Abstract
Phytochrome interacting factors (PIFs) are nuclear basic helix-loop-helix (bHLH) transcription factors that negatively regulate photomorphogenesis both in the dark and in the light in Arabidopsis. The phytochrome (phy) family of photoreceptors induces the rapid phosphorylation and degradation of PIFs in response to both red and far-red light conditions to promote photomorphogenesis. Although phys have been shown to function under blue light conditions, the roles of PIFs under blue light have not been investigated in detail. Here we show that PIF1 negatively regulates photomorphogenesis at the seedling stage under blue light conditions. pif1 seedlings displayed more open cotyledons and slightly reduced hypocotyl length compared to wild type under diurnal (12 hr light/12 hr dark) blue light conditions. Double-mutant analyses demonstrated that pif1phyA, pif1phyB, pif1cry1, and pif1cry2 have enhanced cotyledon opening compared to the single photoreceptor mutants under diurnal blue light conditions. Blue light induced the rapid phosphorylation, polyubiquitination, and degradation of PIF1 through the ubi/26S proteasomal pathway. PIF1 interacted with phyA and phyB in a blue light-dependent manner, and the interactions with phys are necessary for the blue light-induced degradation of PIF1. phyA played a dominant role under pulses of blue light, while phyA, phyB, and phyD induced the degradation of PIF1 in an additive manner under prolonged continuous blue light conditions. Interestingly, the absence of cry1 and cry2 enhanced the degradation of PIF1 under blue light conditions. Taken together, these data suggest that PIF1 functions as a negative regulator of photomorphogenesis under blue light conditions and that blue light-activated phys induce the degradation of PIF1 through the ubi/26S proteasomal pathway to promote photomorphogenesis.
PLANTS modulate their growth and development in response to the surrounding light environment. Plants can track the intensity, color, direction, duration, and overall day/night cycles of incoming light signals through an array of photoreceptors. These photoreceptors include phytochromes (phys) that primarily respond to the red and far-red regions of the light spectrum; phototrophins (phot), cryptochromes (cry), and the ZTL/FKF1/LKP2 family of F-box proteins to monitor the UVA-blue light region; and an unidentified photoreceptor to respond to the UV-B light (Lin and Shalitin 2003; Chen et al. 2004; Schaefer and Nagy 2006; Demarsy and Fankhauser 2008). The coordinated function of these photoreceptors helps optimize growth and development throughout the plant's life cycle.
In Arabidopsis thaliana, five genes (PHYA–PHYE) encode the phy family members (Mathews and Sharrock 1997). phys exist in two photoreversible dimeric forms: a red light-absorbing Pr form (biologically inactive) and a far-red light-absorbing Pfr form (biologically active) (Schaefer and Nagy 2006). All phy family members are activated by red light, while phyA is activated by both red and far-red light signals (Quail 2007b). phy responses have been classified into three modes: very low fluence response (VLFR), low fluence response (LFR), and high irradiance response (HIR). VLFR responses achieve saturation by exposure to a brief pulse of light and are not photoreversible. LFRs are red/far-red reversible responses induced by low light intensities, and HIR responses are intensity-dependent, nonphotoreversible responses to high light intensities (Casal et al. 1998).
phys are differentially regulated at the post-translational level and at subcellular localization in response to light. For example, phyA is unstable under light and is the most abundant phytochrome in dark-grown seedlings, while phyB–phyE are relatively stable under light and are present in light-grown plants (Whitelam and Halliday 2007). Photoactivation of phys triggers a conformational change that induces the phys to be translocated into the nucleus (Fankhauser and Chen 2008). The light-triggered nuclear translocation has been shown to be necessary for the biological functions of both phyA and phyB (Huq et al. 2003; Matsushita et al. 2003; Rösler et al. 2007). However, cytosolic phyA has been shown to regulate negative gravitropism under blue light, as well as red light-induced enhancement of the blue light-mediated phototropism (Rösler et al. 2007). phys interact with a variety of nuclear proteins and initiate a signal transduction pathway that ultimately regulates ∼10% of the genome to promote photomorphogenesis (Jiao et al. 2007; Quail 2007a,b; Whitelam and Halliday 2007).
Within the nucleus, phys interact with a group of constitutively nuclear-localized basic helix-loop-helix transcription factors called phytochrome interacting factors (PIFs) (Castillon et al. 2007; Bae and Choi 2008; Leivar et al. 2008a). PIFs interact with the biologically active Pfr forms of phyA and phyB using two discrete motifs, namely, the active phyB binding motif (APB) and the active phyA binding motif (APA) that are present at the N terminus of PIFs. PIFs have been shown to act as negative regulators of photomorphogenesis both in the dark and in the light (Castillon et al. 2007; Bae and Choi 2008; Leivar et al. 2008a,b; Shen et al. 2008). To remove this negative regulation, the light-activated Pfr forms of phys physically interact with the PIFs, and induce the phosphorylation, polyubiquitination, and degradation of PIFs by the 26S proteasome-mediated pathway, and thereby promote photomorphogenesis. Strikingly, direct physical interactions with phys are necessary but not sufficient for the light-induced phosphorylation and degradation of PIFs (Al-Sady et al. 2006; Shen et al. 2008).
Although phys are best known to function under red and far-red light conditions, they have also been shown to function under blue light conditions (Casal 2000; Lin 2000). The absorption and action spectra for phys show a distinct peak in the blue light region (Vierstra and Quail 1983; Mancinelli 1994; Shinomura et al. 1996; Rockwell et al. 2006). Genetic evidence demonstrated that the phy and cry family members display both synergistic and antagonistic behavior at the seedling and adult stages. Analyses of photoreceptor mutants demonstrated that, under prolonged light exposure, both phyA and phyB regulate blue light-mediated seedling de-etiolation in an overlapping manner with cry1 and cry2 (Casal and Mazzella 1998; Neff and Chory 1998). phys and crys also displayed synergistic action in regulating blue light-induced anthocyanin production and root greening at the seedling stage (Usami et al. 2007). However, phyB has been shown to oppose the cry1/phyA-mediated inhibition of hypocotyl elongation under blue light conditions (Folta and Spalding 2001). phyB and cry2 antagonistically regulate flowering time, while phyA and cry2 promote flowering time under long days (Mockler et al. 1999; Lin 2000). These photoreceptors also function to entrain the circadian clock (Somers et al. 1998), which independently controls seedling de-etiolation and flowering time (Imaizumi and Kay 2006; Nozue et al. 2007; McClung 2008).
In addition to their overlapping physiological roles, members of the phy and cry families have been shown to physically interact with each other in vivo. For example, phyA interacts with cry1 (Ahmad et al. 1998), while phyB binds with cry2 (Mas et al. 2000). phyB, cry1, and cry2 have been shown to interact with a common signaling partner, constitutive photomorphogenic 1 (COP1) (Yang et al. 2001), suggesting that both photoreceptor families might directly inhibit the negative regulator COP1 to promote photomorphogenesis.
Although the physiological roles of phys have been investigated under blue light conditions, the roles of phy signaling factors in blue light are less understood. HFR1, a basic helix-loop-helix (bHLH) factor isolated as a positive regulator of the far-red (FR)-specific pathway, functions positively in a blue light signaling pathway (Duek and Fankhauser 2003). PIF4, a phyB-interacting bHLH factor, negatively regulates blue light signaling (Kang and Ni 2006). However, the molecular details of how PIF4 and/or other PIFs are regulated by blue light are still unknown. Here we show that PIF1, the PIF family member with the highest affinity for both phyA and phyB, functions negatively to repress seedling de-etiolation under blue light conditions. In addition, we show that blue light-activated phys induce the phosphorylation, polyubiquitination, and subsequent degradation of PIF1 through the ubi/26S proteasomal pathway to promote photomorphogenesis.
MATERIALS AND METHODS
Plant growth conditions and phenotypic analyses:
Plants were grown in Metro-Mix 200 soil (Sun Gro Horticulture, Bellevue, WA) under 24 hr light at 24° ± 0.5°. Monochromatic blue light treatments were performed in growth chambers equipped with light-emitting diodes (LEDs) (model E30LED; Percival Scientific, Madison, WI) as described (Shen et al. 2005). Light fluence rates were measured using a spectroradiometer (model EPP2000; StellarNet, Tampa, FL) as described (supporting information, Figure S1) (Shen et al. 2005). For transgenic plants, the 35S:LUC-PIF1 (LP), 35S:LUC-PIF1-3M (LP-3M), and 35S:TAP-PIF1 (TP) lines were generated as described (Shen et al. 2005, 2008; Moon et al. 2008). Seeds were surface sterilized and plated on Murashige–Skoog (MS) growth medium (GM) containing 0.9% agar without sucrose (GM −Suc) as described (Shen et al. 2005). After 3–4 days of stratification at 4° in the dark, seeds were exposed to 3 hr white light at room temperature to induce germination and kept in the dark for 21 hr. The plates were then placed in the dark or under continuous blue light or under diurnal (12 hr light/12 hr dark) blue light conditions for an additional 3 days.
Cotyledon angles were measured by gently placing the seedlings on adhesive tape facing upward. Digital photographs were taken through the dissection microscope and the angle formed between the two cotyledon tips was measured with the angle tool of ImageJ (1.37v, Wayne Rasband, National Institutes of Health). Measurements for hypocotyl length were performed with ImageJ, using the segmented line selections tool.
Protein extraction and Western blotting:
Protein extraction and Western blotting were performed as described (Shen et al. 2008). Briefly, for blue light-mediated degradation, 4-day-old dark-grown seedlings were exposed to a pulse of 10 μmol m−2 or 30 μmol m−2 and kept in the dark for the indicated time periods. For the experiments requiring exposure to continuous blue light, dark-grown seedlings were exposed to 10 μmol m−2 sec−1 of blue light for the indicated time periods before harvesting for protein extraction. Tissue (0.2 g) was collected and ground in 1 ml of extraction buffer [0.1 m Tris-HCl pH 6.8, 20% glycerol, 5% SDS, 0.01 m MG132, 0.2 m DTT, 2 mm PMSF, and 1× proteinase inhibitors (complete mini, 11836170001; Roche, Indianapolis)] and boiled for 2 min. Samples were run on an 8% SDS–PAGE gel and blotted onto PVDF membrane. Another gel was run in parallel as a loading control. The Western blot procedure was carried out according to manufacturer's instructions, using a KPL Protein Detector kit (54-13-50; KPL, Gaithersburg, MD), utilizing 1:5000 dilutions of anti-PIF1 antibody, and 1:2500 anti-tubulin (Sigma-Aldrich, St. Louis) as loading control. Peroxidase-labeled goat anti-rabbit antibody (KPL) in a 1:50,000 dilution was used as secondary antibody. For the immunoblot analyses to detect ubiquitination and phosphorylation, the membranes were blocked with 1× TBST plus 2% nonfat milk buffer followed by incubation with different primary antibodies in 1× TBST plus 0.5% nonfat milk buffer. Anti-ubiquitin (1:700; Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-c-MYC (1:800) (Sigma-Aldrich) were used at 4° overnight. For secondary antibody, peroxidase-labeled goat anti-rabbit antibody (1:4000; Pierce Biotechnology, Rockford, IL) or anti-mouse IgG HRP conjugate (1:3300) (Promega, Madison, WI) was used, and membranes were developed with a SuperSignal West Pico Chemiluminescent substrate kit (Pierce Biotechnology).
Immunoprecipitation and alkaline phosphatase treatment:
Immunoprecipitation and alkaline phosphatase treatment were performed essentially as described (Shen et al. 2008). Briefly, for pretreatment with MG132, 4-day-old dark-grown TAP-PIF1 seedlings were transferred into MS −Suc liquid media containing 30 μm MG132 or an equal volume of solvent control DMSO and incubated in the dark for 5 hr. Total proteins were extracted from ∼0.4-g seedlings (either kept in darkness or treated with pulses of blue light followed by dark) with 1 ml denaturing buffer [100 mm NaH2PO4, 10 mm Tris (pH 8.0), 100 mm NaCl, 8 m urea, 0.05% Tween-20, 1× Protease inhibitor cocktail (F. Hoffmann-La Roche, Basel, Switzerland), 2 mm PMSF, 10 μm MG132, 25 mm β-GP, 10 mm NaF, 2 mm Na-orthovanadate, and 100 nm calyculin A] and cleared by centrifugation at 14,000 rpm for 15 min at 4°. TAP-PIF1 was immunoprecipitated from supernatants with Ni-NTA magnetic agarose beads (QIAGEN, Valencia, CA) following incubation for 3 hr at 4°. After washing, the pellet was resuspended in 100 μl calf intestinal alkaline phosphatase (CIAP) reaction buffer and then treated either with 100 units CIAP (F. Hoffmann-La Roche) or the same amount of boiled CIAP or without enzyme for 60 min at 37°. Pellets were washed with PBS buffer, heated at 65° in 1× SDS–Laemmli buffer for 5 min, and subjected to Western blot analysis with anti-c-MYC or anti-ubiquitin antibody as described above.
Luciferase assay:
Luciferase assays were performed as described (Shen et al. 2005, 2008). Briefly, samples were collected in liquid nitrogen and total protein was extracted using 1× Luciferase Cell Culture Lysis Reagent (CCLR) (Promega) with 2 mm PMSF and 1× complete protease inhibitor cocktail (F. Hoffmann-La Roche). For cycloheximide chase assays, 4-day-old dark-grown seedlings were pretreated with 50 μm cycloheximide in MS −Suc liquid medium for 3 hr in the dark as described (Shen et al. 2005). After pretreatment, the seedlings were exposed to a pulse of blue light (30 μmol m−2) and kept in darkness before harvesting for the time points indicated in Figure 7C.
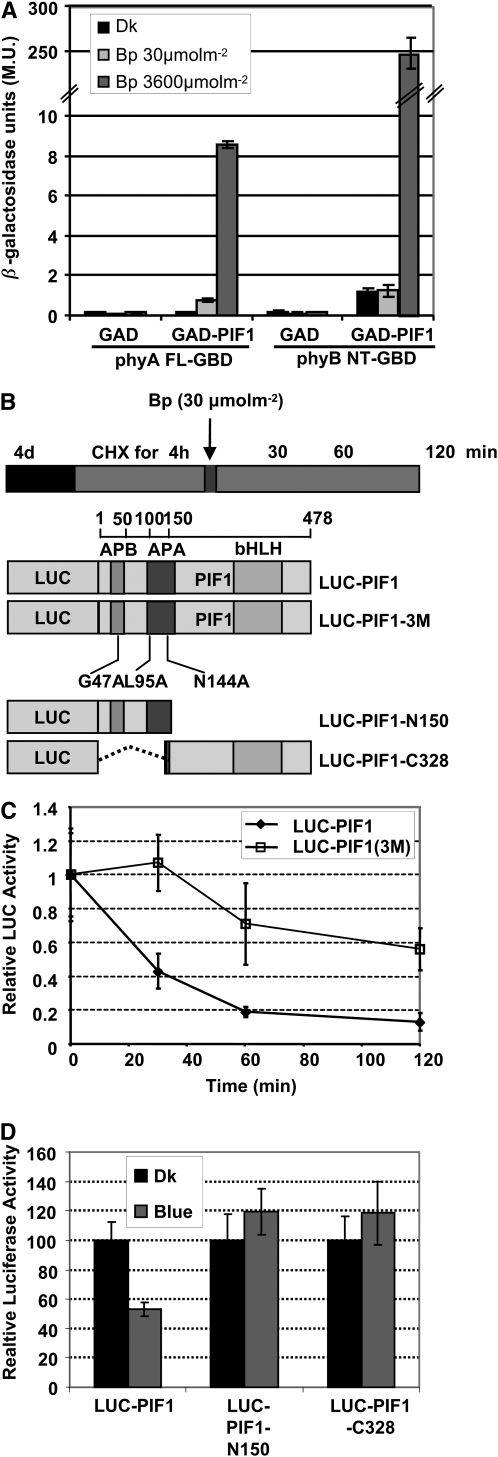
Figure 7.—
Interactions with the Pfr form of phyA and phyB are necessary for the light-induced degradation of PIF1. (A) PIF1 interacts with phyA and phyB-NT in a blue light-dependent manner in quantitative yeast two-hybrid assays. LacZ assays were performed in triplicate and the data represent mean ± SE. Yeast cells were exposed to pulses of blue light as indicated and then incubated in the dark for an additional 3 hr before performing the LacZ assay. M.U., Miller units. phyB-NT is the N-terminal half (1–621 amino acids) of phyB. (B) Top, design of the cycloheximide chase assays. Bottom, schematic representation of the full-length, truncated, and missense mutant forms of LUC-PIF1 fusion proteins used in the experiment. (C) Relative Luciferase activity for phy-interaction-deficient mutants was measured in 4-day-old dark-grown seedlings pretreated with cycloheximide (CHX) in the dark for 3 hr, exposed to a pulse of blue light (30 μmol m−2), and then incubated in the dark for the indicated time (min). Assays show the kinetics of degradation of LUC-PIF1-3M compared to wild-type LUC-PIF1. LUC-PIF1-3M is deficient in both phyA and phyB interaction. Means ± SE of three biological replicates are shown. (D) Relative Luciferase activity for the truncated versions of PIF1 fusion proteins compared to the wild-type LUC-PIF1 fusion protein. Four-day-old dark-grown seedlings were exposed to a pulse of blue light (30 μmol m−2) and then incubated in the dark for 60 min before harvesting for protein extraction and Luciferase assays.
Light-dependent yeast two-hybrid assays:
Light-dependent yeast two-hybrid assays were performed as described (Shimizu-Sato et al. 2002), except the yeast cells were exposed to pulses of blue light (30 or 3600 μmol m−2). Briefly, yeast cells (Y187) transformed with different constructs were grown overnight in synthetic dropout media with 25 μm phycocyanobilin in the dark. After adding YPAD media, these cultures were either kept in the dark or exposed to a pulse of blue light and returned to darkness for an additional 3 hr before assaying for LacZ reporter activity.
Isolation of RNA and RT–PCR:
Total RNA was isolated using an RNeasy Plant mini kit (QIAGEN) from 4-day-old wild-type Col-0 and pif1-2 mutant seedlings treated for different time periods under blue light (25 μmol m−2 sec−1). For RT–PCR, total RNA was treated with DNase I to remove genomic DNA. One microgram of total RNA was reverse transcribed using the RT–PCR kit from Invitrogen (Carlsbad, CA), and the first-strand cDNA was used as a template for PCR amplification. For semiquantitative gene expression, cDNAs were diluted to 40 μl with water and 1 μl of diluted cDNA was used for PCR amplification of PIF1 (forward 5′-CGAGATAACCGGTACATCGTCATC-3′ and reverse 5′-CATCATTGGCATCATTCCAC-3′), HY5 (forward 5′-GCTGCAAGCTCTTTACCATC-3′ and reverse 5′-AGCATCTGGTTCTCGTTCTG-3′), CHS (forward 5′-TCGGTCAGGCTCTTTTCAGT-3′ and reverse 5′-TGTCGCCCTCATCTTCTCTT-3′), and UBQ10 (forward 5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′ and reverse 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′) fragments using gene-specific primers. The UBQ10 fragment was used as a control to normalize the amount of cDNA used. For all cDNAs, the exponential range of amplification cycles for each gene was determined experimentally. Then 26 (PIF1), 27 (HY5), 24 (CHS), and 27 (UBQ10) cycles were used for the RT–PCR experiments. Two biological repeats were carried out for each gene. PCR products were separated on an agarose gel with ethidium bromide and imaged under UV light with an Alpha Innotech Imager.
RESULTS
PIF1 is a negative regulator of seedling de-etiolation under diurnal blue light conditions:
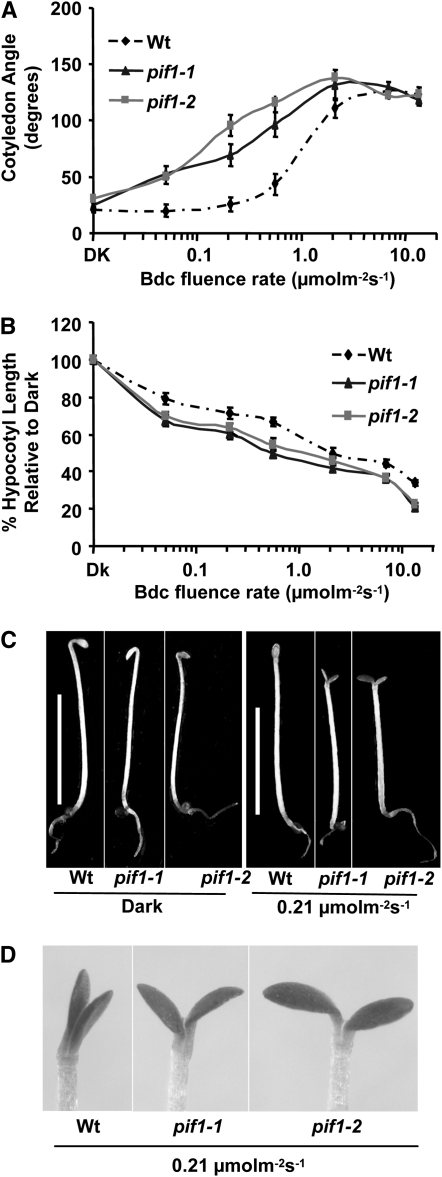
Because phys function under blue light conditions and PIF1 is the strongest interactor of both phyA and phyB, we investigated whether PIF1 plays any role in blue light signaling pathways. Under continuous blue light conditions, de-etiolation phenotypes of the pif1 mutant seedlings were similar to that of the wild type (Figure S2). However, under diurnal blue light conditions, both alleles of pif1 mutant seedlings displayed a hypersensitive phenotype compared to wild-type seedlings. Fluence rate response curves demonstrated that the angle between the cotyledons is significantly higher for pif1 seedlings compared to wild-type seedlings especially at lower fluence rates (Figure 1, A, C, and D). Hypocotyl lengths for the pif1 seedlings were also slightly shorter than those for the wild-type seedlings (Figure 1, B and C). However, the cotyledon areas of pif1 mutant and wild-type seedlings were similar under these conditions (data not shown). These data suggest that PIF1 negatively regulates blue light signaling under diurnal conditions.
Figure 1.—
pif1 seedlings are hypersensitive to blue light-induced seedling de-etiolation. Fluence-rate response curves are shown of mean cotyledon angles (A) and hypocotyl lengths (B) of wild-type (Col-0) and pif1 alleles grown for 4 days under either dark or diurnal (12 hr light/12 hr dark) blue light conditions. Data are presented as mean ± SEM (n ≥ 30, three replicates). Wt, wild-type Col-0. (C) Photographs of seedlings grown under diurnal (12 hr light/12 hr dark) blue light conditions (0.21 μmol m−2 sec−1) and dark conditions for 4 days. White bar, 5 mm. (D) Enlarged photographs of the apical regions of wild-type, pif1-1, and pif1-2 seedlings grown under conditions described in C.
The absence of PIF1 in phyA, phyB, cry1, and cry2 mutant backgrounds enhances photomorphogenic responses of these single photoreceptor mutants under diurnal blue light conditions:
To investigate whether the hypersensitive phenotypes observed for pif1 single mutants were phy or cry dependent, double-mutant combinations of phyApif1, phyBpif1, cry1pif1, and cry2pif1 were created by crossing the null allele of pif1 (pif1-2) with different photoreceptor mutants. Seedling de-etiolation phenotypes including hypocotyl lengths and cotyledon angles were measured under a range of continuous and diurnal blue light conditions. Under diurnal blue light conditions, all four photoreceptor single mutants (phyA, phyB, cry1, and cry2) displayed hyposensitive responses in suppression of hypocotyl elongation and expansion of cotyledon angles compared to the wild-type seedlings under a range of blue light intensities (Figure 2, A–L). Strikingly, the pif1 mutant suppressed all the above phenotypes of the phyA, phyB, cry1, and cry2 single mutants in varying degrees under these conditions (Figure 2, A–L). The cotyledon angles of the phyApif1 and cry1pif1 double mutants were similar to that of the wild-type seedlings under a wide range of blue light intensities (Figure 2, A–C and G–I). Under continuous blue light conditions, all four photoreceptor mutants displayed hyposensitive phenotypes in response to increasing light intensities (Figure S3). Under these conditions, pif1 suppressed the cotyledon angle phenotypes of the phyA and phyB mutant completely, but did not suppress the cotyledon angle phenotypes of the cry1 and cry2 mutants. pif1 suppressed the long hypocotyl phenotype of the phyB mutant, but did not suppress the long hypocotyl phenotypes of the phyA, cry1, and cry2 mutants under these conditions. These data suggest that PIF1 might function under multiple photoreceptors in suppressing the blue light-induced photomorphogenesis at the seedling stage.
Figure 2.—
The absence of PIF1 in phyA, phyB, cry1, and cry2 mutant background enhances photomorphogenic responses under diurnal (12 hr light/12 hr dark) blue light conditions. Fluence-rate response curves of mean cotyledon angles (A, D, G, and J) and hypocotyl lengths (B, E, H, and K) of wt (Col-0), pif1, pif1phyA (A–C), pif1phyB (D–F), pif1cry1 (G–I), and pif1cry2 (J–L) grown for 4 days under either dark or diurnal (12 hr light/12 hr dark) blue light conditions are shown. Hypocotyl lengths were normalized by setting the dark values to 100. Data are presented as mean ± SEM (n ≥ 30, three replicates). (C, F, I, and L) Photographs of seedlings of different genotypes grown under diurnal (12 hr light/12 hr dark) blue light conditions (1.2 μmol m−2 sec−1) for 4 days are shown. White bar, 5 mm.
Blue light-regulated gene expression is unaffected in pif1 seedlings compared to wild type:
Blue light regulates a distinct set of genes including HY5 and CHS in a light-dependent manner (Ma et al. 2001; Jiao et al. 2003). To investigate whether PIF1 plays a role in blue light-induced gene expression, we performed RT–PCR analysis on HY5 and CHS (Ma et al. 2001; Jiao et al. 2003). The results show that the expression of these genes is similar in both pif1 and wild-type seedlings under blue light conditions. However, both HY5 and CHS are expressed at a slightly higher level in dark-grown pif1 seedlings compared to wild-type seedlings (Figure 3). These data suggest that PIF1 is not involved in the blue light-induced expression of HY5 and CHS. By contrast, PIF1 might reduce the expression of these genes in the dark to repress photomorphogenesis.
Figure 3.—
Blue light-regulated gene expression is unaffected in pif1 mutant seedlings. Semi-quantitative RT–PCR assays for HY5 and CHS are shown, using total RNA isolated from wild type and pif1 mutants grown in the dark and dark-grown seedlings exposed to blue light (25 μmol m−2 sec−1) for 3 and 6 hr.
PIF1 is post-translationally regulated under blue light through the ubi/26S-proteasome pathway:
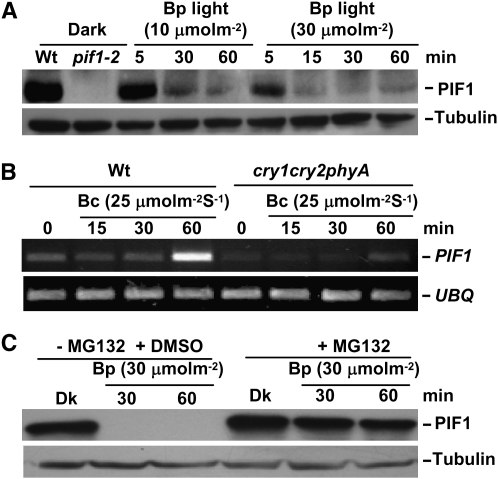
PIF1 functions as a negative regulator of both red and far-red light-mediated seedling de-etiolation processes (Huq et al. 2004; Oh et al. 2004; Shen et al. 2005). Red light and far-red light induce degradation of PIF1 to remove this negative regulation (Shen et al. 2005, 2008; Oh et al. 2006). Since PIF1 also functions as a negative regulator under blue light conditions, we investigated whether PIF1 is degraded under blue light conditions. Western blots using an anti-PIF1 antibody demonstrated that native PIF1 is rapidly degraded in response to a pulse of blue light (Figure 4A). A reduced PIF1 level might be due to a rapid reduction in transcription and/or instability of the PIF1 mRNA under blue light conditions. To determine if the PIF1 mRNA level was reduced in blue light, we measured PIF1 mRNA levels from total RNA isolated from seedlings exposed to blue light for different time periods, using semiquantitative RT–PCR assays. Results show that the expression of PIF1 under blue light is similar to that in the dark up to 30 min. However, PIF1 expression is induced after 1 hr of blue light exposure, and this induction is decreased in phyAcry1cry2 seedlings compared to wild-type seedlings (Figure 4B). These data suggest that blue light induces rapid post-translational degradation of PIF1 to promote photomorphogenesis at the seedling stage.
Figure 4.—
Blue light induces rapid degradation of PIF1 through the ubi/26S proteasomal pathway. (A) Native PIF1 is rapidly degraded after exposure to a pulse of blue (Bp) light conditions. Four-day-old dark-grown seedlings were exposed to Bp light (10 or 30 μmol m−2) and then incubated in the dark for the time indicated before harvesting for protein extraction. As controls, protein extracts from dark-grown wild-type and pif1 seedlings are included in the first two lanes, respectively. Approximately 30 μg of total protein in each lane were separated on an 8% polyacrylamide gel, transferred to PVDF membrane, and probed with anti-PIF1 antibody. A similar blot was probed with anti-tubulin antibody. The bands corresponding to PIF1 and tubulin are labeled. (B) PIF1 is slightly induced under blue light conditions. RT–PCR analyses are shown of PIF1 mRNA levels extracted from 4-day-old dark-grown seedlings or 4-day-old dark-grown seedlings exposed to continuous blue light (25 μmol m−2 sec−1) for the durations indicated. UBQ10 was used a control for the RT–PCR assays. (C) Blue light-induced degradation of PIF1 is mediated through the ubi/26S proteasomal pathway. Four-day-old dark-grown seedlings were pretreated with or without MG132 (30 μm) for 5 hr before being exposed to Bp light (30 μmol m−2) and then incubated in the dark for the durations indicated.
To investigate whether blue light-induced degradation of PIF1 is mediated by the ubi/26S proteasomal pathway, we measured the PIF1 protein level of extracts prepared from seedlings pretreated with and without MG132 (a proteasome inhibitor) in the presence and absence of blue light exposure. Results show that MG132 strongly inhibited the blue light-induced degradation of PIF1 (Figure 4C), suggesting that PIF1 degradation under the blue light conditions is mediated through the ubi/26S proteasomal pathway.
Blue light induces rapid phosphorylation and ubiquitination of PIF1:
Because PIF1 is rapidly phosphorylated and polyubiquitinated prior to degradation under both red (R) and FR light conditions (Shen et al. 2008), we investigated whether PIF1 is also phosphorylated and ubiquitinated under blue light conditions. Seedlings expressing a 35S:TAP-PIF1 fusion protein were exposed to a pulse of blue light (3600 μmol m−2) followed by incubation in darkness for 1 hr. Protein extraction, immunoprecipitation, and subsequent Western blotting show PIF1 migrated as a diffuse band with a higher mobility shift than PIF1 isolated from dark samples, suggesting that PIF1 is post-translationally modified under blue light (Figure 5A). To test whether this modification was due to the addition of phosphate groups, TAP-PIF1 was immunoprecipitated from samples exposed to blue light and treated with CIAP. After CIAP treatment, the diffuse band is reduced to a sharp single band of lower molecular weight, indicating the removal of the phosphates. Performing this experiment with boiled CIAP showed no effect on the diffuse band. These results demonstrate that PIF1 is phosphorylated in response to blue light.
Figure 5.—
Blue light induces rapid phosphorylation and ubiquitination prior to degradation of PIF1. (A) Blue light induces phosphorylation of PIF1. TAP-PIF1 was immunoprecipitated from protein extracts prepared using 4-day-old dark-grown 35S:TAP-PIF1 seedlings kept in the dark or exposed to Bp (30 μmol m−2 sec−1 × 2 min = 3600 μmol m−2) followed by dark incubation. The immunoprecipitated pellets from the Bp-exposed samples were dissolved in buffer and incubated without (−) or with (+) native calf intestine alkaline phosphatase (CIAP) or with boiled CIAP (+B). Samples were then separated on 6.5% SDS–PAGE gels and Western blots probed with anti-MYC antibody. Asterisks denote cross-reacting bands. (B) Blue light induces ubiquitination of PIF1. TAP-PIF1 was immunoprecipitated from protein extracts prepared using 4-day-old dark-grown seedlings either kept in the dark (Dk) or exposed briefly to Bp light (30 μmol m−2 sec−1 × 2 min = 3600 μmol m−2). The immunoprecipitated samples were then separated on 6.5% SDS–PAGE gels and probed with anti-ubiquitin (Ubi) or anti-MYC antibodies. Arrows indicate ubiquitinated forms of PIF1.
To investigate whether PIF1 is ubiquitinated in response to blue light signals, Western blots of immunoprecipitated TAP-PIF1 samples were probed using anti-ubi antibody. Figure 5B shows that TAP-PIF1 is ubiquitinated under blue light conditions. Both anti-myc (specific to TAP-PIF1) and anti-ubi antibodies detected high molecular weight bands, which are enhanced in the presence of the proteasomal inhibitor MG132. These ubiquitinated forms are present only in the light-exposed samples, but not in the dark samples. These results along with Figure 4C suggest that PIF1 is ubiquitinated and degraded under blue light conditions through the ubi/26S proteasomal pathway.
phyA is responsible for PIF1 degradation under pulses of blue light while the absence of cry1 and cry2 destabilizes PIF1 under prolonged blue light:
Crys and phys are predominantly responsible for regulating seedling de-etiolation under blue light conditions. To investigate which photoreceptor induces PIF1 degradation under blue light, we performed Western blot analyses of native PIF1 levels in monogenic and multiple photoreceptor mutant combinations. Results show that while cry1 and cry2 are not necessary for PIF1 degradation, phyA is responsible for the complete degradation of PIF1 under pulses of blue light (Figure 6A). However, prolonged exposure to continuous blue light induced strong degradation of PIF1 in the phyA background, suggesting other photoreceptors are also involved in PIF1 degradation under blue light conditions (Figure S4). To investigate whether cry1 and cry2 participate in blue light-induced degradation of PIF1 under prolonged light conditions, we performed Western blots of protein extracts from phyA and phyAcry1cry2 triple-mutant seedlings. Interestingly, results show that the PIF1 level is reduced in the phyAcry1cry2 compared to the phyA single-mutant seedlings, suggesting that the absence of both cry1 and cry2 destabilizes PIF1 under these conditions (Figure 6B). To estimate the relative contribution of cry1 and cry2 in PIF1 degradation, we performed Western blots of protein extracts from phyA, phyAcry1, phyAcry2, and phyAcry1cry2 seedlings grown under continuous blue light. PIF1 is slightly less stable in phyAcry1 and phyAcry2 compared to the phyA single mutant (Figure 6C). However, PIF1 is completely degraded in the phyAcry1cry2 triple mutant compared to either phyAcry1 or phyAcry2, suggesting that the absence of both cry1 and cry2 synergistically destabilizes PIF1 under blue light.
Figure 6.—
phyA is necessary for PIF1 degradation while cry1 and cry2 stabilize PIF1 under blue light conditions. (A) phyA mediates PIF1 degradation after exposure to a pulse of blue light. Four-day-old dark-grown seedlings were exposed to a pulse of blue light (Bp, 10 μmol m−2) and then incubated in the dark for the durations indicated before being harvested for protein extraction. (B) PIF1 is less stable in phyAcry1cry2 seedlings compared to phyA seedlings under continuous blue light conditions. Four-day-old dark-grown seedlings were exposed to continuous blue light (Bc, 10 μmol m−2 sec−1) and then incubated in the dark for the durations indicated. (C) Absence of cry1 and cry2 synergistically destabilizes PIF1 under continuous blue light conditions. Four-day-old dark-grown seedlings were exposed to continuous blue light (Bc, 10 μmol m−2 sec−1) and then incubated in the dark for the durations indicated before being harvested for protein extraction. (D) phyA plays a dominant role during the initial light exposure while phyB and phyD regulate PIF1 stability under prolonged light exposure. Western blots showing native PIF1 levels in phyA, phyAB, and phyABD mutant backgrounds are shown. Four-day-old dark-grown seedlings were exposed to continuous blue light (Bc, 10 μmol m−2 sec−1) for the time indicated before harvesting for protein extraction. All three genotypes are in the Ler ecotype.
PIF1 is degraded under blue light in a phy-dependent manner:
Due to increased degradation of PIF1 in phyAcry1cry2 seedlings compared to that in phyA seedlings under prolonged blue light conditions, we focused our attention on single and higher-order phy mutant seedlings. A Western blot of protein extracts from phyA, phyAB, and phyABD seedlings exposed to continuous blue light demonstrated that PIF1 is slightly more stable in the phyAB double-mutant background compared to the phyA single-mutant background (Figure 6D). In addition, PIF1 is completely stable in the phyABD triple-mutant background under these conditions. These data suggest that all three photoreceptors (phyABD) are necessary for the blue light-induced degradation of PIF1 in an additive manner.
PIF1 interacts with phyA and phyB in a blue light-dependent manner:
Because PIF1 is degraded under blue light in a phy-dependent manner, we investigated whether PIF1 can interact with phyA and phyB under blue light, using the light-dependent yeast two-hybrid assays as described (Shimizu-Sato et al. 2002). Results show that PIF1 can interact with the full-length phyA and the N-terminal half of phyB (phyB-NT) in a blue light-dependent manner (Figure 7A). Exposure of 30 μmol m−2 of blue light induced interaction of PIF1 with phyA significantly higher than that with the dark controls. However, exposure of 3600 μmol m−2 of blue light induced strong interactions between PIF1 and either phyA or phyB-NT. These data suggest that PIF1 binds to both phyA and phyB under blue light conditions.
Direct interactions with phys are necessary for the blue light-induced degradation of PIF1:
Previously, we demonstrated that three amino acids (G47, L95, and N144) in PIF1 are critical for interaction with the Pfr forms of phyA and phyB (Shen et al. 2008). Moreover, phy interaction is necessary for PIF1 degradation under red light conditions, since a triple-mutant form of PIF1 fusion protein (LUC-PIF1-3M) that has reduced affinity for both phyA and phyB (Figure 7B) showed reduced degradation compared to wild-type LUC-PIF1 fusion protein (Shen et al. 2008). Using these transgenic lines, we determined the blue light-induced degradation pattern of the triple-mutant form of PIF1 and compared that to the wild-type LUC-PIF1 degradation pattern using a cycloheximide chase assay as previously described (Shen et al. 2008). Results show that in blue light, the rate of degradation of LUC-PIF1 is much higher compared to the LUC-PIF1-3M degradation rate (Figure 7, B and C), suggesting that phy interaction is necessary for the blue light-induced degradation of PIF1.
To investigate whether phy interaction is sufficient for the blue light-induced degradation of PIF1, we measured the level of two truncated LUC-PIF1 fusion proteins (1–150 amino acids necessary for PIF1 interaction with phys and 151–478 amino acids necessary for DNA binding and dimerization) in the dark and blue light conditions. Results showed that both isolated regions of PIF1 are stable under blue light conditions (Figure 7D), suggesting that phy interaction is not sufficient for the blue light-induced degradation of PIF1.
DISCUSSION
Although PIFs are best characterized for their roles in red/far-red light signaling pathways, they have not been characterized under blue light conditions. In this study, we provide genetic, biochemical, and photobiological evidence that PIF1 is a negative regulator of blue light-mediated de-etiolation of Arabidopsis seedlings. Two alleles of monogenic pif1 seedlings displayed significantly larger cotyledon angles and slightly shorter hypocotyls compared to wild-type seedlings under a range of fluence rates of blue lights applied diurnally (Figure 1). Although the hypocotyl lengths of both pif1 alleles were slightly shorter than those of the wild-type seedlings in the dark as has been described previously (Huq et al. 2004; Shen et al. 2008), both pif1 alleles did not display any cotyledon opening when grown in the dark for 4 days under these conditions. These data suggest that PIF1 functions as a negative regulator of the blue light signaling pathways.
However, analyses of the double mutants between pif1 and either phy or cry single mutants revealed a more complex relationship. The absence of pif1 in a phyA or phyB or cry1 or cry2 single-mutant background suppressed the respective photoreceptor mutant phenotypes either completely or partially under blue light conditions (Figure 2, Figure S3). For example, the phyA single mutant displayed a strong hyposensitive phenotype under diurnal blue light conditions, while a phyApif1 double mutant displayed an almost wild-type phenotype under these conditions (Figure 2A). The relatively weak pif1 phenotype in comparison to strong phyApif1 or phyBpif1 or cry1pif1 or cry2pif1 double-mutant phenotypes under blue light suggests that PIF1 might be a very subtle negative regulator of the blue light-mediated developmental processes. The negative role of PIF1 might be so subtle that its effect is very weak under normal strong photocurrents in the wild-type background. However, the negative effect of PIF1 is more penetrable when the photocurrent is reduced in any of the single photoreceptor mutant backgrounds.
An alternative hypothesis is that PIF1 and all other PIFs might function negatively in the dark-grown seedlings as has been demonstrated recently (Leivar et al. 2008b; Shen et al. 2008). In this case, the negative role of PIF1 is very marginal or unpenetrable in the dark-grown monogenic pif1 seedlings, but becomes more penetrant in the presence of light when the level of other PIFs is reduced due to their light-induced degradation. This hypothesis predicts that PIFs might be degraded in response to blue light signals, as previously observed under red/far-red light conditions (Castillon et al. 2007; Shen et al. 2008). To test this hypothesis, we determined the PIF1 level in the dark-grown seedlings and dark-grown seedlings exposed to blue light conditions. Strikingly, PIF1 is rapidly degraded under these conditions through the ubi/26S proteasomal pathway (Figure 4). In addition, as observed under red and far-red light conditions, PIF1 is phosphorylated, polyubiquitinated, and subsequently degraded under blue light conditions (Figure 5). Because PIF1 is degraded in response to a single pulse of blue light in a phyA-dependent manner (Figure 6A), it is possible that this degradation is through the VLFR of phyA, as previously observed under far-red light conditions (Shen et al. 2005). Taken together, these data are consistent with the proposal that PIF1 functions negatively in the dark to repress photomorphogenesis, and the blue light signals induce rapid degradation of PIF1 to remove this negative regulation and thereby promote photomorphogenesis.
The observation that the pif1 mutant displays a hypersensitive phenotype under diurnal conditions (Figures 1 and 2, Figure S2, Figure S3), but not under continuous blue light, is striking. Previous results also demonstrated that the pif1 mutant is hypersensitive to red and far-red light applied diurnally, but not under continuous light (Oh et al. 2004; Shen et al. 2005). Although PIF1 mRNA is not regulated by circadian clock or diurnal conditions (data not shown), PIF1 protein level reaccumulates in the subsequent dark period after rapid degradation under red light and is also slightly diurnally regulated (Shen et al. 2005). It is possible that this diurnal regulation of PIF1 protein level might be one of the molecular bases for the differential phenotypes observed for the pif1 mutant under diurnal as opposed to continuous blue light conditions.
The data presented here also demonstrate that PIF1 interacts with phyA and phyB in a blue light-dependent manner (Figure 7A). phyA plays a dominant role under pulses of blue light, while phyB and phyD regulate PIF1 levels under prolonged blue light conditions in an additive manner (Figure 6D). A reduced level of blue light-induced degradation of a mutant form of PIF1, which has lower affinity for both phyA and phyB, suggests that direct physical interactions with phys are necessary for PIF1 degradation under blue light conditions (Figure 7, B and C). Moreover, independent expression of two separate regions of PIF1 (1- to 150-amino-acid region necessary for phy interaction and 151- to 478-amino-acid region necessary for DNA binding and dimerization) as Luciferase fusion proteins in transgenic plants demonstrated that these isolated regions are not degraded under blue light conditions (Figure 7D). Therefore, phy interaction is necessary, but not sufficient for PIF1 degradation under blue light conditions. Combined, these data along with previous results suggest that PIF1 and other PIFs function as negative regulators of photomorphogenesis in the dark, and phys activated by all three monochromatic lights induce rapid degradation of PIFs to promote photomorphogenesis (Al-Sady et al. 2006; Shen et al. 2007, 2008; Lorrain et al. 2008).
Although crys are the primary photoreceptors for the blue light-induced seedling de-etiolation, they were not necessary for the blue light-induced degradation of PIF1. By contrast, the data show that the absence of both cry1 and cry2 destabilizes PIF1 under blue light conditions (Figure 6, A–C). Although other bHLH proteins have been shown to interact with cry1 and cry2 under blue light (Liu et al. 2008), PIF1 did not show interaction with cry1 and cry2 in both yeast two-hybrid assays and in vivo co-immunoprecipitation assay (data not shown). It is unclear how cry1 and cry2 stabilize PIF1 under blue light conditions. One possibility is that the physical interaction between crys and phys might titrate away phyA and phyB from direct interaction with PIF1. Alternatively, both phy and cry signaling pathways share the same downstream components that are necessary for PIF1 degradation. Therefore, in the absence of cry1 and cry2, higher levels of phys and/or phy signaling components induce increased degradation of PIF1 under blue light conditions. Moreover, the functional significance of PIF1 stabilization by crys is also unknown. Although phys and crys have been shown to function antagonistically in controlling flowering time, phenotypic analyses of monogenic and double mutant plants did not reveal any role of PIF1 in controlling flowering time (data not shown). Because there are multiple PIFs in Arabidopsis, it is possible that higher-order pif mutants would be necessary to uncover the roles, if any, of PIFs in controlling flowering time.
In conclusion, although phys are best known as red/far-red light sensing photoreceptors, our data and those of others establish broader and more direct roles of phys in regulating both morphological and molecular phenotypes under blue light signaling pathways. Therefore, phys might control photomorphorphogenesis under a broad spectrum of light conditions, while crys, phots, and the ZTL/FKF1/LKP2 family might regulate photomorphogenesis specifically under blue light conditions (Figure 8). Elucidation of the mechanisms by which these photoreceptors act synergistically and/or antagonistically to optimize photomorphogenic development awaits further investigation.
Figure 8.—
Simplified model of Arabidopsis photoreceptor function in the light regulation of photomorphogenesis. The cry1, cry2, phot1, phot2, and ZTL/FKF1/LKP2 family of photoreceptors perceive and respond to the blue region of the light spectrum, while phys perceive and respond to all three (blue, red, and far-red) light signals. All five phys perceive and respond to red light signals, while phyA, phyB, phyD, and possibly phyC/phyE respond to blue light signals. phyA is the sole photoreceptor for perceiving and responding to the far-red light signals. phys and crys may function synergistically and/or antagonistically to optimize photomorphogenesis under blue light signals.
Acknowledgments
We thank Alan Lloyd, Jennifer Moon, and Guy Thompson for critical reading of the manuscript; Gary Whitelam for sharing phy mutants; Bruce Downie for sharing anti-PIF1 antibody; and Chentao Lin for sharing cry1, cry2, phyAcry1, phyAcry2, and phyAcry1cry2 mutant seeds. This work was supported by National Science Foundation grant IOS-0822811 to E.H.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.099887/DC1.
References
- Ahmad, M., J. A. Jarillo, O. Smirnova and A. R. Cashmore, 1998. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1 939–948. [DOI] [PubMed] [Google Scholar]
- Al-Sady, B., W. Ni, S. Kircher, E. Schafer and P. H. Quail, 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Bae, G., and G. Choi, 2008. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311. [DOI] [PubMed] [Google Scholar]
- Casal, J. J., 2000. Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem. Photobiol. 71 1–11. [DOI] [PubMed] [Google Scholar]
- Casal, J. J., and M. A. Mazzella, 1998. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 118 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, J. J., R. A. Sanchez and J. F. Botto, 1998. Modes of action of phytochromes. J. Exp. Bot. 49 127–138. [Google Scholar]
- Castillon, A., H. Shen and E. Huq, 2007. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. [DOI] [PubMed] [Google Scholar]
- Chen, M., J. Chory and C. Fankhauser, 2004. Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Demarsy, E., and C. Fankhauser, 2008. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12 69–74. [DOI] [PubMed] [Google Scholar]
- Duek, P. D., and C. Fankhauser, 2003. HFR1, a putative bHLH-transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J. 34 827–836. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and M. Chen, 2008. Transposing phytochrome into the nucleus. Trends Plant Sci. 13 596–601. [DOI] [PubMed] [Google Scholar]
- Folta, K. M., and E. P. Spalding, 2001. Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J. 28 333–340. [DOI] [PubMed] [Google Scholar]
- Huq, E., B. Al-Sady and P. H. Quail, 2003. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35 660–664. [DOI] [PubMed] [Google Scholar]
- Huq, E., B. Al-Sady, M. Hudson, C. Kim, K. Apel et al., 2004. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., and S. A. Kay, 2006. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 11 550–558. [DOI] [PubMed] [Google Scholar]
- Jiao, Y. L., H. J. Yang, L. G. Ma, N. Sun, H. Y. Yu et al., 2003. A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol. 133 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., O. S. Lau and X. W. Deng, 2007. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Kang, X., and M. Ni, 2006. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., E. Monte, B. Al-Sady, C. Carle, A. Storer et al., 2008. a The Arabidopsis Phytochrome-Interacting Factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., E. Monte, Y. Oka, T. Liu, C. Carle et al., 2008. b Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., 2000. Photoreceptors and regulation of flowering time. Plant Physiol. 123 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., and D. Shalitin, 2003. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Liu, H., X. Yu, K. Li, J. Klejnot, H. Yang et al., 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322 1535–1539. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., T. Allen, P. D. Duek, G. C. Whitelam and C. Fankhauser, 2008. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. [DOI] [PubMed] [Google Scholar]
- Ma, L., J. Li, L. Qu, J. Hager, Z. Chen et al., 2001. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli, A. L., 1994. The physiology of phytochrome action, pp. 211–269 in Photomorphogenesis in Plants, edited by R. E. Kendrick and G. H. M. Kronenberg. Kluwer Academic Publishers, The Netherlands.
- Mas, P., P. F. Devlin, S. Panda and S. A. Kay, 2000. Functional interaction of phytochrome B and cryptochrome 2. Nature 408 207–211. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and R. A. Sharrock, 1997. Phytochrome gene diversity. Plant Cell Environ. 20 666–671. [Google Scholar]
- Matsushita, T., N. Mochizuki and A. Nagatani, 2003. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424 571–574. [DOI] [PubMed] [Google Scholar]
- McClung, C. R., 2008. Comes a time. Curr. Opin. Plant Biol. 11 514–520. [DOI] [PubMed] [Google Scholar]
- Mockler, T. C., H. Guo, H. Yang, H. Duong and C. Lin, 1999. Antagonistic actions of the Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Moon, J., L. Zhu, H. Shen and E. Huq, 2008. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. M., and J. Chory, 1998. Genetic interactions between phytochrome A, phytochrome B and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, K., M. F. Covington, P. D. Duek, A. A. Lorrain, C. Fankhauser et al., 2007. Rhythmic growth explained by coincidence between internal and external cues. Nature 448 358–361. [DOI] [PubMed] [Google Scholar]
- Oh, E., J. Kim, E. Park, J. I. Kim, C. Kang et al., 2004. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., S. Yamaguchi, Y. Kamiya, G. Bae, W.-I. Chung et al., 2006. Light activates the degradation of PIL5 to promote seed germination through gibberellin in Arabidopsis. Plant J. 47 124–139. [DOI] [PubMed] [Google Scholar]
- Quail, P. H., 2007. a Phytochrome interacting factors, pp. 81–105 in Light and Plant Development, edited by G. Whitelam and K. Halliday. Blackwell Publishing, Oxford.
- Quail, P. H., 2007. b Phytochrome-regulated gene expression. J. Integr. Plant Biol. 49 11–20. [Google Scholar]
- Rockwell, N. C., Y.-S. Su and J. C. Lagarias, 2006. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler, J., I. Klein and M. Zeidler, 2007. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104 10737–10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, E., and F. Nagy, 2006. Photomorphogenesis in Plants and Bacteria. Springer, Dordrecht, The Netherlands.
- Shen, H., J. Moon and E. Huq, 2005. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize seedling photomorphogenesis in Arabidopsis. Plant J. 44 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen, H., L. Zhu, A. Castillon, M. Majee, B. Downie et al., 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME INTERACTING FACTOR 1 depends upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., R. Khanna, C. M. Carle and P. H. Quail, 2007. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato, S., E. Huq, J. M. Tepperman and P. H. Quail, 2002. A light-switchable gene promoter system. Nat. Biotechnol. 20 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., A. Nagatani, H. Hanzawa, M. Kubota, M. Watanabe et al., 1996. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D. E., P. F. Devlin and S. A. Kay, 1998. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Usami, T., T. Matsushita, Y. Oka, N. Mochizuki and A. Nagatani, 2007. Roles for the N- and C-terminal domains of phytochrome B in interactions between phytochrome B and cryptochrome signaling cascades. Plant Cell Physiol. 48 424–433. [DOI] [PubMed] [Google Scholar]
- Vierstra, R. D., and P. H. Quail, 1983. Purification and initial characterization of 124-kilodalton phytochrome from avena. Biochemistry 22 2498–2505. [Google Scholar]
- Whitelam, G., and K. Halliday, 2007. Light and Plant Development. Blackwell Publishing, Oxford.
- Yang, H.-Q., R.-H. Tang and A. R. Cashmore, 2001. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]