility of our HAT–FLAG purification system, we decided to purify tagged HP1 and associated proteins because our previous work using a yeast two-hybrid system had demonstrated that HP1 interacts directly with DIM-2 (Honda and Selker 2008). The HAT–FLAG-tagged hpo gene was inserted at the his-3 locus under the control of its endogenous promoter in a hpo null mutant. We confirmed correct integration by Southern blotting and selected a homokaryotic transformant for further experiments (data not shown). The DNA methylation defect of the hpo strain was found to be completely complemented by the HP1–HAT–FLAG construct (data not shown), indicating that the fusion protein was fully functional. We then purified tagged HP1 and associated proteins as diagrammed in Figure 5A and detailed in materials and methods. Briefly, cell extracts prepared from strains expressing HP1–HAT–FLAG were applied to TALON affinity resin, and the HAT–FLAG-tagged HP1 and its associated proteins were eluted with imidazole and subsequently incubated with anti-FLAG M2 affinity gel. The purified material was then eluted in buffer containing the FLAG peptide, separated by SDS–PAGE, and visualized by Coomassie blue staining (Figure 5B). Each stained band was excised and subjected to mass spectrometry for protein identification. The strong band that corresponds to the predicted molecular weight of HP1 was confirmed to be this protein (Figure 5B); the multiple peptide coverage by mass spectrometry illustrates that the HAT–FLAG-tagged protein was efficiently isolated (Figure 5C). We also found several HP1-associated proteins, including DIM-2 (Figure 5, B and C), which strongly supports our previous conclusion that DIM-2 and HP1 interact directly (Honda and Selker 2008). This demonstrates the utility of our HAT–FLAG TAP system for Neurospora.
Figure 5.—
Purification of HP1-associtated proteins using the HAT–FLAG purification system. (A) Purification and characterization scheme. Cell extracts prepared from strains containing HP1–HAT–FLAG (N3278) were subject to a two-step affinity purification (Co2+ metal affinity purification followed by anti-FLAG affinity purification). (B) HP1 and its associated proteins isolated by the two-step affinity purification system were separated by SDS–PAGE and visualized by Coomassie blue staining. Molecular weights are indicated on the left. DIM-2 and HP1 were identified in excised gel fragments corresponding to bands indicated on the right and subjected to MS analysis. (C) Identification of DIM-2 and HP1 by liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis. The amino acid sequences, mass, and the isoelectric point (pI) of DIM-2 and HP1 are shown. Peptides detected by LC/MS/MS are indicated in boldface type, and the sequence coverage (percentage of protein sequence detected) is shown.
Verification of the interaction between HP1 and DIM-2 using our tagging, targeting, and knock-in systems:
After identifying candidate proteins associated with a TAP-tagged protein by affinity chromatography, confirmation of their association can be sought by carrying out Co-IP assays on extracts of cells expressing the proteins. To verify interaction between HP1 and DIM-2, we used our epitope-tagging system. We prepared strains expressing GFP-tagged HP1 (HP1–GFP) and/or 3xFLAG-tagged DIM-2 (DIM-2–FLAG) under the control of endogenous promoters, targeted either to his-3 or to their native loci. To guard against artifacts of overexpression, targeting to his-3 was carried out in strains with null mutations of the corresponding genes at their native loci. Western blotting showed that the tagged proteins were expressed equally from his-3 and the endogenous loci (Figure 6, left). All the strains showed normal distribution of DNA methylation, indicating that the tagged proteins were functional (data not shown). We found that HP1 specifically co-immunoprecipitated with DIM-2 in strains expressing both tagged proteins, regardless of whether they were expressed from their native loci or from sequences targeted to his-3 (Figure 6). This demonstrates the utility of our epitope tag expression vectors and of the knock-in system for Neurospora.
DISCUSSION
In this study we developed tools to epitope-tag proteins for expression from heterologous or native promoters in Neurospora targeted either to the his-3 locus or to native loci. To demonstrate the utility of our HAT–FLAG TAP system, we tagged HP1 and highly purified it and associated proteins for identification by mass spectroscopy. This identified the DNA methyltransferase DIM-2 as a partner of HP1, consistent with expectations from our previous yeast two-hybrid study (Honda and Selker 2008). We then used our tagging systems to confirm the interaction between HP1 and DIM-2 by Co-IP assays in strains expressing FLAG-tagged DIM-2 and GFP-tagged HP1 from his-3 and/or native loci under the control of the native promoters. Several other proteins also copurified with HP1, and preliminary work suggests that they, too, represent actual partners of HP1 (S. Honda, T. K. Khlafallah, Z. A. Lewis and E. U. Selker, unpublished data). In a similar study, we have used the HAT–FLAG system to identify proteins associated with the histone methyltransferase DIM-5 (S. Honda, K. K. Adhvaryu, Z. A. Lewis and E. U. Selker, unpublished data; Tamaru and Selker 2001; Tamaru et al. 2003). It is worth noting that the DIM-5-associated proteins identified in this way were not identified in a yeast two-hybrid screen using DIM-5 as bait and prey libraries (S. Honda and E. U. Selker, unpublished data). One possible reason for such a failure of the yeast two-hybrid screen is that the interactions may be indirect. Alternatively, interactions may rely on other associated proteins or on activation by post-translational modifications. Thus the HAT–FLAG tag system potentially allows one to simultaneously isolate all components of protein complexes in a simple, two-step affinity purification.
Although we constructed systems to tag both N and C termini of proteins, we typically first test C-terminal tags, expressed under the control of endogenous promoters, because this generally works and is easy, whereas N-terminal tagging requires additional subcloning. We have tested 13 proteins with C-terminal epitope tags. Eleven of the 13 were genetically functional, and their expression could be confirmed by Western blotting with antibodies specific for the epitope tags (S. Honda, K. K. Adhvaryu, T. K. Khlafallah, Z. A. Lewis and E. U. Selker, unpublished data). The two that were not fully functional when tagged at the C terminus were successfully expressed with an N-terminal epitope tag. In our experience, protein function is not generally hindered by epitope tags but differences occur. Overall, we found the following hierarchy of function: 3xFLAG > 3xHA > GFP > 13xMyc. Because tags can potentially interfere with nearby targeting and/or modification sites and can potentially interfere with protein folding, it is necessary to have a variety of the epitope-tagging systems for both N and C termini.
A variety of methods to characterize protein–protein interactions are useful for elucidation of protein functions. Recently, bimolecular fluorescence complementation was utilized in Neurospora to detect protein–protein interactions (Bardiya et al. 2008). It is based on the principle that the interaction of proteins that are separately fused with fragments of a GFP derivative, which do not individually emit fluorescence, can allow for reconstitution of a functional fluorophore if they are sufficiently close. Although the assay is simple and highly sensitive, in many cases it is expected not to work on the basis of our previous observation that approximately one-half of GFP fusions with Neurospora genes are undetectable even when they are overexpressed (Freitag et al. 2004b). In contrast, using the epitope-tagging systems described here, all our attempts to isolate functional and detectable epitope-tagged proteins expressed under the control of their native promoters have been successful. This system works well not only for protein purification and Co-IP assays but also for other purposes such as immunofluorescence and chromatin immunoprecipitation (ChIP) assays. Indeed, we previously demonstrated that HP1 and DIM-2 are localized to methylated regions by ChIP assay using the epitope-tagging system (Honda and Selker 2008). It is noteworthy that one feature of using antibodies against epitopes, instead of against native proteins, is that the tagged protein is more likely to remain functional and accessible while bound by the antibodies.
We demonstrated that epitope-tagged proteins were equivalently expressed from the his-3 locus and their native loci under the control of their native promoters. Both situations worked efficiently for Co-IP assays but each has advantages and disadvantages. Targeting to his-3 is normally limited to one gene per strain. In principle, a heterokaryon can be used to express two different tagged proteins from the his-3 locus but it can be difficult to maintain a balanced heterokaryon, and artifacts are possible (Kawabata and Inoue 2007). Thus we recommend simply crossing strains expressing epitope-tagged proteins from different loci to isolate homokaryotic double or triple knock-in strains. Site-directed mutagenesis is an invaluable technique for studying protein functions, and targeting to his-3 is convenient for introduction of mutant proteins with epitope tags. Indeed, we used this in work directed at defining critical sites for interaction of HP1 and DIM-2 (Honda and Selker 2008).
We demonstrated application of the Cre/loxP system in Neurospora to excise the selectable marker hph, which is the most popular antibiotic resistance marker for Neurospora and other eukaryotes. We observed a somewhat low frequency of complete marker excision (20%, or 2/10), similar to the case in Aspergillus fumigates, which showed a frequency of 25% (5/20) (Krappmann et al. 2005). In contrast, comparable marker rescue in Aspergillus nidulans and S. cerevisiae worked at higher efficiencies (70–80% and 80–90%, respectively) (Guldener et al. 1996; Krappmann et al. 2005). It may be significant that Neurospora has strong genomic defense systems (Galagan and Selker 2004), including DNA methylation, which interfere with transcriptional elongation (Rountree and Selker 1997); however, we did not detect DNA methylation in the integrated cre gene (data not shown). The efficiency difference may be attributable to differences in expression of Cre driven by different promoters. In the A. nidulans and S. cerevisiae systems, the chemically inducible promoters tightly regulate the expression of Cre, while, in the Neurospora and A. fumigates systems, expression of Cre was initiated by introduction of the cre gene by transformation. Use of tightly inducible promoters, such as the quinic acid-2 promoter (Pqa-2) (Giles et al. 1985), may improve the efficiency in Neurospora. Further improvements to this system could include application of dual selectable marker modules such as “tk-blaster,” which combines hph and the fluorodeoxyuridine (FUDR)-sensitive marker thymidine kinase (tk) (Sachs et al. 1997; Pratt and Aramayo 2002), allowing selection of strains that have excised the cassette. Future work may optimize the Cre/loxP system for Neurospora. The Cre/loxP system could potentially be used not only for marker excision but also for advanced engineering of chromosomal rearrangements, such as in building conditional knock-outs in Neurospora.
Characterization and identification of protein complexes is a basic step in deciphering gene functions. We have developed here a combination of genetic and biochemical approaches in Neurospora, and these techniques can be easily applied to other filamentous fungi. We have used these tools for analyzing the relationship between HP1 and DIM-2 to elucidate DNA methylation machinery in Neurospora. These tools will facilitate various analyses in clarifying the fundamental processes in Neurospora and other filamentous fungi. Indeed, our methods have already been successfully adapted to tag and express fusion proteins in Fusarium graminearum (L. Connolly and M. Freitag, unpublished results).
Acknowledgments
We thank Michael Freitag for the construction of pMF270 and pMF276 and Hui Zhong for the generous gift of pHZ76. We also thank Michael Freitag, Zachary Lewis, Keyur Adhvaryu, and Kirsty Jamieson for comments on the manuscript. We thank Ragna Sack of the Friedrich Miescher Insitute (Basel, Switzerland) for MS analysis of HP1-associated proteins. This work was supported by grant GM025690-22 to E.U.S. from the National Institutes of Health.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.098707/DC1.
References
- Aebersold, R., and M. Mann, 2003. Mass spectrometry-based proteomics. Nature 422 198–207. [DOI] [PubMed] [Google Scholar]
- Bardiya, N., W. G. Alexander, T. D. Perdue, E. G. Barry, R. L. Metzenberg et al., 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher, K. R., and P. Kaiser, 2008. A PCR-based strategy to generate yeast strains expressing endogenous levels of amino-terminal epitope-tagged proteins. Biotech. J. 3 524–529. [DOI] [PubMed] [Google Scholar]
- Borjigin, J., and J. Nathans, 1994. Insertional mutagenesis as a probe of rhodopsin's topography, stability, and activity. J. Biol. Chem. 269 14715–14722. [PubMed] [Google Scholar]
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner et al., 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzard, B., 2008. Epitope tagging. BioTechniques 44 693–695. [DOI] [PubMed] [Google Scholar]
- Chaga, G., D. E. Bochkariov, G. G. Jokhadze, J. Hopp and P. Nelson, 1999. Natural poly-histidine affinity tag for purification of recombinant proteins on cobalt(II)-carboxymethylaspartate crosslinked agarose. J. Chromatogr. A 864 247–256. [DOI] [PubMed] [Google Scholar]
- Chang, I. F., 2006. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 6 6158–6166. [DOI] [PubMed] [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt, B. F., G. M. Simon and J. R. Yates, III, 2007. The biological impact of mass-spectrometry-based proteomics. Nature 450 991–1000. [DOI] [PubMed] [Google Scholar]
- Davis, R. H., 2000. Neurospora: Contributions of a Model Organism. Oxford University Press, Oxford.
- Davis, R. H., and D. D. Perkins, 2002. Timeline: Neurospora: a model of model microbes. Nat. Rev. Genet. 3 397–403. [DOI] [PubMed] [Google Scholar]
- Dementhon, K., G. Iyer and N. L. Glass, 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot. Cell 5 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., K. A. Borkovich, M. R. Henn, G. E. Turner, M. S. Sachs et al., 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment, J. V., D. Ramon and A. P. MacCabe, 2006. Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr. Genet. 50 217–224. [DOI] [PubMed] [Google Scholar]
- Freitag, M., and E. U. Selker, 2005. Expression and visualization of red fluorescent protein (RFP) in Neurospora crassa. Fungal Genet. Newsl. 52 14–17. [Google Scholar]
- Freitag, M., P. C. Hickey, T. K. Khlafallah, N. D. Read and E. U. Selker, 2004. a HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13 427–434. [DOI] [PubMed] [Google Scholar]
- Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker and N. D. Read, 2004. b GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., and E. U. Selker, 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20 417–423. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch et al., 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415 141–147. [DOI] [PubMed] [Google Scholar]
- Giles, N. H., M. E. Case, J. Baum, R. Geever, L. Huiet et al., 1985. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol. Rev. 49 338–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., P. Cheng, Y. Yang, L. Wang, K. H. Gardner et al., 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297 840–843. [DOI] [PubMed] [Google Scholar]
- He, Q., P. Cheng and Y. Liu, 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19 1518–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, S., and E. U. Selker, 2008. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora. Mol. Cell. Biol. 28 6044–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, K., K. Suzuki, Y. Ando, C. Takakura and H. Inoue, 2006. Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc. Natl. Acad. Sci. USA 103 14871–14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik, J. W., and C. A. Telmer, 1998. Epitope tagging. Annu. Rev. Genet. 32 601–618. [DOI] [PubMed] [Google Scholar]
- Kasuga, T., and N. L. Glass, 2008. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot. Cell 7 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., J. P. Townsend, C. Tian, L. B. Gilbert, G. Mannhaupt et al., 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33 6469–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata, T., and H. Inoue, 2007. Detection of physical interactions by immunoprecipitation of FLAG- and HA-tagged proteins expressed at the his-3 locus in Neurospora crassa. Fungal Genet. Newsl. 54 5–8. [Google Scholar]
- Kouzminova, E., and E. U. Selker, 2001. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 20 4309–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann, S., O. Bayram and G. H. Braus, 2005. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot. Cell 4 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. W., J. R. Haag and R. Aramayo, 2003. Construction of strains for rapid homokaryon purification after integration of constructs at the histidine-3 (his-3) locus of Neurospora crassa. Curr. Genet. 43 17–23. [DOI] [PubMed] [Google Scholar]
- Lewis, Z. A., A. L. Shiver, N. Stiffler, M. R. Miller, E. A. Johnson et al., 2007. High-density detection of restriction-site-associated DNA markers for rapid mapping of mutated loci in Neurospora. Genetics 177 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and K. A. Borkovich, 2006. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot. Cell 5 1287–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Margolin, B. S., M. Freitag and E. U. Selker, 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44 34–36. [Google Scholar]
- McNally, M. T., and S. J. Free, 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14 545–551. [DOI] [PubMed] [Google Scholar]
- Ninomiya, Y., K. Suzuki, C. Ishii and H. Inoue, 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K. R., K. T. Vo, S. Michaelis and C. Paddon, 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi, S. K., C. Qiu, E. Bernstein, K. Li, D. Jia et al., 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A., and M. Mann, 2000. Proteomics to study genes and genomes. Nature 405 837–846. [DOI] [PubMed] [Google Scholar]
- Pratt, R. J., and R. Aramayo, 2002. Improving the efficiency of gene replacements in Neurospora crassa: a first step towards a large-scale functional genomics project. Fungal Genet. Biol. 37 56–71. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann et al., 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rountree, M. R., and E. U. Selker, 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin, M., C. T. Tuzon, T. S. Fisher and V. A. Zakian, 2007. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast 24 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, M. S., E. U. Selker, B. Lin, C. J. Roberts, Z. Luo et al., 1997. Expression of herpes virus thymidine kinase in Neurospora crassa. Nucleic Acids Res. 25 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman et al., 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36 79–81. [Google Scholar]
- Sung, M. K., C. W. Ha and W. K. Huh, 2008. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast 25 301–311. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., and E. U. Selker, 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., X. Zhang, D. McMillen, P. B. Singh, J. Nakayama et al., 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34 75–79. [DOI] [PubMed] [Google Scholar]
- Tian, C., T. Kasuga, M. S. Sachs and N. L. Glass, 2007. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 6 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne, G. D., 2001. A structural view of Cre-loxP site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 30 87–104. [DOI] [PubMed] [Google Scholar]
- Yang, P., H. M. Sampson and H. M. Krause, 2006. A modified tandem affinity purification strategy identifies cofactors of the Drosophila nuclear receptor dHNF4. Proteomics 6 927–935. [DOI] [PubMed] [Google Scholar]
- Yu, Y., and A. Bradley, 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2 780–790. [DOI] [PubMed] [Google Scholar]