Abstract
Next-generation methods for rapid whole-genome sequencing enable the identification of single-base-pair mutations in Drosophila by comparing a chromosome bearing a new mutation to the unmutagenized sequence. To validate this approach, we sought to identify the molecular lesion responsible for a recessive EMS-induced mutation affecting egg shell morphology by using Illumina next-generation sequencing. After obtaining sufficient sequence from larvae that were homozygous for either wild-type or mutant chromosomes, we obtained high-quality reads for base pairs composing ∼70% of the third chromosome of both DNA samples. We verified 103 single-base-pair changes between the two chromosomes. Nine changes were nonsynonymous mutations and two were nonsense mutations. One nonsense mutation was in a gene, encore, whose mutations produce an egg shell phenotype also observed in progeny of homozygous mutant mothers. Complementation analysis revealed that the chromosome carried a new functional allele of encore, demonstrating that one round of next-generation sequencing can identify the causative lesion for a phenotype of interest. This new method of whole-genome sequencing represents great promise for mutant mapping in flies, potentially replacing conventional methods.
STANDARD practices of genetic mapping typically occur in three phases. First, polymorphisms that distinguish the chromosome carrying the mutation to be mapped from that of the homolog bearing a wild-type allele of that gene must be identified. Second, by genotyping recombinant chromosomes that do or do not carry the mutation of interest, an association between polymorphisms and the mutation can be identified, which can then be used to pinpoint the location of the relevant mutation. Finally, candidate genes within the interval must be identified and regions sequenced to find the causative mutation. Often, these three steps are performed iteratively. In situations where there are few polymorphic markers or candidate genes, this process can be arduous and, depending on the organism, can consume months to years.
New genome-sequencing technologies (Margulies et al. 2005; Bentley 2006; Barski et al. 2007; Sarin et al. 2008; Smith et al. 2008; Valouev et al. 2008) show tremendous promise for reducing the time needed to identify causative mutations. Using these approaches, one may be able to directly identify causative lesions by comparing the nucleotide sequences of wild-type and mutant genomes. Indeed, we have conducted a proof-of-principle experiment to determine the feasibility of such an approach in Drosophila melanogaster. In the course of conducting an EMS-based genetic screen, we identified a chromosome, designated 791, which displayed a fused dorsal appendage phenotype in embryos of homozygous mothers. Such phenotypes usually arise from a defect in the maternal establishment of the dorso-ventral axis. To identify the mutated gene that gives rise to this phenotype, we used a next-generation sequencing platform to directly compare the nucleotide sequence of the original and the mutagenized chromosomes. Because this phenotype is well studied and our mutation is recessive, we could use complementation analysis to test the causative nature of any candidate lesions. However, even if other mutants with similar phenotypes were not already known, the small number of candidate loci identified could have been easily tested by transformation rescue. Importantly, this approach also improved our understanding of the global effects of EMS mutagenesis. Here we demonstrate how whole-genome sequencing technologies can be used to discover causative mutations and how these technologies can shed light on processes such as EMS mutagenesis and gene conversion at a genomic level.
MATERIALS AND METHODS
DNA preparation for sequencing:
DNA for sequencing was prepared from wandering third instar larvae that were homozygous for either A15 (the target chromosome) or 791 (the mutagenized chromosome). Homozygosity was determined by selection against TM6b,Tb balancer chromosomes. Wandering third instar larvae were chosen for three reasons: first, at this stage they have begun gut evacuation, which minimizes contaminating DNA from the yeast food source; second, they can be easily bleached to remove surface contamination; and third, larval salivary glands contain polytene chromosomes that are enriched for euchromatic over heterochromatic sequences. Since heterochromatic sequences are not easily assembled, especially for the short read lengths generated by Illumina sequencing, we favored minimizing their contribution to the sequencing runs.
DNA was prepared from 10 larvae that had been briefly rinsed in 50% bleach followed by water and frozen at −80° for at least 1 hr. Larvae were then homogenized in 500 μl of 10 mm Tris–HCl (pH 8.0), 20 mm EDTA, 0.1% SDS, and 5 μg of RNase A and incubated at room temperature for 10 min. A total of 5 μl of Proteinase K (20 mg/ml) and 40 μl of 10% SDS were then added and the homogenate was incubated at 65° for 1 hr, followed by 95° for 5 min. A total of 125 μl of 5 m ammonium acetate was added, tubes were incubated on ice for 10 min and spun for 10 min, and supernatant was collected and extracted once with phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform. DNA was precipitated by the addition of 2× volumes of cold ethanol, and the pellet was rinsed once with 70% ethanol. The pellet was resuspended in 50 μl of 10 mm Tris–HCl, pH 8.5.
Illumina whole-genome sequencing:
Genomic DNA (5 μg) from either A15 or 791 homozygous larvae was sheared to ∼800 bp using sonication. We then performed end repair, added “A” bases to the 3′-end of the DNA fragments, ligated adapters, and purified and size selected ligated products. Clusters were generated on the Illumina cluster station according to the manufacturer's protocol. Single read sequencing was done for 36 cycles (36 bp) on an Illumina Genome Analyzer I instrument. One flow cell was run for each library. Seven lanes were run for the A15 background strain, and seven lanes were run for the 791 mutant. The eighth lane of each flow cell was used for a Phi-X control.
Illumina data analysis and SNP detection:
Data analysis was done using a combination of commercially available software, open source software, and custom programs. Images from the Illumina Genome Analyzer were processed using the Illumina Analysis Pipeline version 0.3.0 (Firecrest, Bustard) to generate FASTQ sequence files. Reads (36 bp) that passed through the Gerald chastity filter were aligned uniquely to the reference genome sequence using the eland alignment tool. All quality filtered and uniquely aligning reads were provided to the MAQ package (Li et al. 2008; http://maq.sourceforge.net) using default settings. MAQ was used to align reads to the ensembl 49.44 release of the D. melanogaster genome (http://mar2008.archive.ensembl.org/Drosophila_melanogaster). A15 and 791 consensus sequences from MAQ for the third chromosome were then compared in a pairwise fashion. Criteria used when comparing references were a minimum read depth of 4, a homozygous consensus call, and a minimum consensus quality score of 22. Nonmatching, threshold passing pairs were then annotated. When a pair's chromosomal position was determined to land in a transcript and the resulting translated protein change was nonsynonymous, the SIFT program (Ng and Henikoff 2002) was used to predict the impact as deleterious or tolerated. All subsequent secondary analysis was performed using custom scripts and the R programming language.
Sanger sequencing validation:
Primers of 18–27 bp and temperatures of 57°–63° were designed to amplify ∼700-bp products, including at least 350 bp on either side of the putative SNP. M13 universal primer tags were appended to the 5′-end of each primer to aid in sequencing the reaction setup (forward M13 primer: TGT AAA ACG ACG GCC AGT; reverse M13 primer: AAC AGC TAT GAC CAT G). PCR reactions (15 μl) for each pair of primers were set up in 384-well plates, using TAQ Gold (Applied Biosystems). A standard PCR protocol was used for all regions: 10 min at 95°, 30 sec at 95°, 30 sec at 60°, 1 min at 72° (for a total of 30 cycles), and then 10 min at 72° followed by an 8° hold. Unincorporated primers, nucleotides, and salts were removed on a Biomek FX using AMPure cleanup (Agencourt). In a 384-well plate, 2 μl of each eluted PCR product was added to 8 μl of a Big Dye Terminator v3.1 sequencing cocktail (Applied Biosystems), including either the forward or reverse M13 sequencing primer. The same sequencing PCR cycle was used for all regions: 10 sec at 96°, 5 sec at 50°, and 4 min at 60°, followed by an 8° hold. Reactions were purified on the Biomek FX using CleanSEQ cleanup (Agencourt) and sequenced on an Applied Biosystems 3730xl sequencer. Vector NTI software (Invitrogen) was used to assemble, view the data, and detect SNPs.
RESULTS
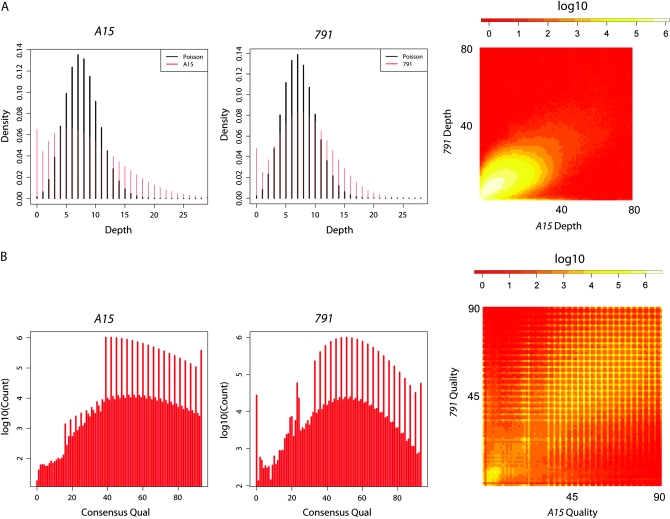
Illumina fragment libraries were made from genomic DNA isolated from homozygous larvae, carrying either the original third chromosome (designated A15) or the EMS-mutagenized third chromosome (designated 791), which carries a lesion that causes a fused dorsal appendage phenotype. Each library was run on a single flow cell on an Illumina Genome Analyzer using the single read protocol. Approximately 30 million filtered and uniquely aligning reads of 36 bp were generated for each sample. This produced 1.1 Gb of sequence for the original stock and 1.0 Gb of sequence for 791, giving 8.7× and 8.3× genome coverage, respectively (Table 1). From this set of data, we limited our analysis to the third chromosome since this chromosome was the target of mutagenesis. Sequencing coverage was not Poisson distributed. Instead, the variance in the distribution of coverage was greater than predicted by a Poisson distribution, and there was an excess of zero coverage bases (Figure 1A) for both sequence runs. If these deviations were due to an underlying random process (albeit a non-Poisson process) that was independent across samples, we would expect to see little correlation in sequence depth between the two samples. However, this was clearly not the case. Figure 1A shows a frequency heat map for pairwise coverage across the two samples. There is a clear correlation between the sequencing depth at any particular base in one run with the sequencing depth in the other. Furthermore, the zero coverage class in one sample is quite coincident with the zero coverage class in the other sample. This correlation indicates that sequence depth is non-independent across samples, suggesting a certain bias for some parts of the genome to not be sampled in each sequencing run. This bias could be due to bias in the sequencing process or due to the contribution of polytene chromosomes to the pool of DNA collected from third instar larvae. Polytene chromosomes likely vary in the extent to which they contribute sequences from different genomic regions. If coverage were truly independent across samples, the removal of low coverage data from the analysis would also be independent across samples. This would multiply the false-negative rate since a site with reasonable coverage in one sample would frequently have low coverage in another and thus be eliminated from analysis.
TABLE 1.
Run statistics
| No. of reads (in millions) | Base pairs (in millions) | Genome coverage | % error rate | Chromosome 3 base-pairs pass filter (%) | Both runs (%) [expected] | |
|---|---|---|---|---|---|---|
| A15 | 30 | 1080 | 8.7× | 0.84 ± 0.05 | 39,604,870 (75.5) | 37,165,510 (70.9) [61.8] |
| 791 | 29 | 1040 | 8.3× | 1.14 ± 0.07 | 42,910,551 (81.8) |
Statistics for A15 and 791 Illumina Genome Analyzer runs. The last column indicates the number of bases of the third chromosome that pass through the quality filter from both runs. The percentage of coverage expected, given the independence between runs for nucleotides to pass through the filter, is also given in the last column.
Figure 1.—
Coverage and quality analysis of the third chromosome from A15 and 791 runs. (A) Distribution of nucleotide coverage depth for the original A15 third chromosome and for the 791 mutagenized third chromosome. The heat map indicates pairwise coverage. (B) Distribution of MAQ consensus nucleotide quality scores for A15 and 791 for nucleotides of the third chromosome. Scores are shown only for consensus nucleotides that were not ambiguous and had a depth of at least 4. Heat map indicates pairwise quality.
Considering only sites that had nonambiguous consensus calls and a minimum sequence depth of 4 (since consensus quality scores are expected to be less meaningful for low coverage bases), we also characterized the distribution of MAQ quality scores for each nucleotide of the consensus. For both samples, the distribution of quality scores is variable at or below a score of ∼20 (Figure 1B). However, above this threshold, the distribution of quality scores appears more continuous (aside from the fact that the MAQ consensus quality score algorithm appears to give a somewhat punctate distribution of values). As with coverage depth, consensus quality scores are somewhat correlated across bases (Figure 1B). A very-high-quality consensus base in one sample is more likely to be a higher quality in the other sample. This again indicates non-independence in quality across runs—some bases are more likely than others to be read as high quality by Illumina sequencing. This is also expected when coverage across bases is correlated between runs and when bases with higher coverage have higher quality scores. For the same reason as with coverage, this non-independence makes a threshold quality cutoff for SNP determination less likely to have a drastic influence on the false-negative rate.
To identify EMS-induced mutations, we chose an approach of directly comparing the consensus sequences of the two chromosomes generated using the MAQ software program. An alternative approach would be to identify all SNPs relative to the completely sequenced reference for each genome and identify the EMS-induced mutations on the basis of the comparison of these two lists of SNPs. This method is problematic, however, since there is a great deal of natural variation that is expected to distinguish the unmutagenized chromosome from the reference genome. Even a very low false-negative or false-positive rate of SNP identification for each genome relative to the reference would lead to a large excess of putative SNPs unique to one genome that, in fact, would not be SNPs between A15 and 791.
Using a threshold that considered only nonambiguous consensus bases from both chromosomes that had a minimum read depth of 4 and a quality score of 22, we covered 70.9% of both third chromosomes. Table 1 shows that this fraction is greater than expected if the chance of a given nucleotide passing this filter is independent across samples. Furthermore, since low-coverage bases are more likely to be low-quality bases (data not shown), a portion of the genome that we removed from analysis is enriched for bases that are predisposed to being low quality. This is supported by the fact that bases excluded from analysis are enriched for repeats. While 7.6% of the third chromosome is masked by RepeatMasker, 20.6% of the bases not meeting the threshold is masked by RepeatMasker. Thus, a portion of the third chromosome that is not included in the analysis is, due to repetitiveness, unlikely to contribute to any whole-genome-sequencing SNP detection approach in Drosophila, even in the face of greater sequencing depth. Furthermore, since the primary goal is to identify mutations in genes with unique function, unidentified SNPs located within repeat sequences, such as transposons, are not likely to be SNPs of interest. The portion of the third chromosome not included in the analysis due to low coverage could have been decreased by running additional lanes of sample. When we added the results of a test run from another full flow cell of 791 reads (with somewhat lower quality and not included in this analysis), we increased the threshold of shared coverage between A15 and 791 from 70.9 to 74.8%. Thus, the percentage coverage does not increase drastically with data from additional flow cells. This is not surprising since only 80% of a complex eukaryotic genome can be uniquely mapped with short 36-bp reads (Whiteford et al. 2005). Using this threshold, we identified 165 candidate SNPs that distinguished the mutagenized third chromosome from the unmutagenized chromosome. We successfully performed Sanger sequencing on 125 of these SNPs and verified 103, giving a false-positive rate of 17.6%. For a complete list of all 103 verified SNPs, see supporting information, Table S1. Visual inspection indicated that a number of false positives were of low complexity and repeated regions while others were likely due to sequencing errors or potential PCR amplification errors during library preparation. If we apply this respective false-positive rate to the entirety of the 165 candidate SNPs, we would yield ∼136 true SNPs for the 70.9% of the genome covered after filtering. This yields ∼1 mutation/273 kb. Considering that a 45-mm dose of EMS was used, this is consistent with previous reports of an ∼1 mutation/380 kb and an ∼1 mutation/480 kb found with a 25-mm dose of EMS (Cooper et al. 2008).
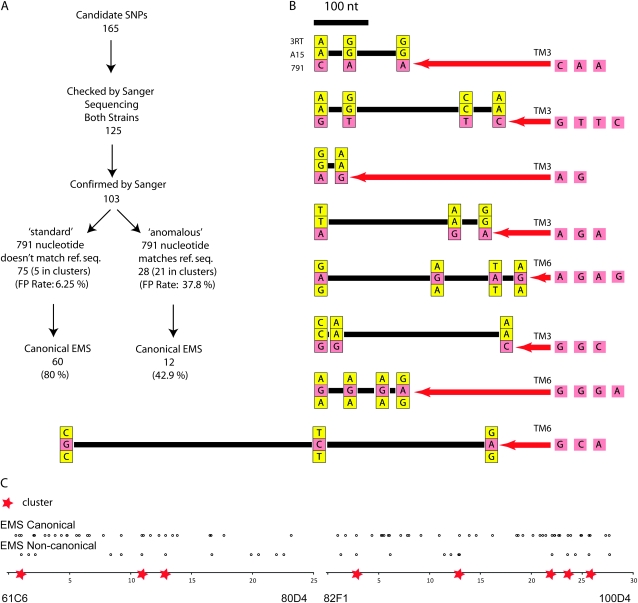
We found that the verified SNPs could be placed in two different categories (Figure 2A). The first category was designated “standard” for SNPs that distinguished the unmutagenized and mutagenized chromosome and for which the nucleotide on the mutagenized chromosome differed from the reference sequence. Seventy-five nucleotides fell into this class. The second category was designated “anomalous” for SNPs that differed between the unmutagenized and mutagenized chromosomes but for which the mutagenized chromosome had the same sequence as the reference genome. Twenty-eight nucleotides fell into this class. Interestingly, the false-positive rate was much higher for this class of SNPs (37.8%) than for the standard class (6.25%). The probability of a nucleotide differing between the unmutagenized chromosome and the reference sequence reverting to the reference sequence is exceedingly small; therefore the verified anomalous SNPs warranted further investigation (see discussion on anomalous SNPs below).
Figure 2.—
Analysis of SNPs between the original A15 and the mutagenized 791 chromosomes. (A) Classification, verification, and confirmation information for initial set of 165 candidate SNPs. (B) Gene conversion clusters. For each SNP cluster, the 3RT nucleotide is shown above, the A15 is shown in the middle, and the 791 nucleotide is shown below. Yellow indicates identity with the 3RT nucleotide, and red indicates a nucleotide that is different from the 3RT nucleotide. The relevant balancer sequence is shown to the right of each cluster, with the inferred gene conversion event indicated by a red arrow. Relative spacing of SNPs is shown with a scale bar. (C) Distribution of verified variants along the third chromosome, EMS canonical G/C-to-A/T differences above, and noncanonical EMS differences below. Gene conversion clusters of mutations are indicated by red stars.
Of the 75 verified standard class lesions, 80% were G/C-to-A/T transitions, which are known to arise from EMS-mediated alkylation of guanine. This is consistent with the proportion observed in other comprehensive analyses of EMS-induced mutations in Drosophila: 70–76% (Cooper et al. 2008), 100% (Bentley et al. 2000), and 84% (Winkler et al. 2005). It also confirms the observation that the mutation profile under EMS dramatically differs from Arabidopsis, which shows >99% G/C-to-A/T transitions (Greene et al. 2003; Cooper et al. 2008). Finally, annotation of these 75 verified standard SNPs indicated that 58 were in noncoding regions, 9 were nonsynonymous, 2 were nonsense, and the remaining were silent (Table 2). The two nonsense mutations were in the genes encore and His2AV. Nonsynonymous mutations were found in the following genes: CG5146, CG3996, prospero, Spt3, CG7839, CG32091, CG32425, Cad99C, and RhoGAP100F.
TABLE 2.
Annotation of verified nucleotide changes
| Total | Noncoding | Synonymous | Nonsynonymous | Nonsense | |
|---|---|---|---|---|---|
| All | 103 | 82 | 10 | 9 | 2 |
| Standard | 75 | 58 | 6 | 9 | 2 |
| Anomalous | 28 | 24 | 4 | 0 | 0 |
Lesions classified as “standard” are more likely to have arisen by EMS mutagenesis and have functional consequence. Lesions classified as “anomalous” are more likely to have arisen by gene conversion and to not have functional consequence. Five standard lesions (three noncoding, one synonymous, and one nonsynonymous mutation in the Cad99C gene) were located in the gene conversion clusters (see Figure 2, B and C) and have been shown to be gene conversion events.
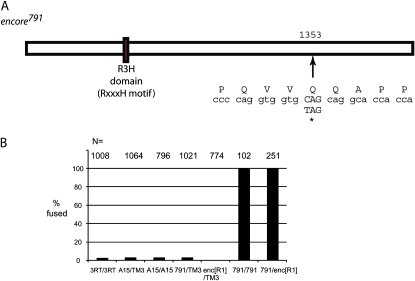
Importantly, one of the EMS-induced mutations was a nonsense mutation in the gene encore (Figure 3A). encore plays a role in the regulation of cyclin E during oogenesis and encodes for a protein that is 1823 amino acids in length (PA isoform) (Hawkins et al. 1996, 1997; Van Buskirk et al. 2000; Ohlmeyer and Schupbach 2003). The lesion that we identified, designated encore791, results in the replacement of glutamine 1353 with a stop codon (Figure 3A). Mutations in the gene encore are known to have an effect on dorsal appendage formation similar to that observed in the embryos of 791 homozygous mothers. A complementation test performed with mothers raised at the sensitive temperature of 18° revealed that the 791 chromosome failed to complement the encoreR1 allele for the fused dorsal appendage defect. This reveals that the 791 mutation is a new hypomorphic allele of encore (Figure 3B). Thus, using a whole-genome sequencing approach, we have identified the causative mutation underlying the fused dorsal appendage phenotype associated with the 791 chromosome.
Figure 3.—
Annotation of encore. (A) A C-to-T transition turns the 1353 glutamine codon to a premature stop. (B) Complementation test of encore791 lesion. Embryos of mothers raised at 18° were assayed for the fused dorsal appendage phenotype. 3RT indicates the target chromosome from which the A15 chromosome was derived.
Strikingly, the mutation profile differed dramatically between the verified standard class SNPs and those that were verified and classified as anomalous (Figure 2A). Only 42.9% of the latter class were G/C-to-A/T transitions. Moreover, we noted that the anomalous SNPs were highly clustered. Defining clusters as SNPs that are <500 bp apart from one another, 21 of 28 verified anomalous SNPs resided in a total of eight clusters (Figure 2, B and C). Annotation of the verified SNPs also indicated a strong difference in the spectrum of impact between the two classes (Table 2). Unlike the verified standard class SNPs, none of the verified anomalous SNPs changed protein function, and all either were in noncoding regions or were silent. This difference in impact is significant between the two classes (Fisher's exact test, P < 0.05). In aggregate, these data indicate that the most likely source of the anomalous lesions is gene conversion off a segregating balancer. This is the most parsimonious explanation as gene conversion is expected to produce what appear to be continuous tracts of mutations that are not canonical G/C-to-A/T EMS-induced transitions, but rather apparent reversions to an alternate sequence.
To determine whether or not these clusters of mutations were in fact due to gene conversion events, using segregating balancer chromosomes (TM6b,Tb and TM3,Sb) as donors, we sequenced the cluster regions on the balancer chromosomes in heterozygous adults. In addition, we performed Sanger sequencing of flies homozygous for a third chromosome designated 3RT from which 791 and A15 had been generated. With these additional data, we found that all of the clustered mutations in fact were identical to a corresponding set of polymorphisms that distinguished a segregating balancer chromosome from the original 3RT chromosome (Figure 2B). This included five SNPs that were originally classified as standard but also resided within the clusters. Moreover, we found that where either A15 or 791 possessed a unique sequence, this sequence corresponded to the balancer that it had been maintained over, namely TM6b,Tb in the case of A15 and TM3,Sb in the case of 791. Thus, we conclude that these lesions arose from gene conversion events that transferred sequence information from balancer to balanced chromosomes. The minimal length of these gene conversion tracts ranged from 12 to 724 bp, with a mean of 245 bp. Since 21 of the 21 clustered anomalous mutations arose from apparent gene conversion events with balancer chromosomes, we conclude this to be the most parsimonious explanation for the reversion of anomalous lesions to the reference sequence. Considering the entire set of 103 differences between the A15 and 791 chromosomes, 33 differences (28 anomalous mutations + 5 standard mutations residing in the clusters) can thus be attributed to gene conversion occurring within either of the balanced stocks.
DISCUSSION
New technologies for whole-genome sequencing have tremendous potential in aiding the search for mutations of interest. By identifying, in one round of sequencing, encore as the gene whose defect caused the fused dorsal appendage phenotype associated with the 791 chromosome, we have demonstrated a proof of concept that next-generation sequencing can be a powerful method for identifying lesions that produce phenotypes of interest. This study was done on an Illumina Genome Analyzer I (GAI) with a single-read 36-bp protocol using the original chemistry and version 0.3.0 of the Illumina Analysis Pipeline. The current version of the Illumina Genome Analyzer platform (GAII with paired-end module, analysis pipeline v1.3 and chemistry v3) is capable of much longer reads of >100 bp and can generate almost 20 Gb/run compared with ∼1 Gb/run reported in this study. In addition, paired-end reads make reading through repetitive regions possible. On the basis of the current performance statistics of the platform, we predict that >90% of the Drosophila genome can be sequenced to >20× coverage with just several lanes of a flow cell. This economy of scale makes large throughput whole-genome sequencing in flies economically feasible for most Drosophila researchers.
It is important to note that this approach unifies several different aspects of genetics research. Historically, fine-scale mapping was done in an iterative process that required narrowing down a region of interest and identifying new markers that could identify recombination events within successively smaller regions. However, using the approach outlined here, one may be able to identify candidate lesions that can immediately be tested for their role in a given phenotype. Even without alleles that enable the complementation test, overlapping deficiencies and transformation rescue experiments can be used to identify causative lesions. We expect that additional confirmation by these methods will be fairly straightforward since, with a 45 mm dose of EMS, we recovered only 11 lesions that affected coding sequence, 10 of which were obviously EMS-induced candidates with one lesion in the Cad99C gene, likely resulting from a gene conversion event off a balancer. Moreover, even in the face of no obvious causative lesion, future researchers will be able to use the EMS-induced SNPs themselves as mapping markers. This will eliminate the need to recombine mutations of interest onto chromosomes with previously defined SNP markers.
A second aspect of genetics research that is unified with this approach is the generation of new alleles. In the past, an EMS screen would be used to identify genes with a particular phenotype of interest. In this process, however, countless other lesions that might have been of interest to others would be ignored due to lack of an effect on the relevant phenotype. In one iteration of this process, we have identified a total of 82 noncoding mutations, nine new nonsynonymous alleles (one of which was attributed to gene conversion), and two new nonsense alleles. Thus, using a next-generation sequencing approach, future geneticists will effectively be able to merge marker discovery, mapping, and targeted mutagenesis.
But beyond using next-generation sequencing as a genetics tool, this approach also allows deeper insight into fundamental biological processes. The spectrum of EMS-induced lesions is known to differ between flies and other organisms, but the mechanism underlying this difference is not clear. It has been suggested that the mechanism of DNA repair may differ enough between species to explain this difference. We have found evidence that a significant fraction of noncanonical EMS mutations in flies is found in clusters that likely arise through gene conversion. Thus, part of the difference in the mutational spectrum during treatment with EMS may lie in the false attribution of gene conversion events as being induced by EMS. This false inference will be more common with an increasing likelihood of gene conversion off a homolog with distinguishing variants. Drosophila and broader dipterans are especially known for their efficiency in homolog pairing. Even though balancers inhibit crossing over through their multiple inversions, they pair surprisingly well (Gong et al. 2005). Furthermore, there is strong evidence that gene conversion events can occur from balancers to balanced chromosomes (Cooper et al. 2008). Thus, one possible explanation for the difference in mutational profiles after EMS treatment in Arabidopsis and Drosophila is that, while mutagenesis in Arabidopsis typically makes use of inbred lines for which gene conversion will not carry distinguishing variants between homologs, mutagenesis in Drosophila is typically performed using males that are mated to females carrying a balancer chromosome. A gene conversion event off the balancer chromosome within the stock would appear to be an “induced lesion” that is not a canonical EMS-induced mutation.
Acknowledgments
We thank Casey Wimberly, Keith Smith, and Michael Peterson for Sanger sequencing and help with data analysis. We also thank Madelaine Gogol for Sanger sequencing primer design. We are grateful to Dorothy Stanley for editing of the manuscript and help with submission for publication. This research was supported by the Stowers Institute for Medical Research, by an American Cancer Society Postdoctoral Fellowship to J.P.B., and by an American Cancer Society Professorship to R.S.H.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101998/DC1.
Sequence data from this article have been deposited with the Short Read Archive at NCBI under accession no. SRA008255.
References
- Barski, A., S. Cuddapah, K. R. Cui, T. Y. Roh, D. E. Schones et al., 2007. High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Bentley, A., B. MacLennan, J. Calvo and C. R. Dearolf, 2000. Targeted recovery of mutations in Drosophila. Genetics 156 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. R., 2006. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16 545–552. [DOI] [PubMed] [Google Scholar]
- Cooper, J. L., E. A. Greene, B. J. Till, C. A. Codomo, B. T. Wakimoto et al., 2008. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics 180 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., K. S. McKim and R. S. Hawley, 2005. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 1 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, E. A., C. A. Codomo, N. E. Taylor, J. G. Henikoff, B. J. Till et al., 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, N. C., J. Thorpe and T. Schupbach, 1996. encore, a gene required for the regulation of germ line mitosis and oocyte differentiation during Drosophila oogenesis. Development 122 281–290. [DOI] [PubMed] [Google Scholar]
- Hawkins, N. C., C. Van Buskirk, U. Grossniklaus and T. Schupbach, 1997. Post-transcriptional regulation of gurken by encore is required for axis determination in Drosophila. Development 124 4801–4810. [DOI] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader et al., 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P. C., and S. Henikoff, 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer, J. T., and T. Schupbach, 2003. Encore facilitates SCF-ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development 130 6339–6349. [DOI] [PubMed] [Google Scholar]
- Sarin, S., S. Prabhu, M. M. O'Meara, I. Pe'er and O. Hobert, 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. R., A. R. Quinlan, H. E. Peckham, K. Makowsky, W. Tao et al., 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 18 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev, A., J. Ichikawa, T. Tonthat, J. Stuart, S. Ranade et al., 2008. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk, C., N. C. Hawkins and T. Schupbach, 2000. Encore is a member of a novel family of proteins and affects multiple processes in Drosophila oogenesis. Development 127 4753–4762. [DOI] [PubMed] [Google Scholar]
- Whiteford, N., N. Haslam, G. Weber, A. Prugel-Bennett, J. W. Essex et al., 2005. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 33 e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, S., A. Schwabedissen, D. Backasch, C. Bokel, C. Seidel et al., 2005. Target-selected mutant screen by TILLING in Drosophila. Genome Res. 15 718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]