Abstract
The D1 protein is a high mobility group A (HMGA)-like nonhistone chromosomal protein with primary localization to certain AT-rich satellite DNA sequences within heterochromatin. The binding of D1 to euchromatic sequences is less studied and the functional significance of its chromosomal associations is unclear. By taking advantage of existing P-insertion alleles of the D1 gene, I generated D1 null mutations to investigate the phenotypic effect of loss of the D1 gene. In contrast to a previous report, I determined that the D1 gene is not essential for viability of Drosophila melanogaster, and moreover, that loss of D1 has no obvious phenotypic effects. My tests for an effect of D1 mutations on PEV revealed that it is not a suppressor of variegation, as concluded by other investigators. In fact, the consequence of loss of D1 on one of six variegating rearrangements tested, T(2;3)SbV, was dominant enhancement of PEV, suggesting a role for the protein in euchromatic chromatin structure and/or transcription. A study of D1 protein sequence conservation highlighted features shared with mammalian HMGA proteins, which function as architectural transcription factors.
THE Drosophila genome, like that of other eukaryotes, exists in the form of chromatin, a complex of DNA and an assortment of DNA-binding proteins. Histone proteins facilitate the organization of DNA into nucleosomal fibers, and together with nonhistone chromosomal proteins compact, organize, and regulate the activity of the genome. Differential targeting of nonhistone chromosomal proteins is important for generating distinct chromatin domains, and both the genomic distribution and functions of such proteins continue to be a rich area of investigation. In Drosophila, genetic and biochemical studies have successfully identified proteins specific for or highly enriched in the heterochromatic regions of the genome (James and Elgin 1986; reviewed by Grigliatti 1991; Cortes et al. 1999; De Felice et al. 1999; reviewed by Schotta et al. 2003). For example, both methodologies converged in the identification of HP1 (heterochromatin protein 1, encoded by the Su(var)205 gene; Sinclair et al. 1983; James and Elgin 1986). This protein, found in animals, plants, and fungi, associates with nucleosomes having lysine 9 of histone H3 methylated, a characteristic of heterochromatin (Bannister et al. 2001). However, the activities of many heterochromatin-associated proteins have yet to be defined. An example of this is the D1 (Drosophila protein 1) protein, which binds to a subset of highly repetitive DNAs, called satellite DNAs, that are found in heterochromatin.
The D1 protein was first identified >30 years ago (Alfageme et al. 1974), but its function remains elusive. It is a nonhistone chromosomal protein that shares many structural similarities with high mobility group (HMG) proteins (Rodriguez Alfageme et al. 1980). These include its solubility in 5% perchloric acid, high fraction of charged amino acids, properties for extraction from chromatin, and relative nuclear abundance. The cloning and sequence analysis of the D1 gene showed that it is most similar to the high mobility group A (HMGA) family of proteins (Ashley et al. 1989). Both D1 and the HMGA proteins possess several copies of the AT-hook DNA binding motif, which confers upon them the ability to bind to short uninterrupted AT tracts (Levinger 1985a; Ashley et al. 1989; Reeves and Nissen 1990). While HMGA proteins have only three copies of this motif, there are 10 AT hooks in the D1 protein, which is more than three times larger. Biochemical studies have shown that D1 associates with two AT-rich satellite DNAs, in vivo and in vitro (Levinger and Varshavsky 1982a; Levinger 1985a,b). It shows greatest affinity for the simple 1.672 g/cm3 satellite, which has the pentamer AATAT as its primary repeat. It also binds to the complex 1.688 g/cm3 satellite, which is chiefly a 359-bp repeat and 69% AT in composition. In agreement with this work, a heterochromatic localization was observed for D1 in both mitotic and interphase diploid cells by immunostaining (Renner et al. 2000; Aulner et al. 2002). As revealed by immunostaining to salivary gland polytene chromosomes, D1 shows a less predominant localization to euchromatic sites, which could reflect its binding to interspersed AT tracts (Alfageme et al. 1976; Rodriguez Alfageme et al. 1980).
Mutant alleles of the D1 gene have not been isolated in phenotype-based genetic screens. However, two P-insertion alleles of D1 were recently obtained in P-element mutagenesis experiments conducted as part of the Berkeley Drosophila Genome Project (BDGP) functional annotation of the Drosophila genome (Rorth 1996; Bellen et al. 2004). Chromosomes bearing these P insertions were reported to be homozygous lethal (http://flystocks.bio.indiana.edu), suggesting that the D1 gene was essential for viability. These strains could thus represent a starting point for the genetic and molecular characterization of D1 function. To this end, I examined the lethality of the P-insertion lines, carried out genetic screens to isolate D1 null mutants, and performed genetic tests to study the effects of loss of D1 protein. Although other investigators reported that the D1 gene is essential (Aulner et al. 2002), the studies described herein demonstrated that D1 is not required for viability or fertility. In addition, it is not a suppressor of position effect variegation.
MATERIALS AND METHODS
Drosophila stocks and culture conditions:
Stocks were maintained at 25° on cornmeal-malt medium described as standard medium by the Bloomington Drosophila Stock Center (BDSC) (http://flystocks.bio.indiana.edu). The P{EP}D1EP473 stock was obtained from the BDSC, although it is no longer available through that facility. The wm51b, wmmcT wm4f, and wm4Ta stocks were a gift of P. Talbert and S. Henikoff (Talbert and Henikoff 2000). K. Ahmad generously provided the bwD and Byron stocks. The Df(3R)BSC24 deletion was created at the BDSC using the hybrid element insertion (HEI) strategy (Gray et al. 1996; Preston et al. 1996) with P-element insertions P{EP}EP3243 (3R:4,757,601) and P{EP}EP707 (3R:5,220,293) (Parks et al. 2004). The Exelixis deletion Df(3R)Exel6152 was synthesized using FLP recombinase and the FRT-bearing transposon insertions P{XP}d04033 (3R:4,983,798) and P{XP}CG8420d04746 (3R:5,073,203) (Golic and Golic 1996; Parks et al. 2004). Other mutations and strains utilized in this study are described in FlyBase (Tweedie et al. 2009).
P-element excision:
The P{EP}D1EP473 and P{EPgy2}D1EY05004 insertions within the D1 gene were mobilized using P transposase to determine if excision of the P element restored homozygous viability to the chromosome. Males bearing the P insertion heterozygous with the TMS, Sb P{ry+ Δ2-3}99B P-transposase chromosome were backcrossed to w; P{EP}D1EP473/TM3, Sb or y1 w67c35; D1EY05004/TM3, Sb Ser females as appropriate. The progeny were screened for either Sb+ individuals, produced by reversion of the lethal mutation, or white-eyed males, produced by loss of expression of the P-element marker gene, w+. The same process was carried out for a P{EP}D1EP473 chromosome that had undergone P-element-mediated male recombination to replace the third chromosome left arm and DNA proximal to the P insertion on the right arm (recombinant 70).
The P element that remained at the deletion site on the Df(3R)D1C12 chromosome was mobilized by crossing st1 Df(3R)D1C12/TM3, Sb stAP1 e females to w/Y; T(2:3)ltx13, Sp ltx13/CyO, H{PDelta2-3}Hop2.1; TM3, Sb stAP1 e males to produce +/CyO, H{PDelta2-3}Hop2.1; st1 Df(3R)D1C12/TM3, Sb stAP1 e dysgenic males, which were then mated to w; TM3, Sb stAP1 e/TM6B, Tb Hu e females. The Df(3R)D1C12w− chromosome was isolated among the w− e+ Cy+ male progeny, and stocked.
PCR analysis:
The presence of P-element sequence in the D1 gene following exposure of the chromosome to P transposase was assessed by isolating genomic DNA and performing PCR as follows. Single fly DNA was isolated according to Gloor et al. (1993). For the P{EP}D1EP473 mobilization experiments, the D1 proximal primer (D1 2171F; 5′-GCGCTTCTTTACCGCAACTT-3′) was used in combination with primer Pry4 (5′-CAATCATATCGCTGTCTCACTCA-3′; BDGP) to assess the presence of the 3′ P end and integrity of the flanking sequence. The D1 distal primer (D1 2965R; 5′-GGCCAGCCGTCTCATGTAGT-3′) was used in combination with primer Plac1 (5′-CACCCAAGGCTCTGCTCCCACAAT-3′; BDGP) to assess the presence of the 5′ P end and integrity of the flanking sequence. For the P{EPgy2}D1EY05004 mobilization experiments, primer D1 2171F was used in combination with primer Plac1 to assess the presence of the 5′ P end and integrity of the flanking sequence. Primer D1 2965R was used in combination with primer Pry2 (5′-CTTGCCGACGGGACCACCTTATGTTATT-3′; BDGP) to assess the presence of the 3′ P end and integrity of the flanking sequence. The D1 2171F and D1 2965R primer combination generated a 794-bp fragment from wild-type D1 sequence.
For w− excision line 2A, the extent of the residual P-element sequence and the integrity of the flanking D1 gene sequence was assessed by PCR amplification using primers Pwht1 (5′-GTAACGCTAATCACTCCGAACAGGTCACA-3′) and D1 2171F, followed by DNA sequencing using the same primers.
The D1 mutant third chromosomes of lines 1A, 4A, and 70-7 were balanced with TM3, P{w+ GAL4-twi.G}2.3, P{ w+ UAS-2xEGFP}AH2.3, Sb1 Ser1 and these flies were crossed to Df(3R)BSC24/TM3, P{w+ GAL4-twi.G}2.3, P{ w+ UAS-2xEGFP}AH2.3, Sb1 Ser1 flies. The D1 mutant/Df(3R)BSC24 progeny were identified as EGFP-negative first instar larvae, and DNA was isolated from single larvae in a 10-μl volume as per Gloor et al. (1993). The D1 PCR primers were as follows: D1 21F (5′-CGAAGCGCACTGAGAAACAC-3′), D1 853F (5′-CATAACCGTCGTTGGCATCA-3′), D1 1605F (5′-TGGTTGCGGAATGTTGAAAT-3′), D1 2171F, D1 3374F (5′-GTGCATCGAGCAGCGATAA-3′), D1 3688F (5′-TGCGTGAACAACCAAGTTAAGC-3′), D1 3941F (5′-CGCTCACTTCCACAGCTTGA-3′), D1 921R (5′-GGACACCAACCAAAGGAGATG-3′), D1 1699R (5′-TGCTTCCACCAAACTTGCAC-3′), D1 2305R (5′-TGAGCGTGTGTTCGTGAGAG-3′), D1 3285R (5′GCAAGTAATTCCCTTTCGGATCT-3′), D1 2965R, D1 4320R (5′-GGACATCACCAACCCAAAGAA-3′). The pumilio primer sets were pum 9494F (5′-TCCCTTTCGGTCCTTTCGT-3′) and pum 9835R (5′-TGTGTGTGCTCTCTCGCTCTT-3′), and pum 6925F (5′-CTCAACATGTTACTACAATGGCTCT-3′) and pum 7624R (5′-CGTGTGGTTCTTTGTGCTG-3′). The DNA integrity of each larval DNA sample was verified by successful amplification using a primer set specific for the BSC24 deficiency chromosome, BSC24 5′ (5′-CAACTCGTCCGCTCCGCACAAC-3′) and Plac1. Positive control DNA was isolated from TM3/Df(3R)BSC24 first instar larvae, identified as EGFP positive and giving rise to the BSC24-specific PCR fragment.
The inversion breakpoint of In(3R)D11A was isolated by inverse PCR according to the protocol of E. J. Rhem, BDGP (http://www.fruitfly.org/about/methods/inverse.pcr.html). Genomic DNA isolated from the line 1A was digested with BamHI, ligated and PCR amplified using primers D1 3941F and D1 2965R, which directed synthesis away from each other. The PCR product was gel purified using the QIAquick Gel Extraction kit (QIAGEN, Valencia, CA) prior to sequencing.
For identification of the pum–D1 deletion chromosome, Df(3R)D1C12, DNA isolated from each recombinant line was analyzed by PCR to show that sequences proximal to the pumKG02259 insertion and distal to the D1EY05004 insertion were present and abutting P-element ends, but that the pumKG02259 distal and D1EY05004 proximal sequences were not detectable. Both transposon insertions were oriented with the 5′ P end centromere proximal. Primer combinations were pum 9494F and Plac1, Pry2 and pum 9835R, D1 2171F and Plac1, and Pry2 and D1 2965R. Subsequent to mobilization of the P element marking the deletion, primers pum 9494F and D1 2965R were used to amplify across the deletion breakpoint. This PCR product was sequenced.
Primers were designed using Primer 3 (http://primer3.sourceforge.net/). For DNA sequence analysis, PCR products were treated with ExoSAP-IT (USB, Cleveland) and used directly for sequence determination at the ISU Molecular Research Core Facility.
Southern analysis:
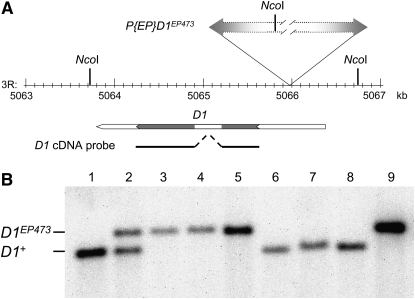
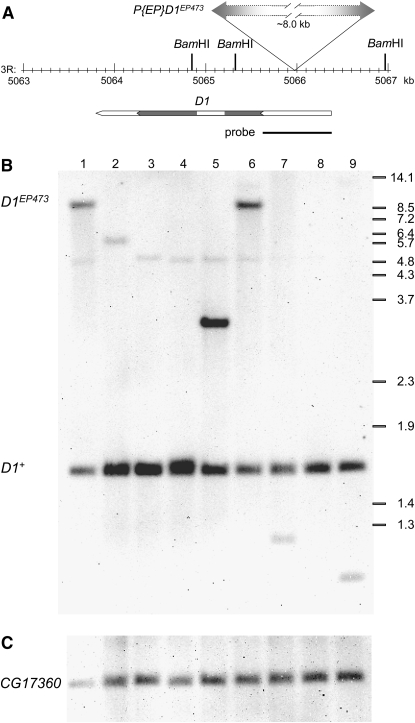
Genomic DNA was isolated using the DNeasy Blood and Tissue kit (QIAGEN). For each strain, DNA derived from ∼10 mg of whole flies was digested with NcoI (Figure 2) or BamHI (Figure 4) and fractionated on an agarose gel. The DNA was transferred to a positively charged nylon membrane and processed using the DIG Luminescent system according to the manufacturer's instructions (Roche Applied Science, Indianapolis). For analysis of the deficiency lines (Figure 2), the PCR DIG probe consisted of the entire D1 coding region, using a cloned D1 cDNA as a template. For analysis of the D1EP473 excision lines (Figure 4), the PCR DIG probes were synthesized using the D1 2171F and D1 2965R primer combination (Part A) or a primer set that amplified CG17360 genomic DNA (5′-TGATGGTTGCTGCTGGTGTT-3′ and 5′-GAGCCCAATATCGGAGATGC-3′; Part B) and a fly genomic DNA template.
Figure 2.—
Southern analysis of deletion strains. (A) The NcoI restriction map of the chromosome 3R genomic region surrounding the D1EP473 insertion site is illustrated. The gene span of D1, with coding region in gray, and position of the D1 cDNA probe used for Southern analysis (solid line) are shown below the map. (B) Genomic DNA was isolated from w1118 (lane 1), D1EP473/+ (lane 2), D1EP473/Df(3R)Exel6152 (lane 3), D1EP473/Df(3R)BSC24 (lane 4), D1EP473/Df(3R)D1C12w− (lane 5), +/Df(3R)Exel6152 (lane 6), +/Df(3R)BSC24 (lane 7), +/Df(3R)D1C12w− (lane 8), and D1EP473 (lane 9), and digested with NcoI. A wild-type D1 locus was expected to yield a 3041-bp genomic fragment whereas the D1EP473 insertion was expected to yield a 3481-bp genomic fragment. Only the D1EP473 allele was observed when the flies were heterozygous for D1EP473 and deletions Df(3R)Exel6152, Df(3R)BSC24, or Df(3R)D1C12w−. In contrast, only the wild-type allele was observed when the flies were heterozygous for D1+ and deletions Df(3R)Exel6152, Df(3R)BSC24, or Df(3R)D1C12w−.
Figure 4.—
Southern analysis of D1EP473 excision lines. (A) The BamHI restriction map of the chromosome 3R genomic region surrounding the D1EP473 insertion site is illustrated. The gene span of D1, with coding region in gray, and position of the region amplified by the D1 proximal and D1 distal primer pair (Figure 1), which was used as a probe for Southern analysis, are shown below the map. (B and C) A representative line from each class of w− excision lines (see Table 1) is included in the Southern blot shown here. Genomic DNA was isolated from D1EP473/TM3, Sb (lane 1), w1118 (lane 2), D1Rev1B/TM3, Sb (lane 3), D11E/TM3, Sb (lane 4), D12A/TM6B, Tb (lane 5), D170-5/TM6B, Tb (lane 6), In(3R)D11A/TM3, Sb (lane 7), Df(3R)14A/TM3, Sb (lane 8), and Df(3R)D170-7/TM3, Sb (lane 9), and digested with BamHI. (B) The D1 probe hybridizes to a ∼1.7-kb genomic fragment for wild-type flies, and an ∼9.7-kb genomic fragment for the D1EP473 allele. A faint cross-hybridizing fragment, polymorphic in w1118, can be detected at 5–6 kb. The approximate positions of the BstEII-digested λ-size markers are shown at right. (C) The blot was rehybridized with a CG17360 probe that recognizes an ∼2.2-kb genomic fragment, to serve as a normalization (loading) control.
P-element-mediated male recombination:
The technique of P-element-mediated male recombination was used to induce the exchange of DNA flanking the P{EP}D1EP473 insertion site (i.e., 3L and proximal 3R, or distal 3R) with that of the homolog, with the potential of recovering deletion alleles among the recombinants. The Gl1 mutation was recombined onto the left arm of the P{EP}D1EP473 chromosome to serve as a dominant marker for detecting recombinants. w1118 females were crossed to P transposase-expressing w/Y; +/CyO, H{PDelta2-3}Hop2.1; Gl1 P{EP}D1EP473/Bsb males, and Gl+ Bsb+ or Gl− Bsb− male recombinant progeny were recovered. Recombinant 70, which retained the P insertion and did not suffer a flanking deletion, was identified within the Gl+ Bsb+ class.
As a means to isolate a deletion between the P{EPgy2}D1EY05004 and P{SUPor-P}pumKG02259 elements, P transposase was expressed in male flies carrying the P insertions in trans. In preparation, a st1 P{SUPor-P}pumKG02259 ca1 chromosome was produced by meiotic recombination to facilitate the subsequent identification of recombinants. The y1 w67c35/Y; +/CyO, H{PDelta2-3}Hop2.1; st1 P{SUPor-P}pumKG02259 ca1/P{EPgy2}D1EY05004 males were obtained by crossing y1 w67c35; P{EPgy2}D1EY05004/TM3, Sb Ser females to +/CyO, H{PDelta2-3}Hop2.1; st1 P{SUPor-P}pumKG02259 ca1/TM3, Ser males and selecting Cy− Sb+ Ser+ progeny. These males were then crossed to st1 Sbsbd-1 es ro1 ca1 females and st− ca+ recombinant progeny, which were expected to include the desired deletion class, were recovered and stocked.
RT–PCR analysis:
Total RNA was isolated from ovaries dissected from females of each genotype, using UltraSpec RNA (Biotecx Laboratories, Houston). Random-hexamer primed cDNA was synthesized from 1 μg of total RNA using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. One-twentieth of the synthesis reaction was used as a template for a 25-μl PCR reaction, unless otherwise indicated. The primers D1 1261F (5′-CCGACTTGTTCTGTGGTGGA-3′) and D1 1803R (5′-CCAGCGATAGCGAGAATGAA-3′) were used to amplify a 224-bp segment of the D1 mRNA. The primers E(var)3-9 2261F (5′-GCCGAACTGCTCCTGTGTCT-3′) and E(var)3-9 2660R (5′-GTCGCTTTGTGGAACGGATT-3′) were used to amplify a 351-bp segment of the E(var)3-9 mRNA, as a control (Weiler 2007). RT–PCR was performed using two sets of independently isolated RNA samples, with identical results.
Stubble variegation assay:
A reciprocal translocation between the second and third chromosomes places the Stubble (Sb) gene, having the Sb1 mutation, under the repressive influence of the chromosome 2 heterochromatin in the T(2;3)SbV strain. Silencing of Sb1 effects a wild-type bristle, while its expression results in the Stubble phenotype. Crosses were performed at 25° between T(2;3)SbV, In(3R)Mo, Sb1 sr1/+; TM3, Ser e females and males heterozygous for the D1 mutant or wild-type control chromosome and the ru1 h1 th1 st1 cu1 sr1 es Pr1 ca1 chromosome. The D1/T(2;3)SbV progeny were identified as Ser+ Pr+. To eliminate the potential influence of sex on variegation, only female progeny were scored. Fourteen bristles: the anterior and posterior sternopleurals, the upper and lower humerals, the anterior and posterior scutellars, and the posterior dorsocentrals, were scored for a Sb− or wild-type phenotype.
D1 protein comparison:
The predicted protein sequences of D1 orthologs in other Drosophila species were obtained from FlyBase (Tweedie et al. 2009) with the exception of that of D. simulans, which was not present. The partial gene sequence of the D. simulans D1 gene was identified by tBLASTn of D. simulans genomic sequence using the D. melanogaster protein as a query (http://insects.eugenes.org/species/blast/). FGENESH+ (http://www.softberry.com) was used to predict the partial protein sequence. The missing N terminus of the protein (33 amino acids) was constructed by translation of the adjoining genomic sequence, assuming two DNA sequencing errors that affected the reading frame and using the D. melanogaster D1 protein sequence as a guide. Prediction of protein motifs was performed against the Pfam database (Finn et al. 2006). Amino acid similarity to D. melanogaster D1 was determined using BLAST2 (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi).
RESULTS
P-element alleles of the D1 gene do not revert to viability:
A genetic approach toward elucidating the function of the D1 gene was undertaken using two P-insertion alleles of the D1 gene, which were recovered in the P{EP} and P{EPgy2} mutagenesis experiments that contributed to the BDGP Gene Disruption Project (Rorth 1996; Rorth et al. 1998; Bellen et al. 2004). Both insertions map to the 5′-untranslated region (UTR) of D1 (Bellen et al. 2004). For both stocks, the third chromosome bearing the P insertion was homozygous lethal, suggesting that the insertions disrupted the D1 gene and that the D1 gene was essential for viability. If true, it should have been possible to mobilize the P elements and revert the lethal phenotype. However, I was unable to recover any homozygous viable chromosomes following the introduction of a P-transposase source. For example, none of ∼1500 progeny produced from a cross between w; P{EP}D1EP473/TM3, Sb females and P{EP}D1EP473/TMS, Sb P{Δ2-3}99B males was found to be Sb+, the phenotype expected for D1+/P{EP}D1EP473 flies. Similarly, none of ∼850 progeny produced from a cross between y1 w67c35; P{EPgy2}D1EY05004/TM3, Sb Ser females and y1 w67c35/Y; P{EPgy2}D1EY05004/TMS, Sb P{Δ2-3}99B males were Sb+. In contrast, a high frequency of white-eyed progeny resulting from loss of expression of the w+ gene carried by either P element was observed. This result indicated that the P element was being mobilized in both experiments.
Chromosomes isolated upon precise excision of the P{EP}D1EP473 (hereafter referred to as D1EP473) and P{EPgy2}D1EY05004 (hereafter referred to as D1EY05004) elements remained homozygous lethal, revealing the presence of extraneous lethal mutations. Presumptive precise excision lines were identified among the w− progeny following P mobilization, by PCR analysis of the genomic DNA encompassing the insertion site (see Figure 1 and materials and methods). DNA sequencing confirmed that the wild-type gene sequence was restored upon excision of D1EP473, for isolate D1Rev1B (see below). Nevertheless, the chromosome bearing the D1Rev1B “revertant” allele was homozygous lethal, as were excision lines derived from the D1EY05004 insertion chromosome. These results indicated that lethal mutations were present on the D1EP473 and D1EY05004 chromosomes, but did not reveal if the insertions themselves conferred lethality.
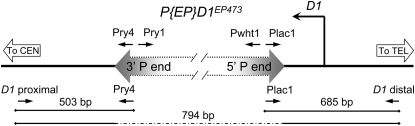
Figure 1.—
PCR analysis of D1 P-insertion alleles. The left–right block arrow represents the P{EP}D1EP473 element inserted within the 5′-UTR of the D1 genomic locus. The relative locations of the PCR primers used to analyze the insertion and its excision derivatives and PCR product sizes are shown. The distance between the D1 proximal and D1 distal primers was too large for PCR amplification with standard Taq polymerase when the entire P{EP} element was present, but yielded a 794-bp product in its absence. The P{EPgy2}D1EY05004 insertion is similarly located in the 5′-UTR but in the opposite orientation. PCR analyses of this element typically utilized the Pry2 primer (adjacent to the Pry4 primer; not shown) instead of the Pry4 primer.
P-element alleles of the D1 gene are not lethal:
The presence of lethal mutations on the D1EP473 and D1EY05004 chromosomes left open the question of whether the P insertions themselves were lethal. To address this issue, the two mutants were crossed to each other as well as to strains bearing deficiencies of the D1 gene. When w; D1EP473/TM3, Sb and y1 w67c35/Y; D1EY05004/TM3, Sb Ser flies were crossed, the expected frequency of D1EP473/D1EY05004 progeny flies was observed (32.1%; n = 134), indicating that the two P-insertion chromosomes did not share lethal mutations. Outcrossing of the D1EP473 strain eventually resulted in the isolation of a homozygous viable D1EP473 chromosome, confirming the viable nature of this allele. Moreover, both D1 P-insertion alleles were hemizygous viable in combination with the large deficiencies Df(3R)BSC24 and Df(3R)Exel6152. The Df(3R)BSC24 chromosome is reported to possess a deletion of ∼463 kb (from pyd to Fsp85D) that includes the D1 gene (Parks et al. 2004; Tweedie et al. 2009). The ∼89-kb Df(3R)Exel6152 deletion has breakpoints proximal (in pumilio) and distal (in CG8420) to D1 (Parks et al. 2004). The D1 gene was not detectable in the deficiency chromosomes by Southern analysis using the entire D1 coding sequence as a probe (Figure 2) or by PCR (data not shown). These results clearly demonstrated that the two D1 P-insertion alleles were not lethal.
Generation of new D1 alleles:
Although the two D1 P-insertion alleles were determined to be hemizygous viable, the possibility existed that, being located within the 5′-UTR of the gene, neither precluded D1 expression. Therefore, the results described above did not rule out that the D1 gene could be essential and that a level of gene product sufficient for viability was produced by the P-insertion alleles. Indeed, both the D1 protein and D1 RNA were detectable in ovarian tissue of D1EP473 flies (Figure 3 and data not shown). To generate an unequivocal D1 null allele, one that lacked D1 coding sequence, two approaches were undertaken (as described in more detail below). The D1EP473 insertion was mobilized with the intention of recovering imprecise excision events that would delete some or all of the D1 gene. Second, P-element-mediated male recombination was performed in flies possessing both a P insertion into the 5′-UTR of the D1 gene and a P insertion downstream of the D1 coding sequence with the aim of recovering a deletion mediated and demarcated by the two P insertions (Parks et al. 2004).
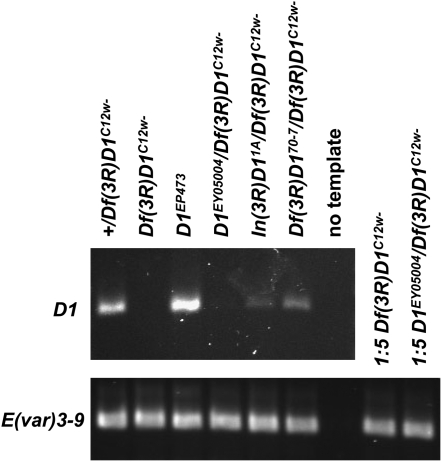
Figure 3.—
RT–PCR analysis of D1 mutant alleles. cDNA samples prepared from equivalent amounts of ovary RNA, for the indicated strains, were used as templates for PCR of the D1 mRNA (primers D1 1261F and D1 1803R) and control E(var)3-9 mRNA (primers E(var)3-9 2261F and E(var)3-9 2260R). Both primer sets spanned an intron, enabling products from potential contaminating genomic DNA to be distinguished. However, no genomic DNA products were observed. Where no (Df(3R)D1C12w−) or very little (D1EY05004/Df(3R)D1C12w−) D1 RT–PCR product was observed, the control E(var)3-9 PCR reaction was performed using fivefold diluted cDNA template, as a means to confirm the integrity of the cDNA template. E(var)3-9 mRNA has been quantified at ∼69% of the level of D1 mRNA in the ovary, using microarray analysis (Chintapalli et al. 2007).
Imprecise excision strategy:
As a means to isolate imprecise excision derivatives of D1EP473, the male progeny of a cross between w; D1EP473/TM3, Sb females and D1EP473/TMS, Sb P{ry+ Δ2-3}99B males were screened for loss of expression of the w+ P{EP} element marker gene. The results of two experiments are described here, the second using a derivative of the original D1EP473 chromosome, recombinant 70, which had been recovered following P-element-mediated male recombination (see materials and methods). A combined total of 18 w− exceptions were stocked. The PCR strategy illustrated in Figure 1 was employed for initial molecular analyses of the exceptional lines. To ascertain if either P-element end did not excise, primers that hybridized to D1 genomic sequence proximal and distal to the insertion site were used in combination with primers that hybridized to the 3′ and 5′ P-element ends, respectively. To detect small deletions of genomic DNA (extending no more than ∼400 bp in either direction from the insertion site), the proximal and distal D1 genomic primer combination was used. Southern analysis was performed using genomic DNA extracted from balanced stocks of each w− line to further investigate the nature of each P-excision event. A probe was synthesized by PCR using the D1 proximal and distal genomic primers (Figure 1). This probe should recognize an ∼1.7-kb fragment for a wild-type D1 allele, such as was present on the TM3, Sb balancer chromosome of each stock, and an ∼9.7-kb fragment for the D1EP473 allele.
The results of the molecular analyses, as shown in Figure 4 and Table 1, suggested that eight w− isolates likely resulted from precise excision, seven w− chromosomes retained some or all of the P{EP} element, and three w− chromosomes possessed D1 deletions or other rearrangements.
TABLE 1.
Molecular analyses of D1EP473 w− excision lines
| Linea | D1 proximal + Pry4 PCR | D1 distal + Plac1 PCR | D1 proximal + D1 distal PCRb | Southernc |
|---|---|---|---|---|
| D1EP473 | 685 bp | 503 bp | 794 bp | ∼9.7 kb/∼1.7 kb |
| 1A | − | − | + | ∼1.3 kb/∼1.7 kb |
| 1B, 1C, 1D, 1F | − | − | + | ∼1.7 kb |
| 1E | − | − | +/∼0.85 kb | ∼1.7 kb doublet |
| 2A | + | − | + | ∼3.2 kb/∼1.7 kb |
| 2B, 2C | − | − | +/∼0.85 kb | ∼1.7 kb doublet |
| 3A | − | − | +/∼0.85 kb | ∼1.7 kb doublet |
| 4A | − | − | + | ∼1.7 kb |
| 70-1, 70-2, 70-3 | − | − | + | ∼1.7 kb |
| 70-5 | + | + | + | ∼9.7 kb/∼1.7 kb |
| 70-6 | − | − | +/∼0.85 kb | ∼1.7 kb doublet |
| 70-7 | − | − | + | ∼1.1 kb/∼1.7 kb |
| 70-9 | − | − | + | ∼1.7 kb |
The PCR primer sets are as illustrated in Figure 1. The presence of a PCR product of size expected for the original D1EP473/TM3, Sb strain (first row) is denoted by a +, the absence of a product by a −, and a product of other size by the estimated size.
Lines that may not be independent isolates are grouped together in a row. Analyses were performed on balanced lines, due to extraneous lethal mutations.
The PCR products derived from one or both homologs. When the intact P{EP} element is present, size limitations preclude amplification of a product from that homolog. However, the wild-type D1 locus of the balancer chromosome yielded a 794-bp product.
The D1 proximal + D1 distal PCR product was used as a probe of BamHI-digested genomic DNA, as illustrated for representative lines in Figure 4. The band(s) derived from one or both homologs. The intensity of the 1.7-kb band for lines 1A, 2A, 4A, 70-5, and 70-7 appeared approximately half that of the 1.7-kb band for all other w− lines.
Precise excision lines:
The PCR data (Table 1) and Southern data (Figure 4 and Table 1) strongly suggested that the D1EP473 element had precisely excised in lines 1B, 1C, 1D, 1F, 70-1, 70-2, 70-3, and 70-9. The PCR assays showed no evidence of P-element sequence at the D1 locus nor the existence of a small deletion. This was consistent with the results of Southern analysis, which revealed a single band of 1.7 kb for each line. DNA was extracted from allele 1B/Df(3R)BSC24 and allele 70-1/Df(3R)Exel6152 flies, and the region encompassing the original insertion site of the P{EP} element was PCR amplified using the D1 proximal and distal genomic primers. Given the absence of D1 gene sequence on the deficiency chromosome, the only PCR template was the revertant allele. Sequence analysis of this PCR fragment confirmed that line 1B (allele D1Rev1B) and line 70-1 (allele D1Rev70-1) resulted from precise excision of the D1EP473 insertion; the D1 gene sequence was restored to wild type.
D1 insertion mutants:
For lines 1E, 2B, 2C, 3A, and 70-6, the D1 locus appeared to possess an extra ∼50 bp upon P-element excision, observed as an additional PCR fragment using the D1 genomic primer set and as a doublet band by Southern (Figure 4 and Table 1). To determine the nature of this insertion, DNA was extracted from line 1E/Df(3R)BSC24 flies and the region encompassing the original insertion site of the P{EP} element was PCR amplified and sequenced using the D1 proximal and distal primers (Figure 1). Consistent with the PCR results, an 8-bp P-target site repeat and 33 bp of additional P-element sequence were found to remain at the insertion site.
Line 2A also retained extra DNA sequence upon P excision, as indicated by the novel band observed by Southern analysis and revealed by PCR to include the P-element 5′ end (Figure 4 and Table 1). As this P-transposase-induced lesion might include a deletion of D1 sequence, sequence analysis of the genomic DNA was carried out. A PCR primer that hybridized to the 5′-P end and directed synthesis proximally (Pwht1; see Figure 1) was used in combination with the D1 proximal primer to amplify across the genomic DNA-P{EP}-element junction. Sequence analysis of the resulting PCR fragment revealed the presence of P-element sequence, but no deletion of D1 coding sequence, associated with the imprecise P excision.
Line 70-5 appeared to retain the P{EP} element despite loss of expression of the w+ marker gene. Both P-element ends were retained, as indicated by PCR analysis, and the electrophoretic migation of the D1 band observed by Southern analysis was consistent with the P{EP} element being intact (Figure 4 and Table 1).
D1 structural mutants:
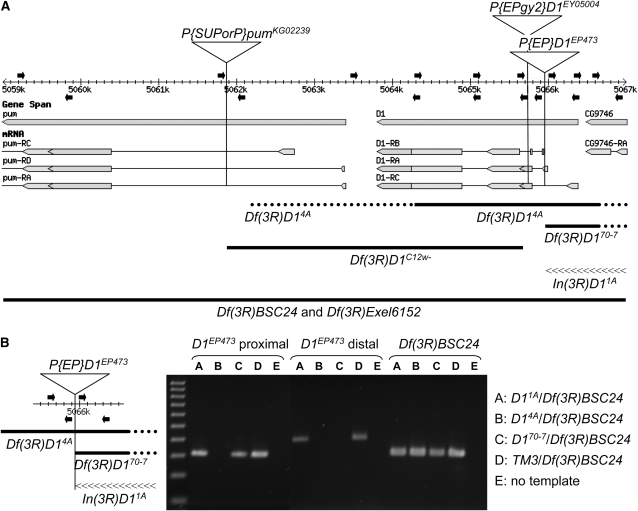
Lines 1A, 4A, and 70-7 distinguished themselves by yielding PCR results that appeared wild type (i.e., no P-element sequence) together with Southern analysis results, suggesting that the w− chromosomes of lines 1A, 4A, and 70-7 were disrupted for the D1 gene (Figure 4 and Table 1). The signal intensity of the 1.7-kb band was about half that of the precise excision lines, and lines 1A and 70-7, but not 4A, exhibited a band of altered size that was recognized by the probe. These results suggested that line 4A possessed a deletion of D1 coding sequence, and that lines 1A and 70-7 were partial deletions or other aberrations. To explore this possibility, PCR was performed on individuals heterozygous for the mutant chromosome and a deficiency for the region, Df(3R)BSC24, using overlapping primer sets that spanned the D1 locus (Figure 5). The inability to amplify a genomic segment, in the context of proper controls, suggested a deletion of some or all of the region. The DNA from single first instar larvae was used, as adult flies were not viable. Sample data are presented in Figure 5B and the results from the most informative of the PCR assays are presented in Table 2. For line 4A, none of the primer sets corresponding to the D1 coding region yielded a product, confirming that line 4A represented a deletion of the D1 gene. The deletion extended into the distal CG9746 gene, disrupting it as well. The deletion did not extend proximally into the pumilio (pum) coding region, but, as the endpoint was not precisely mapped, it could affect expression of three pum transcripts. The DNA sequence proximal to the former D1EP473 insertion site was amplifiable for line 70-7, although distal sequence was not. Similar to line 4A, the deletion extended into the distal CG9746 gene. Figure 5A illustrates how these deletions map to the genomic region. In contrast to the results with lines 4A and 70-7, D1 coding sequence was amplifiable to either side of the former D1EP473 insertion site for line 1A. However, no PCR products were obtained when the predicted amplimer spanned the former insertion site, suggesting the existence of an inversion. To test this hypothesis, inverse PCR was employed to amplify the genomic sequence spanning the putative inversion breakpoint for line 1A (see materials and methods). Sequence analysis of the inverse PCR product confirmed the existence of a small inversion with breakpoints at the site of the D1EP473 insertion and within the CG9746 gene (Figure 5).
Figure 5.—
A map of D1-mutant alleles. (A) The D1 gene and portions of the flanking pumilio and CG9746 genes and their transcripts are illustrated as they map to the 3R genomic sequence (adapted from FlyBase Release 5.1, http://www.flybase.org). Proximal is to the left and distal is to the right. The insertion sites of the three P elements utilized in this study are indicated by vertical lines on the map. Both the pumKG02239 and the D1EY05004 P insertions are oriented with the 5′-P end proximal and the 3′-P end distal. The D1EP473 insertion is oriented with the 3′-P end proximal and the 5′-P end distal. Primers used for PCR analyses of D1 mutants isolated in this study are shown above (forward primers) and below (reverse primers) the map as solid arrows. The precise locations are listed in Table 2. The extents of the deletions are shown below as thick solid lines, with the dotted portions reflecting the uncertainty of the endpoints. The Df(3R)BSC24 (3R:4,757,601-5,220,293) and Df(3R)Exel6152 (3R:4,983,798-5,073,203) deletions extend well beyond this ∼8-kb region. The inverted region of In(3R)D11A (distal breakpoint at position 5,067,087) is illustrated by a linear array of “<” symbols. (B) The PCR data for two primer sets, which amplify genomic segments immediately proximal and distal to the D1EP473 insertion as illustrated at left, is shown for the three D1EP473 excision alleles associated with chromosome rearrangements that were isolated in this study. Genomic DNA was isolated from single first instar larvae of genotypes In(3R)D11A/Df(3R)BSC24, Df(3R)14A/Df(3R)BSC24, and Df(3R)D170-7/Df(3R)BSC24, as hemizygous adults were inviable. A sibling TM3/Df(3R)BSC24 larva served as a positive control for the PCR reaction. A third PCR primer set that hybridized to the P element marking the Df(3R)BSC24 deficiency and the flanking genomic DNA, thus specific for the Df(3R)BSC24 chromosome, was used to confirm the integrity of the DNA preparation, as well as the genotypes. A 100-bp ladder (100–1000 bp) is shown in the first lane.
TABLE 2.
PCR analysis of putative D1 deletions generated by imprecise P excision
| Primer seta | Genomic coordinatesb | Line 1A | Line 4A | Line 70-7 |
|---|---|---|---|---|
| pum 6925F + pum 7624R | 5,059,192–5,059,891 | + | + | + |
| pum 9494F + pum 9835R | 5,061,761–5,062,102 | + | + | + |
| D1 21F + D1 921R | 5,063,450–5,064,350 | + | − | + |
| D1 853F + D1 1699R | 5,064,282–5,065,128 | + | − | + |
| D1 1605F + D1 2305R | 5,065,034–5,065,734 | + | − | + |
| D1 2171F + D1 3285R | 5,065,600–5,065,914 | + | − | + |
| D1 2171F + D1 2965R | 5,065,600–5,066,394 | − | − | − |
| D1 3374F + D1 2965R | 5,066,003–5,066,394 | + | − | − |
| D1 3688F + D1 4320R | 5,066,317–5,066,949 | + | − | − |
| D1 3941F + D1 4320R | 5,066,570–5,066,949 | + | − | − |
The generation of a PCR product is indicated by a plus and the absence of a product by a minus.
The results of overlapping PCR amplifications that did not yield additional information are omitted.
The genomic coordinates of the amplified regions correspond to Flybase Release 5.1 of chromosome 3R. The insertion site for the D1EP473 element is 5,065,965–5,065,972 (8-bp duplication). The D1 gene coding region extends between 5,064,241 and 5,065,626 and the pum coding regions extend between 4,896,667 and 5,059,583.
P-element-mediated male recombination strategy:
As demonstrated by Parks et al. (2004), deletion of the genomic sequence between two P elements on homologous chromosomes can be induced by expressing P transposase. The rare deletion events are recovered among progeny that exhibit recombinant flanking markers. To delete D1 coding sequence, the D1EY05004 insertion within the 5′-UTR of D1 and the P{SUPor-P}pumKG02259 insertion within the first intron of the pum gene were selected. The region separating the two elements is 3871 bp long and includes the entire D1 coding sequence as well as the noncoding first exons of the pum A, D, and C transcripts (Figure 5). The scarlet1 (st1) and claret1 (ca1) mutations were recombined onto the P{SUPor-P}pumKG02259 (hereafter referred to as pumKG02259) chromosome for selection of recombinants. P transposase was expressed in D1EY05004/st1 pumKG02259 ca1 male flies and their progeny screened for the st− ca+ recombinant class that would include the desired deletion events. PCR was used to assess the presence of either or both parental P elements on the recombinant chromosome of progeny flies. Of 22 st− ca+ recombinants, six were determined to have retained a 5′-P end within the pum gene and a 3′-P end within the D1 gene, but have lost the 3′-P end adjacent to pum sequence and 5′-P end adjacent to D1 sequence. This result was consistent with the six recombinant chromosomes possessing a deletion marked by a single, hybrid P element. However, I desired further proof that the desired deletion, rather than another anomalous recombination event, had occurred. The anticipated large size of the hybrid P element made it impractical to show by standard PCR using pum and D1 primers that the two P ends were part of a single transposon that joined distant pum and D1 genomic sequence. Therefore, the hybrid P element was mobilized by expressing P transposase in a putative deletion line, recombinant C12, and progeny showing loss of the w+ marker were isolated. A small PCR product was generated using a primer proximal to the pumKG02259 insertion site and the D1 distal primer for one w− isolate, named C12w− (see Figure 5A). Sequence analysis of this product confirmed that genomic sequence normally separated by almost 4 kb had been juxtaposed by deletion, for chromosome Df(3R)D1C12w−. In addition, no coding sequence was detectable on this chromosome by Southern analysis (Figure 2). Thus, Df(3R)D1C12w− represents a second definitive D1-null allele.
D1-null flies are fully viable and fertile:
As described above, both Df(3R)D14A and Df(3R)D1C12w− were deleted for the D1 coding region and were therefore clearly null alleles. Although the In(3R)D11A and Df(3R)D170-7 alleles suffered inversion or deletion of the D1 regulatory region, respectively, they could conceivably still be expressed. To address this possibility and ascertain if these two alleles were also D1-null, ovarian RNA was isolated from In(3R)D11A/Df(3R)D1C12w− and Df(3R)D170-7/Df(3R)D1C12w− females, and qualitatively assessed by RT–PCR. Ovarian tissue was selected due to the high expression level of D1 in this tissue in wild-type flies (Renner et al. 2000; Aulner et al. 2002). As shown in Figure 3, D1 RNA was detected for both alleles, suggesting that they are hypomorphs. This assay also revealed gene product for the D1EY05004 P-insertion allele. As expected, no D1 RNA was detectable for Df(3R)D1C12w− ovaries.
The generation of two new null alleles of the D1 gene enabled a test of the requirement for the D1 gene product for viability. Flies bearing the targeted D1 deletion, of genotype w; st1 Df(3R)D1C12w−/TM3, Sb, were crossed to w/Y; Df(3R)D14A/TM3, Sb flies for complementation analysis. As shown in Table 3, D1-deficient flies were obtained at expected frequency. The same result was obtained when w; st1 Df(3R)D1C12w−/TM3, Sb flies were crossed to w/Y; Df(3R)Exel6152/TM3, Sb flies or to Df(3R)BSC24/TM3, Sb flies, the two large deficiencies that span the D1 gene (Table 3). These data clearly proved that the D1 gene was not essential for viability.
TABLE 3.
Complementation analysis of D1 null mutants
| Cross | Trial | No. of progeny | Sb+ progeny (%) |
|---|---|---|---|
| w; st1 Df(3R)D1C12w−/TM3, Sb x w/Y; Df(3R)D14A/TM3, Sb | 1 | 189 | 69 (36.5) |
| 2 | 353 | 110 (31.2) | |
| w; st1 Df(3R)D1C12w−/TM3, Sb x w/Y; Df(3R)BSC24, st1 ca1 /TM3, Sb | 1 | 463 | 162 (35.0) |
| w; st1 Df(3R)D1C12w−/TM3, Sb x w/Y; Df(3R)Exel6152/TM3, Sb | 1 | 555 | 194 (35.0) |
| 2 | 601 | 200 (33.3) | |
| 3 | 705 | 249 (35.3) |
D1-null flies did not exhibit any obvious phenotypic abnormalities. In addition, females of genotype Df(3R)D1C12w−/Df(3R)Exel6152 and Df(3R)D1C12w− homozygotes were tested and found to be fertile.
D1 is not required for heterochromatin-mediated repression:
As the D1 gene encodes a nonhistone chromosomal protein that localizes to the heterochromatin, the dominant effect of a D1 loss-of-function mutation on position effect variegation (PEV) was assessed. Modification of the severity of PEV due to a decrease in gene dosage of an unlinked locus has been used to implicate that locus in the determination of chromosome structure (reviewed by Weiler and Wakimoto 1995; Schotta et al. 2003). The D1EP473 insertion was tested for a modifying effect on PEV of three different rearrangements that induce variegation of the white (w) gene. To avoid the potentially confounding effects of the w+ marker gene present within the P{EP} transposon on an assessment of w variegation, the w− D1EP473 derivative allele 70-5 was used for the experiments. The isogenic D1+ third chromosome, D1Rev70-1 served as the control. In the first experiment, In(1)wm4 females were crossed to w/Y; D1EP473w−/TM3, Ser and w/Y; D1+/TM3, Ser males. A visual examination of the male and female progeny of the two crosses revealed no difference in eye pigmentation among the genotypes (when sorted by age and sex; data not shown). As this result differed from that in the published literature (see discussion), the experiment was repeated using two In(1)wm4 stocks from another source, designated In(1)wm4Ta and In(1)wm4f (Talbert and Henikoff 2000). These stocks were molecularly verified as having the wm4 inversion (Talbert and Henikoff 2000). In addition, two other w-variegating alleles, In(1)wm51b and In(1)wmmc, were tested in case there might be rearrangement-specific effects. An additional advantage to testing these four w-variegating alleles was their generally higher level of eye pigmentation in comparison with the lab In(1)wm4 stock used initially, a more extreme variant. Thus, either suppression or enhancement of PEV should have been readily detectable using these alleles. The results of this experiment confirmed and extended those of the first study. Neither suppression nor enhancement of PEV of the w gene by the D1EP473w− P-insertion allele was observed for any of the rearrangements (data not shown).
D1 mutant alleles were similarly tested for the recessive modification of PEV, again using the In(1)wm4 variegating rearrangement. w; Df(3R)D1C12w− females were crossed to In(1)wm4 males bearing the w− D1EP473 derivative allele 70-5, the isogenic Df(3R)D170-7 allele, or the isogenic D1+ allele, D1Rev70-1, each heterozygous with TM3, Sb. Comparative visual examination of the w/In(1)wm4 progeny females revealed no difference in pigmentation between D1+ and D1− genotypes (data not shown). The experiment was repeated by crossing w; Df(3R)D1C12w− females to In(1)wm4/Y; Df(3R)D14A/TM3, Sb and isogenic D1+ In(1)wm4/Y; D1Rev1B/TM3, Sb males, to generate and assess D1-null flies. Consistent with prior results, loss of both D1 alleles did not significantly affect variegation of In(1)wm4 (data not shown).
Assays for modification of PEV were expanded to include rearrangements variegating for the brown (bw) or Stubble (Sb) genes. Sb variegation in the adult bristles is associated with the T(2:3)SbV translocation, which exhibits variable inactivation of the dominant Sb1 allele due to its juxtaposition near heterochromatin (Sinclair et al. 1983). For this experiment, the D1EP473 allele was tested as was the deletion derivative Df(3R)D14A. The precise excision allele D1Rev1B was used as a control. As the third chromosomes of the three strains should only differ at the D1 locus, any differential effect on PEV would be attributable to loss of D1. Enhancement of PEV is viewed as a decrease in Sb− (abnormal) bristles while suppression of PEV is observed as an increase in Sb− bristles. As shown in Table 4, the D1EP473 allele and Df(3R)D14A allele similarly enhanced SbV variegation.
TABLE 4.
Loss of D1 enhances Stubble variegation
| Genotype | Trial | No. of flies | Average no. Sb− bristles (± SD)a | P-valueb |
|---|---|---|---|---|
| +; P{EP}D1EP473/T(2:3)SbV | 1 | 21 | 9.3 ± 1.6 | 0.015 |
| +; Df(3R)D14A/T(2:3)SbV | 1 | 40 | 9.5 ± 1.6 | 0.011 |
| +; D1Rev1B/T(2:3)SbV | 1 | 31 | 10.5 ± 1.7 | |
| +; P{EP}D1EP473/T(2:3)SbV | 2 | 50 | 8.5 ± 2.1 | <0.001 |
| +; Df(3R)D14A/T(2:3)SbV | 2 | 46 | 8.8 ± 2.1 | 0.001 |
| +; D1Rev1B/T(2:3)SbV | 2 | 40 | 10.2 ± 1.7 |
As described in materials and methods, 14 bristles were scored per fly.
For each trial, the mean number of Sb− bristles for the two D1 mutants was compared with that of the D1Rev1B control using a Student's' t-test.
A potential role for D1 in trans-inactivation or para-inactivation was assessed using the variegating rearrangements bwD and Dp(2;2)Byron (Henikoff et al. 1995). The bwD allele is a large insertion of heterochromatin into the bw coding region, which can variably repress expression of a wild-type bw allele on the homolog by chromosome pairing (Slatis 1955; Henikoff and Dreesen 1989). In the case of the Byron bwD bw+ duplication, the heterochromatic block causes variegation of the bw+ genes in cis and in trans. Females of genotype bwD; st or Byron sp/CyO; st were concurrently crossed to st1 Df(3R)D1C12w−/TM3, Sb st and st1 P{SUPorP}pumKG02239/TM3, Sb st males. The st1 P{SUPorP}pumKG02239 chromosome is the progenitor for Df(3R)D1C12w−. The male and female (Cy+) progeny of each pair of crosses were compared between genotypes, with the Sb− progeny serving as an internal control. No differences in eye pigmentation were observed in the progeny due to haploinsufficiency for D1 (when sorted by age and sex; data not shown).
Conservation of the D1 protein:
As described above, no noticeable phenotypes were manifest by D1-null flies that might hint at the function of the D1 protein. Furthermore, the results indicated that D1 is not a modifier of PEV. To potentially gain insight into D1 function, in the absence of phenotypic data, a comparative genomics approach was applied. Homologs of the D. melanogaster D1 protein appeared limited to the Drosophila genus, using standard protein similarity search tools. The D1 homolog was identified within the genomes of the other 11 sequenced Drosophila species (see materials and methods; Richards et al. 2005; Clark et al. 2007). The amino acid identity and similarity to D. melanogaster D1, as shown in Table 5, was low as compared to the median identity for D. melanogaster homologs in each species (Heger and Ponting 2007). Multiple sequence alignment of the 12 proteins revealed that similar amino acids almost exclusively localized to the AT-hook DNA-binding motifs. Each protein had 10 ± 1 predicted AT hooks similarly distributed throughout the protein, as illustrated in Figure 6. There were no additional functional motifs predicted for any of the proteins, a characteristic of a subset of AT-hook proteins including the HMGA family (Aravind and Landsman 1998). While the amino acid sequence was not well conserved, per se, the preponderance of charged amino acids was. Positively and negatively charged residues accounted for between 34.7 and 37.4% of the total (Table 5).
TABLE 5.
Comparison of D1 proteins of Drosophila
| Speciesa | Length (aa) | % identity | % similarity | % Asp + Glu | % Arg + Lys |
|---|---|---|---|---|---|
| D. melanogaster | 355 | — | — | 18.3 | 17.7 |
| D. simulans | 352 | 88 | 92 | 18.2 | 17.6 |
| D. sechellia | 353 | 90 | 94 | 18.7 | 18.1 |
| D. yakuba | 354 | 81 | 86 | 18.6 | 17.2 |
| D. erecta | 360 | 74 | 81 | 18.6 | 16.9 |
| D. ananassae | 353 | 47 | 56 | 17.3 | 17.6 |
| D. pseudoobscura | 350 | 41 | 51 | 18.9 | 18.6 |
| D. persimilis | 350 | 41 | 51 | 18.9 | 18.6 |
| D. willistoni | 411 | 35 | 48 | 19.5 | 16.1 |
| D. mojavensis | 415 | 40 | 51 | 19.3 | 17.3 |
| D. virilis | 441 | 42 | 51 | 19.3 | 16.6 |
| D. grimshawi | 417 | 35 | 45 | 19.2 | 18.0 |
The order of species reflects the phylogeny (http://insects.eugenes.org/species/).
Figure 6.—
AT-hook organization of the D1 proteins of 12 Drosophila species. The D1 protein sequences are drawn to relative scale as rectangles. The AT-hook motifs predicted by Pfam (http://pfam.janelia.org/) are illustrated as shaded boxes, with light shading indicating matches of lower confidence. The proteins are ordered to reflect the evolutionary relatedness of the species (http://insects.eugenes.org/species/).
DISCUSSION
The experiments described herein revealed that the D1 gene of D. melanogaster was not required for viability. The homozygous lethality of the original chromosomes bearing either of two different P insertions in the D1 gene was determined to be due to other lethal mutations on the chromosomes. Indeed, the D1EP473 and D1EY05004 P-insertion alleles were found to be hemizygous viable. However, neither P-element insertion disrupted the D1 coding region, and both D1 protein and RNA were readily detected for the D1EP473 allele (Figure 3 and data not shown). Consequently, to determine if the D1 gene were essential, it was necessary to generate a null allele. Several genetic approaches were undertaken, including the imprecise P-element excision strategy and P-element-mediated male recombination HEI strategy described herein, to isolate D1 mutant alleles that removed the D1 coding region, as such alleles would be unarguably null for function. These approaches were successful in generating two small deletion alleles. Thus, the requirement for D1 for viability could be unequivocally assessed. The results of complementation analysis demonstrated that flies heterozygous for the two D1-null alleles were fully viable, being recovered from a cross at expected frequency (Table 3). The different genetic backgrounds of the two D1-null alleles effectively eliminated any phenotypic contribution by second-site mutations. As the D1 gene product is maternally loaded into the oocyte during oogenesis (Renner et al. 2000; Aulner et al. 2002), it was formally possible that the maternal contribution was sufficient for embryogenesis in the absence of zygotic D1 expression in null embryos. However, I observed that D1-null females were fertile, indicating that maternal expression of D1 was not required for oogenesis or early embryogenesis. In support of these conclusions, both Df(3R)D1C12w− and Df(3R)D1C12w−/Df(3R)Exel6152 flies were maintained as stocks for many generations.
It is difficult to reconcile these results with those of Aulner et al. (2002), who reported that excision of the P{EP}D1EP473 transposon restores homozygous viability to the chromosome. In case there might be differences between P{EP}D1EP473 fly cultures, I obtained a copy of the stock from the Szeged Drosophila Stock Center, the likely source for this group. The reversion studies were repeated using this stock with the same results. Precise excision of the transposon did not revert the lethality, and the Szeged P{EP}D1EP473 insertion was lethal in combination with the D1Rev1B allele, a precise excision of the Bloomington Stock Center P{EP}D1EP473 insertion (data not shown). Thus, copies of the stock derived from both locations shared chromosome 3 lethal mutations. The additional observation of Aulner et al. (2002) that a heat-shock promoter-driven D1 cDNA transgene could partially rescue the lethality of D1EP473 homozygotes could potentially be explained by changes in gene expression mediated by ectopic D1 expression (see below).
It is likely that multiple lethal mutations were present on the original P{EP}D1EP473 chromosome. The viability of P{EP}D1EP473/Df(3R)BSC24 flies indicated that the mutations were not closely linked to the P insertion. P-element-mediated male recombination was employed to replace the chromosomal regions to either side of the insertion with that of the homolog. However, homozygous lethality persisted for both classes of single recombinants—those that had replaced 3L and proximal 3R and those that had replaced distal 3R (data not shown). By maintaining the P{EP}D1EP473 insertion heterozygous with the Df(3R)Exel6152 chromosome for many generations, a homozygous viable P{EP}D1EP473 chromosome was eventually recovered.
The dispensability of the D1 gene for development suggests that it has overlapping function(s) with other genes. Functional redundancy was observed for the products of the HMGB genes of Drosophila, HMGZ and HMGD. Although these two proteins do not share sequence similarity with D1, they share some biochemical properties and have similarly been proposed to play an architectural role in chromatin (Grosschedl et al. 1994; Renner et al. 2000; Aleporou-Marinou et al. 2003). The HMGZ HMGD double mutant has only minor phenotypic defects and, surprisingly, no severe phenotypes were revealed in combination with null alleles of one or more other HMGB genes (although these studies were limited by available mutant alleles; Ragab et al. 2006). The mammalian HMGA genes, HMGA1 and HMGA2, are similarly not essential for viability. Developmental abnormalities are associated with loss of either gene, although the null phenotypes are distinct (Zhou et al. 1995; Foti et al. 2005; Fedele et al. 2006). Whereas there are five HMGB genes in Drosophila and two HMGA genes in mammals, D1 appears to be the only HMGA-like gene of Drosophila. At the level of protein architecture, the D1 protein appears unique in having 10 predicted copies of the AT-hook motif. Most of the AT-hook proteins of D. melanogaster have 1 or 2 copies of this motif, with only ASH1 protein having 3. Unlike the D1 protein, these proteins (including ASH1 protein) typically possess additional functional motifs. If the function(s) of the D1 protein rely on its ability to bind to AT tracts, then perhaps proteins with similar DNA binding properties mediated by other motifs share in its activities.
HMGA-like features of D1 proteins:
The D1 proteins of other Drosophila species having sequenced genomes were identified on the basis of protein sequence homology, but sequence similarity rapidly declined with increasing evolutionary distance (Table 5). It was consequently not surprising that D1 protein homologs were not identified in other genera using sequence homology. However, the comparison and alignment of the drosophilid D1 proteins suggested that other features of the protein, such as the density of AT-hook motifs and/or the frequency of charged amino acids, might more appropriately be the defining criteria for a D1 protein family. Perhaps not coincidentally, these are among properties shared with mammalian HMGA proteins. In this regard, it is relevant that the HMGA1 and HMGA2 proteins are only ∼50% similar to each other (in both mouse and human) and that this similarity is primarily in the three AT-hook regions and acidic C terminus (Reeves and Beckerbauer 2001). Indeed, the numerous similarities between the D1 and HMGA proteins suggest that they might share functional, rather than evolutionary, relatedness. In addition to the shared biochemical properties already noted, the D1 protein is predicted to have extensive intrinsic protein disorder (Uversky et al. 2005; data not shown), a demonstrated attribute of HMGA proteins (Lehn et al. 1988; Huth et al. 1997). Both D1 and HMGA proteins are highly post-translationally modified (Zhai et al. 2008; Zhang and Wang 2008). The primary distinction, increased size for D1, is accompanied by a proportional increase in number of AT-hook motifs. Although the HMGA proteins do not have intrinsic transcriptional regulatory activity, they have been shown to regulate the activity of many genes as architectural proteins (Reeves and Beckerbauer 2001). A potential similar gene regulatory role for the D1 protein is supported by the finding, in this study, of decreased Sb1 expression (enhancement of Sb variegation) in a D1-mutant background.
CG9746 is essential:
This work revealed that predicted gene CG9746 is essential for viability. The three D1 mutants obtained through imprecise excision of the P{EP}D1EP473 insertion, two deletions and an inversion, disrupted the neighboring CG9746 gene, as well. All were lethal in combination with the ∼89-kb deficiency Df(3R)Exel6152. In contrast, these D1 mutants were viable in combination with the small Df(3R)D1C12− deficiency, which deleted the D1 gene and noncoding sequences of the pum gene, but not CG9746. The lethality of the In(3R)D11A/Df(3R)Exel6152 mutant flies in particular indicated that CG9746 is an essential gene, as no other gene was affected by this inversion. The sequence of the CG9746 gene predicts that it encodes a protein serine/threonine kinase.
D1 and PEV:
As the D1 protein is enriched in the heterochromatin, I sought to test the hypothesis that mutant alleles of the D1 gene might be haplosuppressors of PEV (reviewed by Weiler and Wakimoto 1995; Schotta et al. 2003). However, I realized that a potential effect of D1 mutations on PEV of In(1)wm4, the variegating rearrangement most commonly used to assess potential PEV modifiers, could reflect the local influence of the block of 359-bp satellite repeat sequence normally present at the heterochromatic base of the X chromosome (Hilliker and Appels 1982). This repeat is a high-affinity binding site for the D1 protein (Levinger and Varshavsky 1982b). For this reason, this study included several w-variegating alleles that were molecularly and cytologically characterized by Talbert and Henikoff (2000). The w locus of the wm51b rearranagement is juxtaposed to the 359-bp satellite block, whereas it is separated from it by rDNA for the In(1)wm4 chromosomes (Tartof et al. 1984; Talbert and Henikoff 2000). In contrast, the 359-bp satellite block was determined to be absent from the In(1)wmmcT inversion chromosome (Talbert and Henikoff 2000). The tests failed to show any dominant effect of mutations in the D1 gene on variegation of w associated with In(1)wm4, In(1)wmmcT, or In(1)wm51b. Hence, my findings contradict those of Aulner et al. (2002) who reported suppression of wm4 variegation by the P{EP}D1EP473 insertion. One difference between experiments of the two laboratories is that the P{EP}D1EP473 insertion tested by Aulner et al. (2002) expressed the w marker gene, whereas both D1 mutant alleles tested herein were w−. My approach of assessing wm4 variegation in a w− background obviates the need for methods to subtract the effects of extraneous w-gene activity and thus makes data interpretation straightforward. A second difference is the possibility that their results were influenced by a maternal effect, as their tests for modification of PEV involved at least one strain bearing the TM6B balancer chromosome, which harbors a mutation in the E(var)3-9 gene (Weiler 2007). Although I was unable to deduce the details of the crosses that were performed, the enhancing effect of an E(var)3-9 mutation could make a wild-type chromosome appear to have a suppressor phenotype by comparison. A third difference is that the reported suppression of PEV attributed to the P{EP}D1EP473 insertion might actually map elsewhere on the chromosome. In this report, the P{EP}D1EP473w− insertion is compared to an isogenic D1+ control.
The isolation of new D1 mutant alleles that were viable in combination (this report) enabled a test for potential recessive effects of D1 mutations on PEV. To this end, D1-null females were crossed to wm4 males bearing isogenic D1+ or D1− chromosomes, and the w variegation of the D1+ and D1− female progeny was compared. Neither D1EP473w−/Df(3R)D1C12w− nor Df(3R)D14A/Df(3R)D1C12w− females showed suppression (or enhancement) of wm4 variegation in comparison to their respective controls (data not shown). Hence, D1 is not a recessive modifier of wm4.
Additional tests for dominant modification of PEV by D1 mutant alleles were performed using the brown-variegating rearrangements bwD and Byron, and the Stubble-variegating rearrangement, T(2;3)SbV. The former set of crosses assess for the potential requirement for D1 in trans-inactivation and both trans-inactivation and para-inactivation of bw, respectively. For these tests, flies bearing the D1-null chromosome Df(3R)D1C12w− were compared to those bearing the D1+ progenitor chromosome. No effect was observed on bw variegation for either rearrangement (data not shown). In contrast, both the P{EP}D1EP473 and the Df(3R)D14A chromosomes were found to enhance Sb variegation in comparison to the isogenic D1Rev1B wild-type control chromosome (Table 4).
Collectively, the assays for an effect on PEV by D1 mutant alleles indicated that D1 is not a modifier of PEV. Variegation associated with five of the six rearrangements tested was not affected by mutation of D1. Several of these assays included D1-null alleles. In addition, Aulner et al. (2002) noted seeing no significant effect of D1EP473 on PEV for two other rearrangements that they tested, not included in this study. Enhancement of Sb variegation, the only effect observed in these studies, most likely reflects a role for the D1 protein in promoting transcription of the Sb gene. It has previously been postulated that D1 could regulate gene expression via binding to AT-rich promoter elements (Levinger 1985b). As noted above, a gene regulatory function has been clearly demonstrated for HMGA proteins (Reeves and Beckerbauer 2001). The number and spacing of AT tracts required for D1 binding has not been established. However, there are 25 AT tracts extending five or more bases within 1 kb 5′ of the transcription start of the Sb gene. Thus, positive regulation of Sb gene expression by the D1 protein is a formal possibility.
Acknowledgments
K.S.W. acknowledges the research efforts of numerous undergraduate students at Idaho State University, in particular Annie Bankhead, who participated in genetic screens to isolate D1 mutants. Kami Ahmad, Paul Talbert, and Steve Henikoff are thanked for providing fly strains as are the Bloomington and Szeged Drosophila Stock Centers. This work was supported in part by a National Science Foundation grant (award nos. MCB0131604 and MCB0531808).
References
- Aleporou-Marinou, V., H. Marinou and T. Patargias, 2003. A mini review of the high mobility group proteins of insects. Biochem. Genet. 41: 291–304. [DOI] [PubMed] [Google Scholar]
- Alfageme, C. R., A. Zweidler, A. Mahowald and L. H. Cohen, 1974. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J. Biol. Chem. 249: 3729–3736. [PubMed] [Google Scholar]
- Alfageme, C. R., G. T. Rudkin and L. H. Cohen, 1976. Locations of chromosomal proteins in polytene chromosomes. Proc. Natl. Acad. Sci. USA 73: 2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and D. Landsman, 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26: 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, C. T., C. G. Pendleton, W. W. Jennings, A. Saxena and C. V. Glover, 1989. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J. Biol. Chem. 264: 8394–8401. [PubMed] [Google Scholar]
- Aulner, N., C. Monod, G. Mandicourt, D. Jullien, O. Cuvier et al., 2002. The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation. Mol. Cell. Biol. 22: 1218–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli, V. R., J. Wang and J. A. Dow, 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. B. Eisen, D. R. Smith, C. M. Bergman, B. Oliver et al., 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Cortes, A., D. Huertas, L. Fanti, S. Pimpinelli, F. X. Marsellach et al., 1999. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 18: 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice, B., G. Pontecorvo and M. Carfagna, 1999. Identification of a new gene encoding pericentromeric dodeca-satellite binding protein in Drosophila melanogaster. FEBS Lett. 455: 31–35. [DOI] [PubMed] [Google Scholar]
- Fedele, M., V. Fidanza, S. Battista, F. Pentimalli, A. J. Klein-Szanto et al., 2006. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 66: 2536–2543. [DOI] [PubMed] [Google Scholar]
- Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich et al., 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti, D., E. Chiefari, M. Fedele, R. Iuliano, L. Brunetti et al., 2005. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat. Med. 11: 765–773. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and M. M. Golic, 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., M. M. Tanaka and J. A. Sved, 1996. P-element-induced recombination in Drosophila melanogaster: hybrid element insertion. Genetics 144: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigliatti, T., 1991. Position-effect variegation–an assay for nonhistone chromosomal proteins and chromatin assembly and modifying factors. Methods Cell. Biol. 35: 587–627. [PubMed] [Google Scholar]
- Grosschedl, R., K. Giese and J. Pagel, 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10: 94–100. [DOI] [PubMed] [Google Scholar]
- Heger, A., and C. P. Ponting, 2007. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 17: 1837–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and T. D. Dreesen, 1989. Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc. Natl. Acad. Sci. USA 86: 6704–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., J. M. Jackson and P. B. Talbert, 1995. Distance and pairing effects on the brownDominant heterochromatic element in Drosophila. Genetics 140: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., and R. Appels, 1982. Pleiotropic effects associated with the deletion of heterochromatin surrounding rDNA on the X chromosome of Drosophila. Chromosoma 86: 469–490. [DOI] [PubMed] [Google Scholar]
- Huth, J. R., C. A. Bewley, M. S. Nissen, J. N. Evans, R. Reeves et al., 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4: 657–665. [DOI] [PubMed] [Google Scholar]
- James, T. C., and S. C. Elgin, 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehn, D. A., T. S. Elton, K. R. Johnson and R. Reeves, 1988. A conformational study of the sequence specific binding of HMG-I (Y) with the bovine interleukin-2 cDNA. Biochem. Int. 16: 963–971. [PubMed] [Google Scholar]
- Levinger, L., 1985. a Nucleosomal structure of two Drosophila melanogaster simple satellites. J. Biol. Chem. 260: 11799–11804. [PubMed] [Google Scholar]
- Levinger, L. F., 1985. b D1 protein of Drosophila melanogaster. Purification and AT-DNA binding properties. J. Biol. Chem. 260: 14311–14318. [PubMed] [Google Scholar]
- Levinger, L., and A. Varshavsky, 1982. a Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc. Natl. Acad. Sci. USA 79: 7152–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger, L., and A. Varshavsky, 1982. b Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell 28: 375–385. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab, A., E. C. Thompson and A. A. Travers, 2006. High mobility group proteins HMGD and HMGZ interact genetically with the Brahma chromatin remodeling complex in Drosophila. Genetics 172: 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, R., and L. Beckerbauer, 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519: 13–29. [DOI] [PubMed] [Google Scholar]
- Reeves, R., and M. S. Nissen, 1990. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265: 8573–8582. [PubMed] [Google Scholar]
- Renner, U., S. Ghidelli, M. A. Schafer and J. R. Wisniewski, 2000. Alterations in titer and distribution of high mobility group proteins during embryonic development of Drosophila melanogaster. Biochim. Biophys. Acta 1475: 99–108. [DOI] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Alfageme, C., G. T. Rudkin and L. H. Cohen, 1980. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma 78: 1–31. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, R. Dorn and G. Reuter, 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14: 67–75. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A. R., R. C. Mottus and T. A. Grigliatti, 1983. Genes which suppress position effect variegation in Drosophila melanogaster are clustered. Mol. Gen. Genet. 191: 326–333. [Google Scholar]
- Slatis, H. M., 1955. A reconsideration of the Brown-Dominant position effect. Genetics 40: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P. B., and S. Henikoff, 2000. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics 154: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof, K. D., C. Hobbs and M. Jones, 1984. A structural basis for variegating position effects. Cell 37: 869–878. [DOI] [PubMed] [Google Scholar]
- Tweedie, S., M. Ashburner, K. Falls, P. Leyland, P. McQuilton et al., 2009. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37: D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky, V. N., C. J. Oldfield and A. K. Dunker, 2005. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18: 343–384. [DOI] [PubMed] [Google Scholar]
- Weiler, K. S., 2007. E(var)3–9 of Drosophila melanogaster encodes a zinc finger protein. Genetics 177: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- Zhai, B., J. Villen, S. A. Beausoleil, J. Mintseris and S. P. Gygi, 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., and Y. Wang, 2008. High mobility group proteins and their post-translational modifications. Biochim. Biophys. Acta 1784: 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., K. F. Benson, H. R. Ashar and K. Chada, 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774. [DOI] [PubMed] [Google Scholar]