Abstract
The Drosophila Hox gene, Sex combs reduced (Scr), is required for patterning the larval and adult, labial and prothoracic segments. Fifteen Scr alleles were sequenced and the phenotypes analyzed in detail. Six null alleles were nonsense mutations (Scr2, Scr4, Scr11, Scr13, Scr13A, and Scr16) and one was an intragenic deletion (Scr17). Five hypomorphic alleles were missense mutations (Scr1, Scr3, Scr5, Scr6, and Scr8) and one was a small protein deletion (Scr15). Protein sequence changes were found in four of the five highly conserved domains of SCR: the DYTQL motif (Scr15), YPWM motif (Scr3), Homeodomain (Scr1), and C-terminal domain (CTD) (Scr6), indicating importance for SCR function. Analysis of the pleiotropy of viable Scr alleles for the formation of pseudotracheae suggests that the DYTQL motif and the CTD mediate a genetic interaction with proboscipedia. One allele Scr14, a missense allele in the conserved octapeptide, was an antimorphic allele that exhibited three interesting genetic properties. First, Scr14/Df had the same phenotype as Scr+/Df. Second, the ability of the Scr14 allele to interact intragenetically with Scr alleles mapped to the first 82 amino acids of SCR, which contains the octapeptide motif. Third, Scr6, which has two missense changes in the CTD, did not interact genetically with Scr14.
THE Homeotic selector (Hox) genes are required for patterning the anterior–posterior axis of all bilateral animals (Lewis 1978; Carroll 1995). In Drosophila melanogaster, the Hox genes establish segmental identity in the embryo by controlling the spatial expression of target genes (Capovilla et al. 1994). Although much is known about the requirement of HOX activity in development, there is little known about internal domain structure of these proteins and how these transcription factors are regulated. In fact, the analysis of the functional domains of HOX proteins has proven difficult and often contradictory (Zhao et al. 1996; Galant et al. 2002; Hittinger et al. 2005; Tour et al. 2005). For example, the insect specific QA motif of the HOX protein Ultrabithorax (UBX) is required for full Ubx repression of limb development in Drosophila when UBX is ectopically expressed (Galant and Carroll 2002; Ronshaugen et al. 2002). Noninsect UBX homologs lack a QA motif and lack the ability to suppress limb development when ectopically expressed in Drosophila; however, limb repression can be conferred to these noninsect UBX homologs by fusing the QA motif to the carboxyl termini (Galant and Carroll 2002). These ectopic expression experiments suggested that the QA motif was essential for UBX activity; therefore, it was surprising that a deletion of the QA motif within the Ubx locus produced only a subtle phenotype (Hittinger et al. 2005). The observation of differential pleiotropy in the Ubx and Antennapedia (Antp) loci offers a potential explanation for these difficulties: HOX proteins are made up of small independently acting peptide elements that alone make only a small contribution to HOX activity (Carroll 2005; Hittinger et al. 2005; Prince et al. 2008). Uniform pleiotropy is the same relative behavior of a set of alleles in a locus on two or more phenotypic characteristics, whereas, differential pleiotropy is a distinct relative behavior. Differential pleiotropy has been described in Ubx (Hittinger et al. 2005). Analysis of the Ubx-ΔQA allele revealed a differential requirement for the QA motif in the development of various UBX-dependent tissues. This preferential requirement for the QA motif in a subset of tissues is an example of differential pleiotropy (Hittinger et al. 2005). In addition, the YPWM motif of the HOX protein Antennapedia (ANTP), a motif that has been conserved across evolution in most HOX proteins, exhibits differential pleiotropy by being required for the formation of ectopic wing tissue but not the formation of ectopic leg tissue (Prince et al. 2008).
The sequence of Scr mutant alleles allowed the analysis of the requirement of highly conserved motifs of the HOX protein, Sex combs reduced (SCR). Like Ubx, Scr is haplo-insufficient, making it an excellent gene for identifying small changes in SCR activity because even subtle changes in levels of protein function are registered in SCR-dependent phenotypes. SCR function is essential for the development of labial derivatives, such as the adult proboscis and larval salivary glands, and for establishing the identity of the adult prothoracic legs (Lewis et al. 1980b; Struhl 1982; Panzer et al. 1992; Percival-Smith et al. 1997). SCR activity is required with a second HOX protein, Proboscipedia (PB), for the formation of the proboscis but does not require PB for the formation of the sex comb bristles or the salivary gland (Kaufman 1978; Struhl 1982). The SCR protein has five highly conserved regions (Curtis et al. 2001). The octapeptide, YPWM motif and homeodomain (HD) are well conserved across evolution and are found in all SCR homologs. The MvDYTQLQPQRL sequence (DYTQL motif) and the carboxy-terminal domain (CTD) are insect and SCR specific. From our analysis of an Scr antimorphic allele, we suggest that the octapeptide of SCR participates in protein complex formation required for the formation of sex combs and pseudotrachea, and that the CTD inhibits protein complex formation by masking the octapeptide. In addition, analysis of the proboscis phenotype of viable Scr alleles in the presence of one or two copies of the pb locus suggests that the DYTQL motif and the CTD of SCR mediate a genetic interaction with pb.
MATERIALS AND METHODS
DNA sequencing of Scr mutant alleles:
The fly strains used were obtained from the Bloomington Stock Center (Indiana University, Bloomington, IN) with the exception of Scr13A, which was provided by Gary Struhl (Struhl 1982). The genotypes of the stocks used were: y w; FRT82B Scr1 P{w+}/TM6B, Tb1, P{walLy}, y w; FRT82B pb27Scr2pp cn P{w+}/TM6B, Tb1, P{walLy}, y w; Scr3pp/TM6B, Tb1, P{walLy}, lab4Scr4pp/TM3, Sb1, y w; Scr5red1e1/TM6B, Tb1, P{walLy}, y w; Scr6pp/TM6B, Tb1, P{walLy}, Scr7pp/TM6B, Tb1, y w; Scr8rnroe-1rn1 pp/TM6B, Tb1, P{walLy}, Scr11red1e1/TM3,Sb1, Scr13e1/TM3, Sb1, y w; FRT82B Scr13A Ubx1e/TM6B, P{walLy}, y w; kniri-1Scr14e1/TM6B, Tb1, P{walLy}, y w; kni ri-1 Scr15 e1/TM6B, Tb1, P{walLy}, kni ri-1Scr16 e1/TM3, Sb1, Scr17/TM6B, Tb1, w; Ki pb34pp/TM6B, Tb1, y w; Ki pb34pp/TM6B, Tb1, P{walLy}, y w (Lewis et al. 1980b; Struhl 1982; Lindsley and Zimm 1992; Percival-Smith et al. 1997).
To sequence the DNA of Scr alleles, two approaches were used to collect material for DNA extraction. For null Scr alleles, embryos were collected on apple juice plates and allowed to develop to the first instar larval stage. DNA was extracted from unhatched eggs containing larvae with deformed mouth skeletons characteristic of homozygous Scr null mutants (Pattatucci et al. 1991). For viable alleles, DNA was extracted from imagos hemizygous for the Scr allele and a complete deletion of the Scr locus (pb34). The one exception was Scr1 where DNA was extracted from heterozygous imagos. Coding region DNA from exon 2 and exon 3 was amplified for all alleles and sequenced at the Robarts DNA Sequencing Facility (London, Ontario, Canada). The sequence of the 3′- and 5′-untranslated regions of Scr5, Scr7, and Scr8 was also determined. See supporting information for the primers used (Table S1).
Phenotypic characterization of viable Scr mutants:
Imagos of the genotype Scrx/Scr+, Scrx/pb34, Scrx/Scr14, and Scrx/Scr13A were collected. Either these imagos were critical point dried and the heads and prothoracic legs mounted for scanning electron microscopy or the first legs were pulled off and suspended in Hoyer's mountant to count the number of sex comb bristles under bright field optics (Wieschaus and Nusslein-Volhard 1986). To count the number of salivary gland cells, salivary glands were dissected from non-Tubby third instar larvae with the genotypes Scrx/pb34, Scrx/Scr14, and Scrx/Scr13A. The glands were fixed and the DNA of the salivary gland nuclei was visualized with DAPI. For analysis of cold-sensitive phenotypes Drosophila were reared at 18° and 23°.
Analysis of Scr larval cuticles:
First instar larvae were collected from apple juice plates, dechorionated, devitellinized, and mounted in 50% Hoyer's mountant/50% lactic acid (Wieschaus and Nusslein-Volhard 1986).
Quantitative real-time PCR:
Total RNA was extracted from three independent samples of 0- to 14-hr Scrx/pb34 pupae. cDNA was made using Superscript II reverse transcriptase (Invitrogen) and Oligo d(T)15 primers (Invitrogen). To determine transcript levels, real-time PCR was performed in the Corbett Rotor-Gene 3000 real-time cycler using SYBR green detection (Bustin 2000; Karsai et al. 2002) and was analyzed using Rotor-Gene Analysis software 6.0 (Corbett). See supporting information for primer sequences (Table S2).
Western blot analysis:
Protein was extracted from three independent samples of 0- to 14-hr Scrx/pb34 pupae. SCR was detected on the Western analysis with a mouse monoclonal SCR antibody (Glicksman and Brower 1988), diluted 1:5. The antibody–antigen complex was detected with an anti-mouse HRP conjugated secondary antibody (Sigma) and the SuperSignal West Femto Chemiluminescent kit (Pierce). An image was collected on a Fluorchem 8900 gel documentation system (Alpha Innotech) and quantified using the AlphaEase Fluorchem Software (v.4.0.1).
Mosaic analysis:
Clones of Scr1 and Scr13A tissue were generated using FLP-mediated mitotic recombination, and were identified by the Sb+ M+ y phenotype (Xu and Rubin 1993; Percival-Smith et al. 1997). Images were acquired using a scanning electron microscope.
Statistical analyses:
All data were statistically analyzed using SPSS v.16.0. Analyses of SCR transcript levels were performed with a one-way analysis of variance (ANOVA), and data for cold-sensitive alleles were analyzed using two-way ANOVAs. Transcript and cold-sensitive sex comb bristle data were log10 transformed before analysis to meet the requirements of homoscedasticity and normality (Zar 1999). If significant differences were detected with an ANOVA, multiple pairwise comparisons were made using a Tukey test. Analyses of SCR protein levels were performed using a Kruskal–Wallace test; if a significant interaction was found, multiple pairwise comparisons were made using a Dunnett T3. All data for the number of rows of pseudotrachea were analyzed using Kruskal–Wallace tests. All salivary gland data were analyzed using one-way ANOVAs, with the exception of Scrx/Scr13A data, which were analyzed using a Kruskal–Wallace test. All sex comb bristle data were analyzed using Kruskal–Wallace tests, with the exception of Scrx/Scr+ data, which were analyzed using a one-way ANOVA.
RESULTS
DNA sequence of Scr mutant alleles:
Identifying mutational changes within the coding region of a gene reveals important functional domains of a protein. The Scr locus in Drosophila is represented by 15 alleles that are not associated with a cytological change (Lewis et al. 1980b; Struhl 1982; Lindsley and Zimm 1992). These Scr mutant alleles were sequenced, and a number of DNA sequence changes were observed and placed into two groups: polymorphisms or mutations (Table 1). The six sequence changes, T15 → C, A171 → G, C180 → T, A345 → G, C747 → T, and A933 → G, resulted in silent mutations that occurred in more than one independently isolated Scr allele and are presumed to be natural polymorphisms. These six polymorphisms occur frequently, with at least two present in every Scr allele sequenced. The Scr mutant alleles were isolated in many independent mutational screens that used isogenic third chromosomes; therefore, it was expected that the pattern of polymorphisms should group according to the screen in which the alleles were isolated. This pattern is observed for alleles Scr2 and Scr3, alleles Scr6 and Scr7, and alleles Scr13, Scr14, and Scr16; however, many alleles do not follow this expected pattern. For example, Scr4 was isolated in the same screen as Scr5 but has a different pattern of polymorphisms (Lewis et al. 1980). Mutant changes were identified as missense, nonsense, or small deletion mutations unique to each sequenced allele (with the exception of alleles Scr5 and Scr8, and alleles Scr13 and Scr16). The alleles sequenced in this study fall into two broad phenotypic categories: lethal and viable alleles. The following detailed phenotypic analysis of these alleles has shown that the Scr locus is represented by null (amorphic), hypomorphic, and antimorphic mutant alleles (Muller 1932).
TABLE 1.
Analysis of Scr mutant allele sequences
| Allele | Polymorphismsa | DNA sequence changesa | Class of mutation, protein changeb | Class | Source |
|---|---|---|---|---|---|
| Scr1c | A171 → Gd C180 → Td | G1093 → A | Missense, E365 → K (Homeodomain) | Hypomorph | Lewis et al. (1980a,b) |
| A345 → Gd C747 → TdA933 → Gd | |||||
| Scr2 | A171 → G; C180 → T; | C340 → T | Nonsense, Q114 → stop | Null | Lewis et al. (1980a,b) |
| A345 → G; C747 → T | |||||
| Scr3 | A171 → G; C180 → T; | C917 → T | Missense, P306 → L (YPWM motif) | Hypomorph | Lewis et al. (1980a,b) |
| A345 → G; C747 → T | |||||
| Scr4 | T15 → C;d A171 → G; | C393 → T | Nonsense, Q131 → stop | Null | Lewis et al. (1980a,b) |
| C180 → T; A345 → G | |||||
| C747 → T; A933 → G | |||||
| Scr5 | T15 → C; A345 → G; | G862 → A | Missense, A288 → T | Hypomorph | Lewis et al. (1980a,b) |
| A933 → G | |||||
| Scr6 | T15 → C; A345 → G | T1184 → A; T1213 → A; | Missense, M395 → K; missense, F405 → I; | Hypomorph | P. Fornili and T. C. Kaufman |
| C747 → T; A933 → G | G1239 → T | Silent (C-terminal domain) | |||
| Scr7 | T15 → C; A345 → G; | No changes | N/A | Hypomorph | P. Fornili and T. C. Kaufman |
| C747 → T; A933 → G | |||||
| Scr8 | T15 → C; C180 → T; | G862 → A | Missense, A288 → T | Hypomorph | P. Fornili and T. C. Kaufman |
| A171→ G; A345 → G | |||||
| C747 → T; A933 → G | |||||
| Scr11 | C180 → T; A345 → G | G1112 → A | Nonsense, W371 → stop | Null | Lambert |
| C747 → T; A933 → G | |||||
| Scr13 | A171 → G; C180 → T; | C439 → T | Nonsense, Q147 → stop | Null | K. A. Matthews |
| A345 → G; C747 → T; | |||||
| A933 → G | |||||
| Scr13A | A345 → G; C747 → T | G781 → T | Nonsense, G261 → stop | Null | Struhl (1982) |
| Scr14 | A171 → G; C180 → T; | C10 → T | Missense, S10 → L (Octapeptide motif) | Antimorph | K. A. Matthews |
| A345 → G; C747 → T; | |||||
| A933 → G | |||||
| Scr15 | C747 → T; A933 → G | del(T246–C350); | Deletion, T83 → P117; | Hypomorph | K. A. Matthews |
| T244 → G | Missense, Y82 → E (DYTQL motif) | ||||
| Scr16 | A171 → G; C180 → T | C439 →T | Nonsense, Q147 → stop | Null | K. A. Matthews |
| A345 → G; C747 → T; | |||||
| A933 → G | |||||
| Scr17 | T15 → C; A171 → G; | del(C2,667,211–T2,663,845)e | Null | M. A. Pultz and T. C. Kaufman | |
| C180 → T; A345 → G; | |||||
| A933 → G |
Each nucleotide change in the transcript is designated in subscript, where A of the ATG in the Celera mRNA sequence is +1.
The position of each amino acid change in the protein is designated in subscript, where the first methionine is +1.
Scr1 is the only allele that has not been confirmed in homozygous flies.
The A171 → G, C180 → T, A345 → G, C747 → T, T15 → C, and A933 → G polymorphisms all result in silent mutations.
Numbering is based on nucleotide position on third chromosome, according to FlyBase (R5.7).
Null alleles:
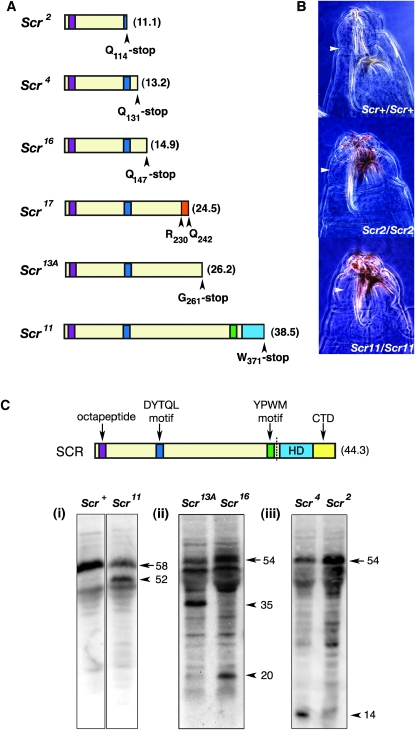
Six of seven null alleles were nonsense mutations, and one was an intragenic deletion mutation. Scr17 was a 3.4-kb deletion that removed the 3′ portion of the second exon, a splice site, and a portion of the second intron. Scr17 has the potential to encode a truncated protein of 242 amino acids (Figure 1A). Scr2, Scr4, Scr11, Scr13A, and Scr16 were all nonsense mutations that would result in the truncation of the SCR protein and loss of conserved SCR protein domains (Figure 1A). Although the loss of these conserved domains may explain the null nature of the alleles, nonsense mutations also affect the stability of the transcripts, resulting in degradation via nonsense-mediated RNA decay (NMD) (Hentze and Kulozik 1999; Alonso and Akam 2003). The signal that triggers NMD is a premature termination codon followed by an intron (Hentze and Kulozik 1999). All Scr nonsense alleles, with the exception of Scr11, met this requirement of NMD; therefore, if NMD were a factor, it was expected that SCR11 alone would be detected in a Western analysis of protein extracted from progeny embryos of heterozygous flies, in a 1:1 ratio with SCRWT protein. However, truncated SCR11, SCR13A, SCR16, and SCR4 proteins were also detected (Figure 1C). This indicates that SCR is not degraded via NMD; therefore, the phenotype observed in Scr nonsense mutants is caused by the deletion of important functional domains from SCR. SCR2 was not detected in the Western analysis and we propose that the epitope for the SCR monoclonal antibody used is missing from SCR2 (Glicksman and Brower 1988). Larval cuticles of Scr11 and Scr2 mutants were compared to determine if there were differences in the phenotypes. Scr11 encodes the longest truncated SCR peptide with a Trp371 to stop codon in the third α-helix of the HD. Scr2 encodes the shortest truncated SCR peptide only encoding a protein containing the octapeptide and part of the DYTQL sequence. No significant difference between the larval cuticle phenotypes of Scr11 and Scr2 mutants were observed, suggesting that deletion of the last 13 residues of the DNA recognition helix of the HD and the CTD results in a Scr null phenotype (Figure 1B; Table S3).
Figure 1.—
Structure, activity, and expression of the polypeptides encoded by Scr null alleles. (A) The colored boxes indicate conserved regions of SCR. Regions conserved in all SCR homologs are the octapeptide motif (purple), the YPWM motif (green), and the homeodomain (HD, cyan). The insect-specific conserved motifs are the DYTQL motif (blue) and the carboxyl terminal domain (CTD, yellow). Scr17 contains an intragenic deletion and the predicted protein contains novel amino acids (orange). Expected molecular weights (kiloDaltons) of each protein are indicated in parentheses beside each peptide. (B) Phase contrast micrographs of first instar larval cuticles of wild-type, Scr2 and Scr11 mutants. In both Scr mutants, the head is severely disrupted and the number of rows of T1 beard denticles is reduced. T1 beards are indicated with an arrowhead. (C) Western analysis of the expression of truncated protein from Scr nonsense alleles. Proteins were resolved on (i) 10%, (ii) 13%, and (iii) 11% SDS-polyacrylamide gels. Above the Western blot a conceptual translation of SCR mRNA is shown where the dotted line indicates the junction between exons 2 and 3. Protein was extracted from 3-to 10-hr embryos laid by wild-type and heterozygous stocks. The arrow indicates the protein expressed from the wild-type Scr locus, and arrowheads indicate the truncated protein products. The relative molecular mass (kiloDaltons) of the SCR peptides is indicated beside the arrows and arrowheads.
Scr1 a lethal hypomorph:
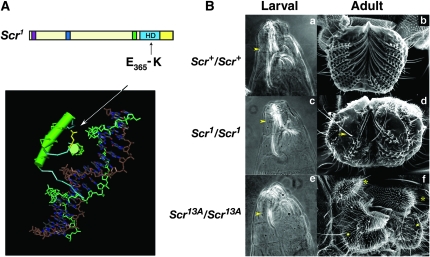
The embryonic lethal Scr1 allele was a missense mutation in a codon of the SCR HD that alters Glu365 to Lys (Figure 2A). Analysis of the affect of the HD missense mutation on the phenotype of marked Scr1/Scr1 embryonic cuticle revealed that this mutation was hypomorphic. In comparison to the cuticle of a homozygous null Scr13A mutant, the larval head structures of Scr1 mutants were not deformed and the T1 beard was not significantly reduced when compared to wild type (P = 0.8; Table S3). Hypomorphy of the HD change was also detected in the adult proboscis. Scr1/Scr1 clones of cells in the proboscis showed a weak proboscis to maxillary palp transformation, but pseudotrachea still formed while Scr13A/Scr13A clones in the proboscis were completely transformed toward maxillary palp identity (Figure 2B; Struhl 1982).
Figure 2.—
Scr1 is a lethal hypomorphic allele. (A) At the top is the primary structure of SCR1 showing the position of the Scr1 missense change; the color scheme for the SCR domains is the same as used in Figure 1. Below is the 3-D structure of the SCR HD–EXD complex bound to the fkh enhancer (Joshi et al. 2007) with EXD removed, leaving the SCR-HD and fkh enhancer, and glutamic acid 365 of SCR is highlighted in yellow in Cn3D 4.1 (NCBI). (B) The larval and adult phenotype of Scr1 and Scr13A. a, c, and e are first instar larval cuticle of wild type, Scr1 and Scr13A mutants, respectively. The arrowheads indicate the T1 beards. b is a wild-type labial palp. d and f are labial palps with clones of Scr1 and Scr13A cells. The arrows indicate non-Stubble, non-Minute maxillary palp bristles. The asterisks indicate normal maxillary palps and the squares highlight probocis toward maxillary palp transformations. Pseudotrachea still form in Scr1 clones but not in Scr13A clones.
Viable alleles:
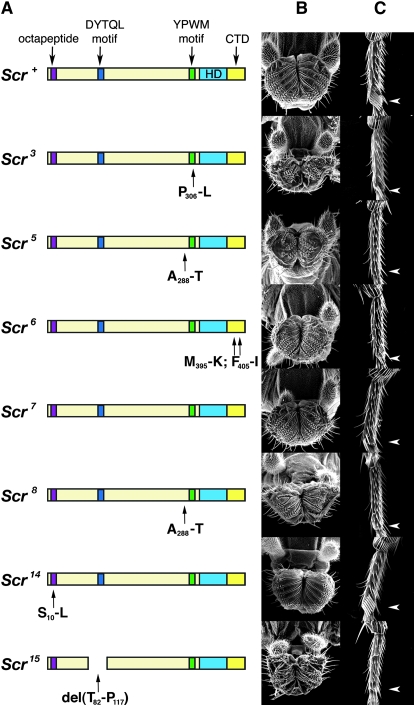
Six of the sequenced viable alleles contained missense mutations, and one allele had a small protein deletion mutation (Table 1). Two alleles resulted in changes in motifs conserved across evolution and found in all SCR homologs, the YPWM (Scr3) and octapeptide (Scr14) motifs. Two alleles resulted in changes in insect-specific regions of SCR, the DYTQL (Scr15) motif, and the CTD (Scr6). Two alleles resulted in the same change in a nonconserved region of SCR (Scr5 and Scr8) and one allele (Scr7) had no changes in the coding region of Scr.
To determine whether the sequence changes identified in the SCR protein were uniformly or differentially pleiotropic, detailed and quantitative analyses of the phenotype of the viable alleles were performed. For these analyses, the phenotype was assessed in imagos or larvae hemizygous for the Scr allele and a deletion that encompassed the Scr and pb loci (pb34). The three phenotypes assayed were the number of sex comb bristles that developed on the prothoracic legs of males, the number of rows of pseudotracheae that developed in the proboscis, and the number of cells per salivary gland (Figure 3; Table 2). Haplo-insufficiency of the Scr wild-type allele was observed for the number of sex comb bristles, which decreased from 9.5 to 6.3 (P < 0.001). Haplo-insufficiency of the Scr wild-type allele was not observed for the number of larval salivary gland nuclei that formed: on average 121.1 nuclei formed in homozygous wild-type larvae, whereas 117.7 nuclei formed in larvae heterozygous for pb34. Although neither Scr (Table 2) nor pb (data not shown) are haplo-insufficient for formation of pseudotracheal rows, the deletion of both loci, in pb34 does result in a significant reduction of the number of rows relative to wild type (P < 0.001).
Figure 3.—
The adult phenotypes in the proboscis and prothoracic leg of viable Scr alleles. (A) The structure of the SCR proteins encoded by the Scr alleles; the color scheme for the SCR domains is the same as used in Figure 1. Scr3, Scr6, Scr14, and Scr15 have amino acid sequence changes in the YPWM motif, CTD, octapeptide motif, and DYTQL motif, respectively. Scr5 and Scr8 have the same change in a nonconserved region of SCR, and Scr7 is wild type. (B) Scanning electron micrographs of the adult labial palps of Scrx/pb34 mutants. (C) Scanning election micrographs of the fifth tarsal segment of the adult prothoracic leg of Scrx/pb34 mutants. The arrowheads indicate the position of the sex combs.
TABLE 2.
Affect of sequence changes in viable hypomorphic Scr alleles on the phenotypes of the proboscis, prothoracic leg, and salivary glands (± SEM)
| Mean no. of Sex comb bristles, /pb34 | Mean no. nuclei per salivary gland, /pb34 | Mean rows of pseudotrachea
|
||
|---|---|---|---|---|
| Allele | /pb34 | /Scr13A | ||
| Scr+/Scr+ | 9.5 ± 0.2 (a) | 121.1 ± 2.2 (a) | 6.0 ± 0.0 (a) | 6.0 ± 0.0 (a) |
| Scr+ | 6.3 ± 0.2 (b) | 117.7 ± 3.9 (a) | 5.3 ± 0.1 (b) | 6.0 ± 0.0 (a) |
| Scr3 | 2.4 ± 0.2 (d) | 82.6 ± 2.2 (c) | 2.2 ± 0.2 (e) | 3.1 ± 0.1 (e) |
| Scr5 | 0.0 ± 0.0 (e) | 110.7 ± 3.1 (a) | 2.6 ± 0.2 (e) | 3.3 ± 0.1 (e) |
| Scr6 | 2.3 ± 0.2 (d) | 105.6 ± 3.9 (a, b) | 4.4 ± 0.1 (c) | 5.9 ± 0.1 (a, b) |
| Scr7 | 3.5 ± 0.2 (c) | 115.8 ± 4.1 (a) | 4.3 ± 0.1 (c) | 5.6 ± 0.1 (b) |
| Scr8 | 0.0 ± 0.0 (e) | 114.2 ± 4.1 (a) | 3.6 ± 0.1 (d) | 3.9 ± 0.1 (d) |
| Scr14 | 7.0 ± 0.3 (b) | 114.8 ± 3.2 (a) | 5.4 ± 0.3 (a, b, c) | 4.8 ± 0.2 (c) |
| Scr15 | 0.4 ± 0.2 (e) | 93.4 ± 2.6 (b, c) | 2.6 ± 0.2 (e) | 5.8 ± 0.1 (a, b) |
Data in the same column with the same letters are not significantly different (P < 0.05).
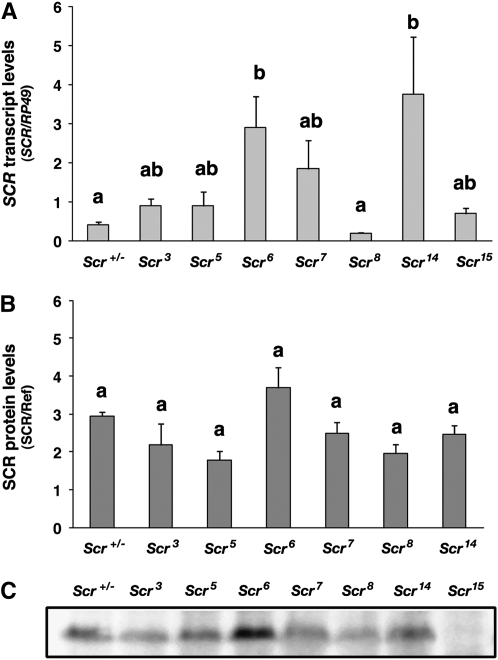
In addition to the phenotypic analysis, pupal SCR transcript and SCR protein levels were determined for the viable Scr alleles. The levels of mutant mRNA were not significantly different from wild type, with the exception of a significant increase in transcript levels in Scr6 (P = 0.02) and Scr14 (P = 0.01; Figure 4A). However, these differences were not translated into increased levels of SCR6 and SCR14 protein expression. All mutant protein accumulated to similar levels (P = 0.07), with the exception of SCR15 (Figure 4B). SCR15 is a small protein deletion not detected on a Western. This is most likely because the epitope for the anti-SCR 6H4.1-s antibody (Glicksman and Brower 1988) is in the region that is either missing or disrupted by the SCR15 deletion, and interestingly this region was also affected in the SCR2 truncated protein (Figure 1A). The β-galactosidase-SCR fusion protein used to generate the monoclonal antibody started at amino acid 79 of the SCR protein; therefore, the epitope must be between amino acids 79 and 130 (SCR4), which is the region affected in both SCR15 and SCR2 (Riley et al. 1987; Glicksman and Brower 1988). Since no significant differences in protein levels were observed, the mutant phenotypes observed are most likely due to a decrease in protein activity, with the exception of SCR7.
Figure 4.—
Pupal transcript and protein expression from viable Scr alleles. (A) Real-time PCR quantification of SCR transcript levels in Scrx/pb34 pupae. SCR transcript levels were normalized to the level of Ribosomal protein-49 (RP49) and are expressed as a ratio of SCR to RP49 in the graph. Standard error of the mean (SEM) is indicated for all measurements and the letters indicate statistical significance. A significant increase was observed in SCR6 and SCR14 transcript levels relative to wild type. (B) Protein expression levels in Scrx/pb34 pupae, quantified from Western blots. The mouse monoclonal anti-SCR 6H4.1-s antibody (Glicksman and Brower 1988) detects, in addition to SCR, some nonspecific proteins and one of these nonspecific bands was used as a loading control. Protein levels are expressed as a ratio of SCR to loading control (SCR/Ref). No significant differences were detected for protein expression levels. (C) Western blot showing expression of SCR from viable alleles. SCR15 is a small protein deletion and a band migrating with SCR+ is not present. In addition, no smaller band corresponding to SCR15 was detected.
Multiple differential pleiotropy:
The sequence analysis of the viable alleles identified protein changes in four of the five conserved sequences of SCR, the exception being the homeodomain. To analyze the pleiotropy of the Scr alleles, we ranked the alleles from weakest to strongest Scr phenotype in each of the three tissues examined. If each region of SCR is uniformly required in all tissues, the same allelic series would be expected for each tissue; however, this was not observed (Table 2). The rank order for the number of salivary gland nuclei was Scr+/− = Scr7 = Scr14 = Scr8 = Scr5 = Scr6 ≥ Scr15 = Scr3; for the number of sex comb bristles the rank order was Scr14 = Scr+/− > Scr7 > Scr3 = Scr6 > Scr15 = Scr8 = Scr5; and for the number of pseudotracheal rows, the order was Scr+/− = Scr14 ≥ Scr6 = Scr7 > Scr8 > Scr15 = Scr5 = Scr3. However, the order for the number of pseudotracheal rows changed when two doses of the pb locus was present to Scr+/− = Scr6 = Scr15 ≥ Scr7 > Scr14 > Scr8 > Scr5 = Scr3. The rank order varied in the tissues examined, and particularly important (indicated in boldface type) was the placement within the order of changes in the DYTQL motif (Scr15), YPWM motif (Scr3), octapeptide (Scr14), and CTD (Scr6), demonstrating a clear differential requirement for the DYTQL and YPWM motifs in the tissues. In addition, the order of the alleles for the number of pseudotracheal rows changed when either one or two pb loci were present; this change is not observed for the number of sex comb bristles (Table 3) or the number of salivary gland nuclei (Table 4). The most interesting shift was that of Scr15 and Scr6. When two pb loci were present both Scr6 and Scr15 exhibited a phenotype not significantly different from wild type (both P = 1.0); however, in the presence of a single pb locus both Scr alleles had significantly less pseudotrachea than wild type (both P < 0.001). The number of pseudotracheal rows observed with Scr5 and Scr8 were not significantly affected by changes in the dose of pb (P = 0.1 and P =1.0, respectively). Scr3 showed a slight yet significant decrease in the number of pseudotracheal rows by the loss of one copy of pb (P = 0.02), whereas, Scr6 and Scr15 showed a stronger significant decrease from a wild-type number of pseudotracheal rows (both P < 0.001). This suggests that changes in the DYTQL motif (Scr15) and the CTD (Scr6) are sensitive to changes in pb dose and that these domains may mediate an interaction with PB required for proboscis formation.
TABLE 3.
Mean number of sex comb bristles on prothoracic legs of males (± SEM)
| Allele | Class | /Scr+ | /Scr14 | /pb34 | /Scr13A |
|---|---|---|---|---|---|
| Scr+ | 9.5 ± 0.2 (a) | 6.9 ± 0.2 (a) | 6.3 ± 0.2 (a) | 6.3 ± 0.1 (a) | |
| Scr3 | Hypo | 8.4 ± 0.2 (b) | 4.7 ± 0.1 (b) | 2.4 ± 0.2 (c) | 2.2 ± 0.1 (c) |
| Scr5 | Hypo | 7.4 ± 0.2 (c, d) | 4.4 ± 0.1 (b) | 0.0 ± 0.0 (d) | 0.0 ± 0.0 (d) |
| Scr6 | Hypo | 8.9 ± 0.3 (a, b) | 6.3 ± 0.2 (a) | 2.3 ± 0.2 (c) | 2.0 ± 0.2 (c) |
| Scr7 | Hypo | 9.4 ± 0.2 (a) | 6.0 ± 0.1 (a) | 3.5 ± 0.2 (b) | 3.5 ± 0.1 (b) |
| Scr8 | Hypo | 7.9 ± 0.2 (b, c) | 4.3 ± 0.1 (b) | 0.0 ± 0.0 (d) | 0.0 ± 0.0 (d) |
| Scr14 | Antimorph | 6.9 ± 0.2 (d, e) | — | 7.0 ± 0.3 (a) | 2.6 ± 0.1 (c) |
| Scr15 | Hypo | 8.4 ± 0.2 (b) | 4.5 ± 0.2 (b) | 0.4 ± 0.2 (d) | 0.5 ± 0.2 (d) |
| Scr1 | Lethal-hypo | 6.9 ± 0.1 (d, e) | 3.3 ± 0.1 (c) | ||
| Scr2 | Null | 6.5 ± 0.1 (e, f) | 2.8 ± 0.1 (c, d) | ||
| Scr4 | Null | 5.9 ± 0.1 (f) | 2.4 ± 0.1 (d) | ||
| Scr11 | Null | 6.3 ± 0.2 (e, f) | 1.5 ± 0.3 (d, e) | ||
| Scr13 | Null | 6.5 ± 0.1 (e, f) | 2.6 ± 0.3 (c, d) | ||
| Scr13A | Null | 6.3 ± 0.1 (e, f) | 2.6 ± 0.1 (c, d) | ||
| Scr16 | Null | 5.9 ± 0.1 (f) | 1.9 ± 0.3 (c, d, e) | ||
| Scr17 | Null | 6.3 ± 0.1 (e, f) | 1.4 ± 0.2 (e) | ||
| pb34 | Df | 6.3 ± 0.2 (e, f) | 7.0 ± 0.3 (a) |
Data in the same column with the same letters are not significantly different (P < 0.05).
TABLE 4.
Mean number of pseudotracheal rows (± SEM) for Scr mutants
| Allele | Class | /Scr14 |
|---|---|---|
| Scr+ | 6.0 ± 0.0 (a) | |
| Scr3 | Hypo | 5.9 ± 0.1 (a) |
| Scr5 | Hypo | 6.0 ± 0.0 (a) |
| Scr6 | Hypo | 6.0 ± 0.0 (a) |
| Scr7 | Hypo | 6.0 ± 0.0 (a) |
| Scr8 | Hypo | 6.0 ± 0.1 (a) |
| Scr15 | Hypo | 5.8 ± 0.1 (a) |
| Scr2 | Null | 3.3 ± 0.1 (e) |
| Scr4 | Null | 3.6 ± 0.1 (d, e) |
| Scr13 | Null | 4.0 ± 0.1 (c, d, e) |
| Scr13A | Null | 4.8 ± 0.2 (b) |
| Scr11 | Null | 5.1 ± 0.1 (b) |
| Scr16 | Null | 4.0 ± 0.3 (b, d, e) |
| Scr17 | Null | 4.0 ± 0.1 (c, d) |
| pb34 | Df | 5.4 ± 0.3 (a, b, c) |
Data in the same column with the same letters are not significantly different (P < 0.05).
Scr14 an antimorphic allele:
Phenotypic analysis of Scr14/pb34 flies and larvae suggested that Scr14 produces the same amount of SCR activity as an Scr+ allele (Table 2). To address the question of how Scr14 was identified in the screens for Scr mutants, the sex comb bristles of all viable alleles over the Scr+ allele were counted (Table 3). The viable alleles Scr3, Scr5, Scr6, Scr7, Scr8, and Scr15 resulted in a number of sex comb bristles that was between that observed with two Scr+ alleles and one Scr+ allele. The relative order of severity in reduction of sex comb bristle number in this heterozygous situation is the same as observed when these alleles are over pb34. However, Scr14, rather than resulting in no reduction in the number of sex comb bristles as might be expected from the data in Table 2, resulted in the strongest reduction of sex comb bristles of all viable alleles. Indeed, there was no significant difference in the number of sex comb bristles between Scr+/pb34, Scr14/pb34, and Scr14/Scr+ (P = 1.0). These results suggested the possibility that Scr14 was an antimorphic allele that alone encoded a fully active protein, but which in combination with SCR protein resulted in a 50% reduction of total SCR activity. To explain the 50% reduction in total SCR activity, we proposed that at least two SCR molecules were required in a protein complex for the complex to have SCR activity, and that in a Scr14/Scr+ heterozygote the 50% of complexes that contained SCR+ and SCR14 were inactive but the 50% of complexes containing either SCR+ or SCR14 were active. To gain greater insight into the interaction of SCR14 with SCR, the phenotypes of Scr14 over all nonsense, hypomorphic lethal, and viable alleles were assessed (Table 3).
Scr14 interactions with Scr alleles:
The initial rationale for crossing all viable and nonsense alleles to Scr14 was to determine whether the protein products of the viable and nonsense alleles would interact with SCR14 and further reduce SCR activity such that <6.3 sex comb bristles, fewer than six pseudotrachael rows, and <110 salivary gland nuclei formed. The reduction was expected for an interaction because the 25% of SCR14 homocomplexes would be fully active but the 25% of homocomplexes that contain just the mutant polypeptides would be either inactive or partially active. As with the SCR+ protein, all complexes containing SCR14 and the SCR mutant protein would be inactive. All nonsense alleles in combination with Scr14 resulted in a reduction in the number of sex combs from 6.3 to an average of 2.2 (Table 3, in italics), and a reduction in the number of pseudotracheal rows from six to an average of 4.1 (Table 5), but no reduction in the number of salivary gland nuclei was observed (Table 4). In one sense these reductions suggest that the nonsense alleles are antimorphic to the Scr14 antimorphic allele. This ability to interact with Scr14 mapped to the first 112 amino acids of SCR encoded by Scr2. The lethal hypomorphic allele Scr1, the missense change in the HD, also reduced the number of sex comb bristles that formed. The viable alleles Scr3, Scr5, Scr8, and Scr15 in combination with Scr14 also significantly reduced the number of sex comb bristles that form below 6.3. Scr15 is particularly important because the encoded product is a deletion of T83–P117, suggesting that the first 82 amino acids of SCR contain a motif important for an interaction with Scr14. The combination of the viable alleles Scr3, Scr5, Scr8, and Scr15with Scr14 all resulted in a reduction of sex comb bristle number consistent with the level of SCR activity exhibited by these alleles over pb34 (Table 3). Scr6 and Scr7 do not result in a significant reduction of sex comb bristle formation (P = 0.8 and P = 0.06, respectively; Table 3 in boldface type). Finally we tested whether a null and the viable alleles would interact with one another as is observed with Scr14. The number of sex comb bristles of all viable alleles over pb34 was not significantly different from the number of sex comb bristles of the viable alleles over the nonsense allele Scr13A (Table 3); therefore, the intragenic interaction is specific to Scr14.
TABLE 5.
Mean number of nuclei per salivary gland (± SEM) for Scr mutants
| Allele | Class | /pb34 | /Scr14 | /Scr13A |
|---|---|---|---|---|
| Scr+ | 117.7 ± 3.9 (a) | 112.8 ± 3.0 (b, c) | 123.5 ± 2.6 (a) | |
| Scr3 | Hypo | 82.6 ± 2.2 (c) | 108.9 ± 3.0 (c) | 76.8 ± 1.6 (c) |
| Scr5 | Hypo | 110.7 ± 3.1 (a) | 128.3 ± 2.4 (a) | 119.4 ± 2.9 (a) |
| Scr6 | Hypo | 105.6 ± 3.9 (a, b) | 126.0 ± 5.9 (a, b) | 109.5 ± 4.1 (a, b) |
| Scr7 | Hypo | 115.8 ± 4.1 (a) | 120.8 ± 3.7 (a, b, c) | 117.8 ± 2.8 (a) |
| Scr8 | Hypo | 114.2 ± 4.1 (a) | 112.8 ± 2.7 (b, c) | 114.7 ± 3.4 (a) |
| Scr14 | Antimorph | 114.8 ± 3.2 (a) | — | 123.6 ± 2.7 (a) |
| Scr15 | Hypo | 93.4 ± 2.6 (b, c) | 110.3 ± 3.1 (b, c) | 99.4 ± 1.5 (b) |
| Scr1 | Lethal-hypo | 116.9 ± 4.7 (a, b, c) | ||
| Scr2 | Null | 109.4 ± 3.3 (c) | ||
| Scr13A | Null | 123.6 ± 2.7 (a, b, c) | ||
| pb34 | Df | 114.8 ± 3.2 (a, b, c) |
Data in the same column with the same letters are not significantly different (P < 0.05).
In summary, nonsense and viable alleles interact with the antimorphic Scr14 allele, and this intragenic interaction was specific to the Scr14 allele. The intragenic interaction between Scr alleles and Scr14 was observed for adult sex comb and pseudotracheae formation, but not for larval salivary gland formation. The hypomorphic allele Scr6 showed no intragenic interaction.
Cold-sensitive alleles
Scr6 was previously characterized as a cold-sensitive allele on the basis of increased lethality at 18° relative to 25° (Pattatucci et al. 1991). Examination of the phenotypes of the proboscis and prothoracic legs at 18° and 23° revealed a significant decrease in the number of rows of pseudotrachea (P = 0.01) and number of sex comb bristles (P < 0.001) that developed in Scr6 mutants at 18° (Table 6). At 18°, Scr6/Scr14 does not significantly reduce the number of sex combs relative to Scr14/Scr+. Therefore, at both 18° and 23° Scr6 does not interact with Scr14.
TABLE 6.
Affect of temperature on proboscis and prothoracic leg phenotypes in cold-sensitive mutants
| Mean no. of pseudotracheal rows
|
Mean no. of sex comb bristles
|
|||
|---|---|---|---|---|
| Genotype | 18° | 23° | 18° | 23° |
| Scr+/pb34 | 5.4 ± 0.1 | 5.3 ± 0.1 | 5.7 ± 0.2 | 6.3 ± 0.2 |
| Scr5/pb34 | 2.4 ± 0.2 | 2.7 ± 0.1 | 0 | 0 |
| Scr6/pb34 | 4.1 ± 0.1* | 4.5 ± 0.1 | 1.1 ± 0.3* | 2.3 ± 0.2 |
| Scr8/pb34 | 2.7 ± 0.1* | 3.6 ± 0.1 | 0 | 0 |
| Scr+/Scr14 | 7.0 ± 0.3 | 6.9 ± 0.2 | ||
| Scr6/Scr14 | 6.1 ± 0.2 | 6.3 ± 0.2 | ||
| Scr8/Scr14 | 3.6 ± 0.1 | 4.3 ± 0.1 | ||
Asterisks denote statistically significant differences for a particular genotype at 18° from values obtained at 23° (P < 0.01).
Scr5 and Scr8 had the same missense DNA change that results in an Ala288 to Thr protein change. These two alleles were isolated in different mutational screens (Lindsley and Zimm 1992) and have very distinct patterns of polymorphisms suggesting that these alleles were independently isolated (Table 1). Scr5 and Scr8 mutants both have a wild-type number of salivary gland cells and no sex comb bristles at 23°; however, despite having the same change these two alleles exhibit distinct phenotypes: Scr5 mutants had an average of 2.6 rows of pseudotrachea and Scr8 mutants had 3.6 rows, which are both significantly different from wild type (P < 0.001) and significantly different from one another (P = 0.01). Also, Scr8 mutants were cold sensitive and Scr5 mutants were not (Table 6). It has been suggested that the translation of SCR mRNA is regulated (Mahaffey and Kaufman 1987); therefore, to test the possibility that translation of SCR8 mRNA may be cold sensitive, the 5′ and 3′ noncoding regions of Scr5 and Scr8 were sequenced but no differences were found between the alleles (data not shown).
DISCUSSION
Scr14 a missense allele in the octapeptide:
Scr14 is a Ser10-to-Leu change in the octapeptide motif of SCR. The octapeptide motif is found in all SCR homologs, and for SCR and murine HOXA5 the octapeptide is required for the formation of ectopic salivary glands in Drosophila (Zhao et al. 1996; Tour et al. 2005). A submotif of the octapeptide, SSYF, is found in the Drosophila HOX proteins Labial, ANTP, Deformed, and UBX, which in the case of UBX is important for function (Tour et al. 2005). Therefore, it was surprising that the Scr14 change of a Ser10 to Leu of the most conserved residue of the octapeptide submotif had little affect on the number of sex comb bristles, pseudotracheal rows, and salivary gland nuclei when hemizygous over a Df. The only strong phenotype was a reduction of the number of sex comb bristles when heterozygous. This suggests that Scr14 is an antimorphic allele, and we propose that SCR14 forms inactive heterocomplexes with SCR+ resulting in a 50% reduction of total SCR activity. This model of inhibition of SCR at the protein level is favored over a mechanism of pairing-dependent repression because all null and hypomorphic alleles that interact with the Scr14 allele are DNA sequence changes that would result in an altered protein product (Southworth and Kennison 2002). These alleles that interact with Scr14 encode proteins that result in a further reduction of total SCR activity. These alleles produce inactive or partially active SCR proteins that interact and inactivate SCR14. In the case of nonsense alleles, the only active complex left is that containing two SCR14 molecules (25%). This ability to interact with SCR14 maps to the first 82 amino acids of SCR, and the only conserved domain in this region of SCR is the octapeptide motif. However, two alleles Scr6 and Scr7 did not interact genetically with Scr14.
Scr7 an allele with no changes in the coding region:
Scr7 mutants had reductions in the number of sex comb bristles, reductions in the number of rows of pseudotrachea, but no reductions in the number of salivary gland cells. Transcript and protein levels in this mutant do not differ significantly from wild type. The Scr7 phenotype may be caused by a subtle change in the pattern of SCR expression at the pupal stage. Therefore, it is possible that this allele may be a regulatory mutant. If Scr7 is a regulatory mutant, it is expected to show a weak genetic interaction with Scr14 because varying the ratio of SCR+ to SCR14 below 1 results in a theoretical maximum loss of 17.5% of total SCR activity (about one sex comb bristle) when at a ratio of 0.5 [number of active complexes = (1/2 − (proportion of SCR+)(proportion of SCR14))total SCR]. This weak effect is due to less inactive SCR14 SCR complexes forming as the expression of SCR decreases.
Scr6 two missense changes in the CTD:
The cold-sensitive Scr6 allele has two missense mutations in the conserved CTD of SCR, both these missense mutations result in amino acid changes of highly conserved amino acids of the CTD (Curtis et al. 2001). The lack of an intragenic interaction between Scr6 and Scr14 suggests that SCR6 does not interact with SCR14 to form an inactive complex. The inability of SCR6 to interact with SCR14 is not due to a lack of CTD function because all proteins expressed from a nonsense allele lack the CTD but are still able to interact with SCR14. We propose that normally the CTD domain has a role in negative regulation of SCR activity by binding the octapeptide, and that the Scr6 missense mutations result in the expression of a protein that is hyperactive for a CTD function of binding and masking the octapeptide. This explains why SCR6 has less activity than SCR; the octapeptide is not available for complex formation. In addition, in SCR6 the octapepide is not available to interact with SCR14. The proposed intramolecular interaction of the octapeptide with the CTD in SCR6 would be temperature sensitive, rendering SCR6 activity cold sensitive.
A possible mechanism of Scr14 antimorphy:
Many models can be proposed for the inactivity of the SCR14 SCR protein complex. But most of these models have difficulty explaining why the genetic interactions of null and hypomorphic alleles are specific to Scr14. One example is an incompatibility model where SCR14 and SCR form an inactive complex because the transcription machinery may not recognize the conformation that the heterotypic octapeptides adopt. In this model and others like it, it is assumed that SCR14 and SCR interact with the same affinity as that between two SCR or two SCR14 molecules. Therefore, complexes would form between the products of nonsense alleles and hypomorphic alleles, and because these complexes only have one HD they would be expected to be less active or inactive. This is not observed because the phenotype of a hypomorphic allele over a deletion is the same as that over a null nonsense allele.
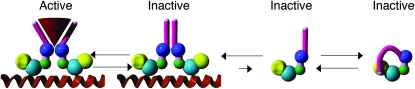
Our favored model is a locked complex model, which to understand first requires presentation of a speculative model for SCR activity. We conjecture that there is a dynamic equilibrium between four forms of SCR during adult sex comb and pseudotrachea formation (Figure 5). Three interactions of the octapeptide mediate the dynamic equilibrium: the octapepide interacting with a component(s) of the transcription machinery, the octapeptide interacting with another octapeptide motif to mediate complex formation of SCR, and the octapeptide interacting intramolecularly with the CTD. In two of the forms SCR is a monomer, and as a monomer SCR is in dynamic equilibrium with a form where the octapeptide is exposed for complex formation with another SCR molecule and a form where the octapeptide interacts with the CTD and is not available for complex formation. The two SCR protein complexes bound to DNA are in dynamic equilibrium between a complex held together by an interaction between two octapeptides, and a complex held together by an indirect interaction of the two octapeptides mediated by a component(s) of the transcriptional machinery. SCR is only active when it is interacting with the transcriptional machinery.
Figure 5.—
Model for SCR activity. The SCR protein is not drawn to scale with the five conserved domains of SCR emphasized: octapeptide (purple), DYQTL (blue), YPWM (green), HD (cyan), and CTD (yellow). SCR exists in dynamic equilibrium between four forms: inactive monomer available for complex formation via the octapeptide, inactive monomer in which the CTD masks the octapeptide, inactive complex bound to DNA, and active complex bound to DNA—stablized by an indirect interaction of two octapeptides with a component(s) of the transcription machinery (red cone).
In the locked complex model, the SCR14 SCR complex is inactive because the two octapeptides are unable to dissociate and interact with the transcriptional machinery and unable to dissociate to form monomers; dynamic equilibrium is lost. The locked complex model also explains why the genetic interactions are specific to Scr14. An interaction between nonsense null alleles and viable alleles is not observed, because all these alleles have a wild-type octapeptide sequence that is in dynamic equilibrium. The complexes that form between a truncated SCR protein and an SCR molecule encoded by a viable allele are transient, falling apart rapidly, and with the additional interaction between SCR protein complex and DNA, the formation of partially active complexes of the SCR proteins expressed from viable alleles is favored. Our model, based on interpretation of genetic evidence, will require biochemical tests of the interaction of the octapeptide with itself and the CTD and of the formation of a locked complex between SCR and SCR14.
Scr3 a missense mutation in the YPWM motif:
Scr3 encodes a protein in which Pro306 is changed to Leu, altering the sequence of the YPWM motif to YLWM. The effect of this change was the most severe of all the viable hypomorphic alleles on the number of pseudotracheal rows and salivary gland cells. The Scr3 allele had a weaker affect on the prothoracic leg identity. The YPWM motif is a highly conserved motif found in all HOX proteins, and is a binding site for two proteins: Extradenticle (EXD) and Bric-à-Brac Interacting Protein 2 (BIP2) (Joshi et al. 2007; Prince et al. 2008). The results are difficult to explain solely as a loss of EXD binding to SCR because although SCR and EXD are essential for salivary gland formation (Ryoo and Mann 1999), EXD is not required for sex comb and pseudotrachea formation (Percival-Smith and Hayden 1998).
The YPWM motif of SCR makes a protein–protein interaction with the hydrophobic pocket of the EXD HD (Joshi et al. 2007); therefore, a mutation in the YPWM motif may be expected to result in no salivary gland formation. Indeed, deleting the YPWM motif of the mammalian homolog, HOXA5, results in an inability to induce ectopic Forkhead (FKH) expression (Zhao et al. 1996); however, this deletion of 16 amino acids includes a His residue important for minor groove interactions by SCR and EXD with the fhk enhancer element (Joshi et al. 2007). A potential explanation for the weak reduction of the salivary gland is that the Pro residue of the YPWM motif is not essential for binding to EXD. In fact, in the Apis mellifera SCR homolog, the YPWM motif is YSWM. Also, the YPWM motif is YKWM and HEWT in the Drosophila HOX proteins Labial and Abdominal-B, respectively. The structure of the vertebrate HOX-EXD (HoxB1, PBX1) homologous heterodimer was solved with a FDWM sequence (Piper et al. 1999). Therefore, the Pro306-to-Leu change may not completely inactivate the YPWM motif.
An explanation for the observation that EXD is not required for pseudotracheae or sex comb development is that the YPWM of SCR interacts with a protein other than EXD. The YPWM motif of ANTP binds BIP2 (Prince et al. 2008). BIP2 is a TATA binding protein-associated factor, associated with the basal transcriptional machinery, that when coectopically expressed with ANTP promotes the formation of ectopic wing tissue in Drosophila (Gangloff et al. 2001; Prince et al. 2008). Since BIP2 is expressed widely throughout all of the imaginal discs of third instar larvae (Gangloff et al. 2001), there is a strong possibility that BIP2 may interact with the YPWM motifs of other HOX proteins such as SCR. If BIP2 binds to the SCR YPWM, the Pro306-to-Leu change observed in the YPWM motif of SCR3 could decrease the ability of these proteins to interact, explaining the proboscis toward maxillary palp transformation and reduction in sex comb bristle number in Scr3 mutants (Figure 3).
Salivary gland formation requires both SCR and EXD for the expression of FKH, which is required for salivary gland formation. The evidence for SCR and EXD binding as a protein complex to a fhk enhancer is extensive at both functional and structural levels (Joshi et al. 2007). However, EXD is not required for the formation of sex comb bristles and pseudotrachea (Percival-Smith and Hayden 1998; Joulia et al. 2006). In addition, the intragenic interaction between Scr14 and Scr alleles is observed for the formation of the sex combs and pseudotrachea, but not salivary gland nuclei. To resolve this inconsistency, we suggest that SCR requires complex formation with itself for sex comb and pseudotracheae formation and complex formation with the HOX cofactor EXD for salivary gland formation. This phenomenon is similar to the observation that UBX does not require EXD for haltered development (Galant and Carroll 2002).
Scr1 a missense mutation in the HD:
The Glu365 residue of the SCR HD is well conserved through evolution and is found in all Drosophila HOX HDs; however, this residue does not mediate important contacts in the crystal structures of SCR, ANTP, and UBX or the NMR structure of ANTP (Joshi et al. 2007; Billeter et al. 1993; Pasner et al. 1999; Fraenkel and Pabo 1998). Glu365 is the first amino acid of the third α-helix of the HD, the helix that makes direct contacts with the major groove of DNA; therefore, the importance of this residue may lie in its position within the HD. A change from an acidic Glu residue to a bulkier, basic Lys residue may affect the structure of the third α-helix and subsequently the ability of the HD to bind DNA. Although the hypomorphic Scr1 allele may suggest that SCR has a HD independent activity like the pair rule protein Fushi tarazu (Hyduk and Percival-Smith 1996), mutational studies have shown that the SCR HD is essential for SCR activity. Berry and Gehring (2000) showed that altering two amino acids in the N-terminal arm of the SCR HD to aspartic acids inactivated the SCR protein. Also Joshi et al. (2007) showed that changing Arg3 of the SCR HD resulted in an inability of the SCR protein to activate the fkh enhancer. The observation that changes in conserved amino acids of the HD result in a hypomorphic allele is not novel; three of four missense alleles with changes in highy conserved positions of the Proboscipedia HD were hypomorphic, only one was null (Tayyab et al. 2004).
Scr15 is a deletion of the conserved DYTQL motif:
Scr15 encodes a 35-amino-acid deletion of Thr83 through Pro117 encompassing the insect-specific DYTQL motif. Although, this change had a strong affect on all three phenotypes assessed, the DYTQL motif is not essential for SCR function, which is similar to the nonessential role of the insect-specific UBX QA motif (Hittinger et al. 2005). However, analysis of differential pleiotropy in flies with one or two copies of pb, suggests that the DYTQL motif and the CTD mediate an interaction with PB in pseudotrachea formation. We have proposed that the CTD encoded by Scr6 is hyperactive; therefore, PB may have a role in overcoming the negative regulation of SCR activity mediated by the CTD. And since Scr15 is the loss of the DYTQL motif, we propose that the DYTQL motif may also have a role assisting in PB overcoming negative regulation of SCR activity possibly mediated by the CTD.
Acknowledgments
We thank Sheila Macfie for her assistance in analyzing statistical data. We thank the reviewers for their comments, which assisted greatly in improving the manuscript. This work was supported by the Natural Sciences and Engineering Research Council of Canada in a Discovery grant to A.P.-S. and a postgraduate scholarship (to L.S.). We thank the Bloomington Stock Center for providing fly stocks and Gary Struhl for providing Scr13A flies. The anti-SCR 6H4.1-s developed by D. Brower and M. A. Glicksman (1988) was obtained from the Developmental Studies Hybridoma Bank and developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.100438/DC1.
References
- Alonso, C. R., and M. Akam, 2003. A Hox gene mutation that triggers nonsense mediated RNA decay affects alternative splicing during Drosophila development. Nucleic Acid Res. 31 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, M., and W. J. Gehring, 2000. Phosphorylation status of the SCR homeodomain determines its functional activity: essential role for protein phophatase 2A,B'. EMBO J. 19 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter, M., Y. Q. Qian, G. Otting, M. Muller, W. Gehring et al., 1993. Determination of the nuclear magnetic resonance solution structure of an Antennapedia Homeodomain-DNA complex. J. Mol. Biol. 234 1084–1093. [DOI] [PubMed] [Google Scholar]
- Bustin, S. A., 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 169–193. [DOI] [PubMed] [Google Scholar]
- Capovilla, M., M. Brandt and J. Botas, 1994. Direct regulation of decapentaplegic by Ultrabithorax and its role in Drosophila midgut morphogenesis. Cell 76 461–475. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., 1995. Homeotic genes and the evolution of arthopods and chordates. Nature 376 479–485. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., 2005. Evolution at two levels: on genes and form. PLoS Biol. 3 1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, C. D., J. A. Brisson, M. A. DeCamillis, T. D. Shippy, S. J. Brown et al., 2001. Molecular characterization of Cephalothorax, the Tribolium ortholog of Sex combs reduced. Genesis 30 12–20. [DOI] [PubMed] [Google Scholar]
- Fraenkel, E., and C. O. Pabo, 1998. Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat. Struct. Biol. 5 692–697. [DOI] [PubMed] [Google Scholar]
- Galant, R., C. M. Walsh and S. B. Carroll, 2002. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129 3115–3126. [DOI] [PubMed] [Google Scholar]
- Galant, R., and S. B. Carroll, 2002. Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415 910–913. [DOI] [PubMed] [Google Scholar]
- Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier et al., 2001. The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)150) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21 5109–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glicksman, M. A., and D. L. Brower, 1988. Expression of the Sex combs reduced protein in Drosophila larvae. Dev. Biol. 127 113–118. [DOI] [PubMed] [Google Scholar]
- Hentze, M. W., and A. E. Kulozik, 1999. A perfect messenger: RNA surveillance and nonsense mediated decay. Cell 96 307–310. [DOI] [PubMed] [Google Scholar]
- Hittinger, C. T., D. L. Stern and S. B. Carroll, 2005. Pleiotropic functions of a conserved insect-specific Hox peptide motif. Development 132 5261–5270. [DOI] [PubMed] [Google Scholar]
- Hyduk, D., and A. Percival-Smith, 1996. Genetic characterization of the homeodomain-independent activity of the Drosophila fushi tarazu gene product. Genetics 142 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joulia, L., J. Deutsch, H. Bourbon and D. L. Cribbs, 2006. The specification of a highly derived arthropod appendage, the Drosophila labial palps, requires the joint action of selectors and signaling pathways. Dev. Genes Evol. 216 431–442. [DOI] [PubMed] [Google Scholar]
- Joshi, R., J. M. Passner, R. Rohs, R. Jain, A. Sosinsky et al., 2007. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai, A., S. Muller, S. Platz and M.-T. Hauser, 2002. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 34 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, T. C., 1978. Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: isolation and characterization of four new alleles of proboscipedia (pb) locus. Genetics 90 579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276 565–570. [DOI] [PubMed] [Google Scholar]
- Lewis, R. A., T. C. Kaufman, R. E. Denell and P. Tallerico, 1980. a Genetic analysis of the Antennapedia gene complex (ANT-C) and adjacent segments 84B-D. Genetics 95 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R. A., B. T. Wakimoto, R. E. Denell and T. C. Kaufman, 1980. b Genetic anlysis of the Antennapedia gene complex (Ant-C) and adjacent chromosomal regions of Drosophila melanogaster. II. Polytene chromosome segments 84A–84B1, 2. Genetics 95 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Mahaffey, J. W, and T. C. Kaufman, 1987. Distribution of the Sex combs reduced gene products in Drosophila melanogaster. Genetics 117 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Furtherstudies on the nature and causes of gene mutations, pp. 213–255 in Proceedings on the Sixth International Congress on Genetics. George Banta Publishing Co., Menasha, WI.
- Panzer, S., D. Weigel and S. K. Beckendorf, 1992. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development 114 49–57. [DOI] [PubMed] [Google Scholar]
- Passner, J. M., H. D. Ryoo, L. Shen, R. S. Mann and A. K. Aggarwal, 1999. Structure of a DNA bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397 714–719. [DOI] [PubMed] [Google Scholar]
- Pattatucci, A. M., D. C. Otteson and T. C. Kaufman, 1991. A functional and structural analysis of the Sex combs reduced locus of Drosophila melanogaster. Genetics 129 423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith, A., and D. J. Hayden, 1998. Analysis in Drosophila melanogaster of the interaction between Sex combs reduced and extradenticle activity in the determination of tarsus and arista identity. Genetics 150 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith, A., J. Weber, E. Gilfoyle and P. Wilson, 1997. Genetic characterization of the role of the two HOX proteins, Proboscipedia and Sex combs reduced, in determination of adult antennal, tarsal, maxillary palp and proboscis identities in Drosophila melanogaster. Development 124 5049–5062. [DOI] [PubMed] [Google Scholar]
- Piper, D. E., A. H. Batchelor, C. Chang, M. L. Cleary and C. Wolberger, 1999. Structure of a HoxB1–Pbx1 Heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96 587–597. [DOI] [PubMed] [Google Scholar]
- Prince, F., T. Katsuyama, Y. Oshima, S. Plaza, D. Resendez-Perez et al., 2008. The YPWM motif link Antennapedia to the basal transcriptional machinery. Development 135 1669–1679. [DOI] [PubMed] [Google Scholar]
- Riley, P. D., S. B. Carroll and M. P. Scott, 1987. The expression and regulation of Sex combs reduced protein in Drosophila embryos. Genes Dev. 1 716–730. [DOI] [PubMed] [Google Scholar]
- Ronshaugen, M., N. McGinnis and W. McGinnis, 2002. Hox protein mutation and macroevolution of the insect body plan. Nature 415 914–917. [DOI] [PubMed] [Google Scholar]
- Ryoo, H. D., and R. S. Mann, 1999. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 13 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth, J. W., and J. A. Kennison, 2002. Transvection and silencing of the Scr homeotic gene of Drosophila melanogaster. Genetics 161 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, G., 1982. Genes controlling segmental specification in the Drosophila thorax. Proc. Natl. Acad. Sci. USA 79 7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayyab, I., H. M. Hallahan and A. Percival-Smith, 2004. Analysis of Drosophila proboscipedia mutant alleles. Genome 47 600–609. [DOI] [PubMed] [Google Scholar]
- Tour, E., C. T. Hittinger and W. McGinnis, 2005. Evolutionary conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development 132 5271–5281. [DOI] [PubMed] [Google Scholar]
- Wieschaus, E., and C. Nusslein-Volhard, 1986. Looking at embryos, pp. 199–228 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL, Oxford.
- Xu, T., and G. M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zar, J. H., 1999. Biostatistical Analysis. Prentice Hall, Englewood Cliffs, NJ.
- Zhao, J. J., R. A. Lazzarini and L. Pick, 1996. Functional dissection of the mouse Hox-a5 gene. EMBO J. 15 1313–1322. [PMC free article] [PubMed] [Google Scholar]