Abstract
The detection of protein–protein interactions through two-hybrid assays has revolutionized our understanding of biology. The remarkable impact of two-hybrid assay platforms derives from their speed, simplicity, and broad applicability. Yet for many organisms, the need to express test proteins in Saccharomyces cerevisiae or Escherichia coli presents a substantial barrier because variations in codon specificity or bias may result in aberrant protein expression. In particular, nonstandard genetic codes are characteristic of several eukaryotic pathogens, for which there are currently no genetically based systems for detection of protein–protein interactions. We have developed a protein–protein interaction assay that is carried out in native host cells by using GFP as the only foreign protein moiety, thus circumventing these problems. We show that interaction can be detected between two protein pairs in both the model yeast S. cerevisiae and the fungal pathogen Candida albicans. We use computational analysis of microscopic images to provide a quantitative and automated assessment of confidence.
THE ability to detect protein–protein interactions rapidly and systematically has driven our understanding of gene function by implicating new proteins in key biological processes and by defining interpathway communication mechanisms (Cusick et al. 2005; Parrish et al. 2006). The clear value of protein–protein interaction information has prompted the development of many different genetic and biochemical approaches to test for interaction (Berggard et al. 2007). Genetic approaches, such as the two-hybrid assay (Fields and Song 1989), facilitate large-scale screening so that diverse protein pairs and growth conditions can be sampled (Parrish et al. 2006; Tarassov et al. 2008). Two hybrid assays are typically carried out in surrogate hosts Saccharomyces cerevisiae or Escherichia coli, which use the universal genetic code. However, many organisms use nonstandard genetic codes (Knight et al. 2001), making surrogate hosts unwieldy for heterologous protein expression. This issue can be overcome with a native host-based protein interaction assay, such as we describe here.
The interaction assay that we present is built upon properties of the highly conserved endosomal sorting complex required for transport (ESCRT). ESCRT comprises 10 subunits, including Snf7/Vps32, that are transiently associated with the cytoplasmic face of endocytic vesicles (reviewed in Hurley and Emr 2006). ESCRT is dissociated through the action of the Vps4 ATPase; Vps4 defects cause accumulation of ESCRT-containing vesicles called class E compartments (Babst et al. 1997, 1998; Obita et al. 2007).
The assay is a test for reassignment of fusion protein localization. One fusion protein has an N-terminal segment chosen by the investigator (Yfg1 protein) fused to the ESCRT subunit Vps32. The Yfg1-Vps32 fusion serves as a bait protein that is targeted to endocytic vesicle surfaces. The second fusion protein has another N-terminal segment chosen by the investigator (Yfg2 protein) fused to GFP. The Yfg2-GFP fusion serves as a prey protein whose targeting to endocytic vesicles depends upon interaction with the bait. The assay is conducted in a vps4 mutant host strain (Babst et al. 1998), which promotes vesicular accumulation of Vps32. A positive interaction between the fusion proteins results in the targeting of GFP to vesicles, yielding bright punctate signals when cells are viewed with fluorescence microscopy. Microscopic images are analyzed through computational methods to arrive at a confidence value for the interaction. Because the assay system captures protein complexes on vesicle surfaces, we refer to it as the vesicle capture interaction (VCI) assay.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions:
S. cerevisiae strain JBY357 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps4Δ∷URA3) was constructed using PCR-directed deletion of the VPS4 gene in parent strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), as previously described (Boysen and Mitchell 2006). Candida albicans strains BWP17 (ura3Δ∷λ imm434/ura3Δ∷λimm434 arg4∷hisG/arg4∷hisG his1∷hisG/his1∷hisG) and SAL2-4F (ura3Δ∷λ imm434/ura3Δ∷λimm434 arg4∷hisG/arg4∷hisG his1∷hisG/his1∷hisG vps4Δ∷dpl200/vps4Δ∷dpl200) have been described (Wilson et al. 1999; Lee et al. 2007).
Plasmids were created using in vivo recombination methods (Ma et al. 1987; Raymond et al. 1999). Plasmid pJB300 was created by integrating the GFP(S65T)-HIS3MX6 cassette from pFA6a-GFP(S65T)-HIS3MX6 into the multiple cloning site (MCS) of pRS314. The cassette was initially amplified using primers pRS314 GFP F and pRS314 GFP R (for all primer sequences, see Table 1) against the pFA6a plasmid template and cotransformed with NotI-/SacI-linearized plasmid pRS314. This strategy integrates the GFP-coding region of the cassette 3′ of the NotI site and allows the majority of the MCS to be retained. Approximately 10 bp of pRS314 sequence (flanking the SacI site, bp +1890 to +1900) was lost during the in vivo recombination event. Plasmids were recovered in E. coli, characterized, and sequenced. Plasmid pJB302, harboring a VPS32 scLEU2 cassette, was derived from pJB300 in two steps. The VPS32 gene was amplified from plasmid with primers GFPtoSNF7 F and GFPtoSNF7 R. The resulting PCR was cotransformed with pJB300 and linearized within the GFP-coding region by digestion with NdeI. The resulting plasmid, pJB301, was converted to Leu+ using primers HIS5toLEU2 F and HIS5toLEU2 R against pRS315 template. The PCR and pJB301, linearized within the HIS3MX6 coding region with SphI digestion, were cotransformed, and plasmids were recovered and characterized. pJB300 and pJB302 were utilized as both PCR templates using the existing pFA6-directed primers for genomic integration and as backbones for subsequent plasmid constructions. Plasmids harboring PBS2, HOG1, SNF1, and SNF4 were constructed using in vivo recombination into pJB300 and pJB302. Primers were constructed to incorporate 1 kb 5′ upstream promoter (or, if necessary, just 3′ of any adjacent coding ORF) and the coding region of interest, less the stop codon. Flanking overhang sequences (from primers 314 cloning F and 314 cloning R; Table 1) were appended to gene-specific primers to allow subsequent integration of PCR into NotI-linearized pJB300 or pJB302 by in vivo recombination. Plasmids were retrieved and characterized as above.
TABLE 1.
Primer sequences
| Primer name | Sequence (5′–3′) |
|---|---|
| pRS314.GFP F | aagcttgatatcgaattcctgcagcccgggggatccactagttctagagcggccgccaccGGTCGACGGATCCCCGGGTT |
| pRS314.GFP R | acgacgttgtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattggaTCGATGAATTCGAGCTCGTT |
| 314 cloning F | ggatccactagttctagagcggccgccacc |
| 314 cloning R | GTTAATTAACCCGGGGATCCGTCGACC |
| GFPtoSNF7 F | ACGCTGCAGGTCGACGGATCCCCGGGTTAATTAACtggtcatcactttttggttgg |
| GFPtoSNF7 R | TCATAAGAAATTCGCTTATTTAGAAGTGGCGCGCCtcaaagccccatttctgcttg |
| HIS5toLEU2 F | TACAGTTCTCACATCACATCCGAACATAAACAACCatgtctgcccctaagaagatc |
| HIS5toLEU2 R | CTTGAAAACAAGAATCTTTTTATTGTCAGTACTCTttaagcaaggattttcttaac |
| GFP.URA3 PGEMT F | GGTGGTGGTTCTAAAGGTGAAGAATTATT |
| GFP.URA3 PGEMT R | TCTAGAAGGACCACCTTTGATTG |
| CaSNF7 F | ACGCTGCAGGTCGACGGATCCCCGGGTTAATTAACTGGGGATATTTTTTTGGAGGA |
| CaSNF7 R | TCATAAGAAATTCGCTTATTTAGAAGTGGCGCGCCTCATAATCCCATTTCAGCTTG |
| CaHIS1 F | TACAGTTCTCACATCACATCCGAACATAAACAACCTGTGGAATTGTGAGCGGATA |
| CaHIS1 R | CTTGAAAACAAGAATCTTTTTATTGTCAGTACTTTCCCAGTCACGACGTT |
| CaPBS2 GFP F | GTTCAATCATTATTGAGAAACAAAGTGAAGGCTCCGGCATTACATAGAGGTGGTTTACAAAAAGTGAATAGAAGCTTTCTTAATAATCATGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| CaPBS2 GFP R | GTGTTTGTGTTAGTTTGTTAGTTTGTTTATTTATTTGTTTGTTTCTATATAATA TACTGTTTATAATACAGCCCAATAACCTGGGCTTCATATTCATCTAGAAGGACCACCTTTGATTG |
| CaHOG1 pRSfus F | GGATCCACTAGTTCTAGAGCGGCCGCCACCTTTCCGTTAAAGTGTCCACTT |
| CaHOG1 pRSfus R | GTT AAT TAA CCC GGG GAT CCG TCG ACC AGC TCC GTT GGC GGA ATC CAA |
| CaSNF1 GFP F | CTAGATGAAGTTGGGTCATTCTCTGCTTATCCTTTCTTACATTTAGCTACTAGA TTAATTATGGAATTAGCCGTAAATAGTCAAAGTGGAGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| CaSNF1 GFP R | GGCGTAAGAAATCCAAAAAATGGGTTGTGAATTTATCATACATATTACATATCTGCTGACATCCAATCTAAGCTAGTACTTACTTACTTTATTTCTAGAAGGACCACCTTTGATTG |
| CaSNF4 pRSfus F | GGATCCACTAGTTCTAGAGCGGCCGCCACCTGAATTGAATGTAAAAGAAGA |
| CaSNF4 pRSfus R | GTTAATTAACCCGGGGATCCGTCGACCATCTTCTCCAAATAATATGTA |
Plasmid pJB407 was constructed by insertion of a GFP-URA3 cassette (Gerami-Nejad et al. 2001) into the pGEMT vector following the manufacturer's instructions (Promega). Subsequent GFP fusions used PCR amplification with gene-specific primers (∼100 bp homologous to the gene of interest plus 23–29 bp homologous to the GFP cassette) followed by PCR-mediated transformation directed to the chromosomal locus of the gene of interest in wild-type strain BWP17 or the vps4 mutant SAL2-4F (Lee et al. 2007).
pJB408, which carries CaSNF7 and CaHIS1 on a pRS314 scTRP1 backbone, was constructed from plasmid pJB300 (see above) using two rounds of in vivo recombination. First, using primers CaSNF7 F and CaSNF7 R, the complete caSNF7 sequence lacking a start codon was amplified and inserted in place of the GFP-coding sequence. Second, using primers CaHIS1 F and CaHIS1 R, the caHIS1 sequence was amplified and inserted into the CaSNF7 plasmid pJB401, with the caHIS1 sequence completely replacing the Schizosaccharomyces pombe his5+ sequence. The resulting plasmid pJB408 contains a 5′ cloning site with a unique NotI site; this site was subsequently used, in conjunction with a homologous linking sequence appended to gene-specific primers, to direct the in vivo recombination of fusions to SNF7. Genes were amplified with either 5′ sequence up to the neighboring gene or with 1 kb, whichever was least. Thereafter, using a unique NruI site in the CaHIS1 sequence, Snf7 fusions were targeted to the chromosomal caHIS1 locus in the GFP+ (wild type and ΔcaVPS4/ΔcaVPS4) strains isolated above.
Yeast growth media (YPD and SC) were of standard composition (Kaiser et al. 1994). All plates and liquid cultures were incubated at 30°.
Microscopy:
Imaging was performed at room temperature on a Nikon Eclipse E800 widefield fluorescence microscope with a Nikon Plan Apo ×100 1.4 objective (Melville, NY) and a Hamamatsu Orca100 digital CCD Camera (Bridgewater, NJ). Images were acquired with OpenLab Improvision software. Staining with N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl)-pyridinium dibromide (FM4-64, purchased from Molecular Diagnostics, Chicago) was performed as described previously (Boysen and Mitchell 2006).
Quantitative image measurement:
Raw fluorescence micrographs of the GFP signal were processed in Matlab 7.4. These 8-bit grayscale images were corrected for background by subtracting the most common pixel value, and then each image was stretched to 64 gray levels. For each of these processed images, four gray-level co-occurrence matrices were calculated to measure horizontal, vertical, left-diagonal, and right-diagonal nearest neighbor occurrences. Thirteen Haralick texture features were calculated from each of these resulting matrices, and these features were averaged in the horizontal/vertical and left-diagonal/right-diagonal directions, giving 26 texture features (Chebira et al. 2007). Additionally, cumulative gray-level frequency features were calculated from the stretched images. For each image, a histogram was calculated on all pixels with intensity greater than zero, and the cumulative frequency of pixels at each of 62 grayscale values was used as features (the cumulative frequency for pixel value 63 is ignored because it is always 1). See http://murphylab.web.cmu.edu/software/ and http://murphylab.web.cmu.edu/data/ for analysis programs and raw image files.
RESULTS
S. cerevisiae VCI assay:
The VCI assay platform was first tested in S. cerevisiae because of the ease of manipulation. We tested two protein-kinase-related complexes, Snf1:Snf4 and Pbs2:Hog1. Snf1 is the S. cerevisiae AMP-activated protein kinase, and Snf4 is its regulatory gamma subunit (Schuller 2003). Snf1:Snf4 interaction was detected in the first published two-hybrid assay (Fields and Song 1989). Pbs2 and Hog1 are the MAPKK and MAPK, respectively, that are required for the S. cerevisiae high osmolarity response (Hohmann 2002). Interaction between Hog1 and Pbs2 is well documented (Posas and Saito 1997) but has never been detected in published two-hybrid assays or other genetic protein–protein interaction tests.
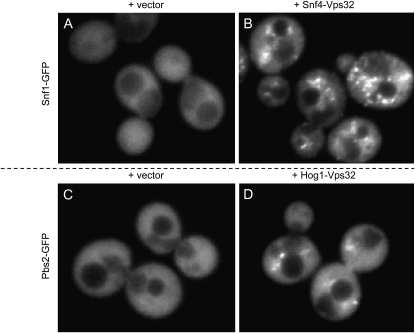
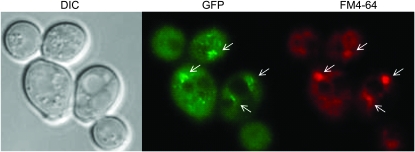
S. cerevisiae vps4Δ cells carrying a Snf1-GFP fusion plasmid, along with the Vps32 fusion vector, gave diffuse cytoplasmic fluorescence (Figure 1A and supporting information, Figure S4). As seen for most cytoplasmic GFP fusions in S. cerevisiae (Huh et al. 2003), there was exclusion from the vacuole. The presence of both Snf1-GFP and Snf4-Vps32 fusion plasmids in the vps4Δ cells yielded punctate fluorescent foci (Figure 1B and Figure S3). Similar foci were observed when the GFP and Vps32 tags were reversed (i.e., Snf4-GFP and Snf1-Vps32; data not shown). The foci colocalized substantially with the membrane dye FM4-64 (Figure 2), which accumulates in the endosome-derived class E compartments in vps4Δ mutant cells (Kranz et al. 2001). Foci were rare and relatively faint with these plasmid combinations in VPS4 cells (see Figure S7 and Figure S8). The two findings—that foci depend upon a vps4 mutation and that they colocalize with FM4-64-stained regions—are consistent with the idea that foci correspond to endosome-associated ESCRT complexes. The fact that GFP foci depend upon the presence of an interacting protein fused to Vps32 argues that the Vps32 fusion protein targets the GFP fusion protein to the endosome.
Figure 1.—
VCI assay for S. cerevisiae Snf1:Snf4 and Pbs2:Hog1. An S. cerevisiae vps4Δ strain was transformed with a Snf1-GFP fusion plasmid and either a Vps32 vector (A) or a Snf4-Vps32 fusion plasmid (B). The vps4Δ strain was independently transformed with a Pbs2-GFP fusion plasmid and either a Vps32 vector (C) or a Hog1-Vps32 fusion plasmid (D). GFP fluorescence images are shown.
Figure 2.—
Comparison of GFP localization and FM4-64 staining. An S. cerevisiae vps4Δ mutant host expressing both Snf1-GFP and Snf4-Vps32 was stained with membrane dye FM4-64. The dye accumulates in endosome-derived class E compartments in vps4 mutants. DIC, GFP, and FM4-64 channels are shown. White arrows indicate regions of GFP and FM4-64 colocalization.
To determine whether VCI assays may be useful for other pairs of proteins, we carried out a similar analysis of Pbs2 and Hog1. Once again, punctate GFP foci were detected only in cells that expressed both Pbs2-GFP and Hog1-Vps32 fusions (compare Figure 1D to Figure 1C and Figure S1 to Figure S2) and were dependent upon a vps4Δ mutation (Figure S5). Formation of GFP foci was dependent upon interacting fusion proteins because no foci were observed when the vps4Δ strain carried Snf1-GFP together with Hog1-Vps32 or Pbs2-GFP together with Snf4-Vps32 (data not shown). Therefore, the VCI assay permits detection of protein–protein interaction for two pairs of S. cerevisiae gene products that were known to exist in complexes.
C. albicans VCI assay:
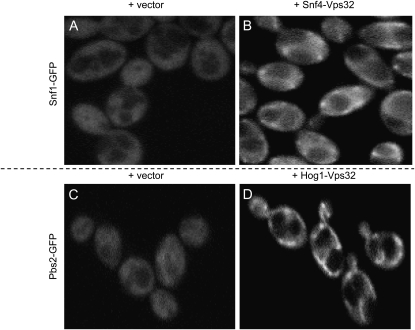
We sought to develop the VCI assay in C. albicans because there is no simple protein–protein interaction assay available for that organism. We chose the C. albicans protein pairs Snf1:Snf4 and Pbs2:Hog1, the orthologs of the S. cerevisiae protein pairs used above. In C. albicans, presence of the Snf1-GFP or Pbs2-GFP with the Vps32 vector yielded diffuse cytoplasmic fluorescence (Figure 3, A and C, and Figure S10 and Figure S12). However, presence of both Snf1-GFP and Snf4-Vps32, or Pbs2-GFP and Hog1-Vps32, yielded punctate GFP foci (Figure 3, B and D, and Figure S9 and Figure S11). The foci occasionally resembled ribbons or whorls, as do some class E compartments (Luhtala and Odorizzi 2004; Russell et al. 2006). The foci were dependent upon the vps4Δ/vps4Δ genotype (data not shown). Interaction was specific because no foci were observed in cells expressing both Pbs2-GFP and Snf4-Vps32 (Figure S13). These observations indicate that the VCI system can detect interactions between two C. albicans protein pairs.
Figure 3.—
VCI assay for C. albicans Snf1:Snf4 and Pbs2:Hog1. A C. albicans vps4Δ/vps4Δ strain was transformed to introduce a Snf1-GFP fusion gene and either the Vps32 vector (A) or a Snf4-Vps32 fusion gene (B). The vps4Δ/vps4Δ strain was independently transformed to introduce a Pbs2-GFP fusion gene and either the Vps32 vector (C) or a Hog1-Vps32 fusion gene (D). GFP fluorescence images are shown.
Computational assessment of VCI images:
Although positive and negative VCI assay images can be distinguished by eye, we sought to develop a computational image analysis strategy to arrive at a confidence level for interaction. We collected random images for strains expressing each prey fusion plus bait vector only (negative class, such as Figure 1, A and C, and Figure 3, A and C) or each prey fusion plus bait fusion (positive class, like Figures 1B, 1D, 3B, and 3D). We evaluated whether or not a classification system can distinguish them (Glory and Murphy 2007) as follows. Each image was processed to produce quantitative features that reflect the degree to which the GFP signal is contained in bright, punctate structures. The simplest approach was to identify punctate structures (if any) and measure the fraction of fluorescence contained in them. However, the heterogeneous nature of the vesicle compartments makes it difficult to identify them directly. We therefore created a series of features that calculate the fraction of fluorescence contained in pixels above a given threshold and used these in conjunction with Haralick texture features, which we previously demonstrated are valuable for analysis of subcellular patterns (Chebira et al. 2007). The features were calculated for each image and used to train support vector machine classifiers (Byvatov and Schneider 2003). The performance of the classifiers was evaluated using leave-one-out cross-validation. In this approach, a classifier was trained on all images except one and then tested on the remaining image; the classification process was repeated until all images had been used for testing. The class assigned by the classifier was then compared to the known class and tabulated (Table 2). For S. cerevisiae VCI assays, we used only 10 images/strain, and the classifier achieved performance of 80–95%. We suspect that performance was limited by the small number of images. For the C. albicans VCI assays, we used 20–25 images/strain and classifier performance was 90–95%. We used multivariate hypothesis tests (Chen et al. 2006) to determine whether the feature distributions of the positive and negative conditions were significantly different. By the Friedman–Rafsky test, all pairs of VCI positive and negative image sets were distinguished with highly significant P-values (Table 2). This computational analysis provides a useful approach to quantifying differences between VCI positive and negative samples.
TABLE 2.
Quantitative assessment of VCI assay results
| GFP fusion | Vps32 fusion | No. of images | Classifier output
|
Classifier accuracy (%) | P-value for positive vs. negative | ||
|---|---|---|---|---|---|---|---|
| Organism | VCI positive | VCI negative | |||||
| S. cerevisiae | Snf1 | Snf4 | 10 | 90.0 | 10.0 | 95.0 | 1 × 10−5 |
| (vector) | 10 | 0.0 | 100.0 | ||||
| S. cerevisiae | Pbs2 | Hog1 | 10 | 90.0 | 10.0 | 80.0 | 0.03 |
| (vector) | 10 | 30.0 | 70.0 | ||||
| C. albicans | Snf1 | Snf4 | 25 | 96.0 | 4.0 | 95.5 | 6 × 10−8 |
| (vector) | 20 | 5.0 | 95.0 | ||||
| C. albicans | Pbs2 | Hog1 | 20 | 90.0 | 10.0 | 90.0 | 3 × 10−6 |
| (vector) | 20 | 10.0 | 90.0 | ||||
DISCUSSION
The VCI assay described here has proven workable for two protein complexes in two organisms. We expect such a native host-based assay to be particularly useful in C. albicans because of its nonstandard genetic code and the lack of any other genetic protein–protein interaction test at present. The assay requires only modest molecular genetic manipulation and is based upon highly conserved eukaryotic machinery. Therefore, it may be useful in many other organisms as well.
The VCI assay has several generally useful features. First, the fusion proteins are expressed from their native promoters, rather than overexpressed, so that natural stoichiometry of interacting proteins can be maintained. Second, real-time imaging may facilitate detection of transient complexes, particularly in response to environmental changes. Third, single-cell assays such as this can be powerful for detecting transient responses in asynchronous or heterogenous populations, such as those engaged in biofilm formation or sporulation. These advantages are shared with protein-fragment-complementation-based interaction assays (Remy and Michnick 2004). However, the VCI assay offers the additional advantage that the native VPS32 coding region is used as one fusion partner, thus eliminating the need for codon changes before implementation in hosts with divergent genetic codes. Many GFP coding regions that function in hosts with variant genetic codes have been described (Ha et al. 1996; Cormack et al. 1997; Hosein et al. 2003).
The need for a vps4 defect in the VCI host strain is a potential limitation of the assay because it may be difficult to disrupt genomic copies of VPS4 in many organisms. However, Vps4 defects can also be achieved through ectopic inactivation strategies, including RNA interference or the use of a dominant-negative VPS4 allele. Dominant-negative VPS4-DN alleles have been used to probe ESCRT function in genetically unwieldy cells, including human cell lines and Leishmania major (Hislop et al. 2004; Besteiro et al. 2006; Taylor et al. 2007). Thus we expect that impairment of Vps4 function will not be a major impediment to VCI assay implementation.
For the assays presented here, computational analysis provides an objective means to compare image sets and support statistical assessment of interaction. In the longer term, computational image analysis yields an avenue for scaling up the VCI assay. It permits use of automated microscopy methods (Glory and Murphy 2007), so that the VCI assay may be implemented with large sample sets, such as large numbers of protein pairs or time points. Indeed, automated microscopy has been used to define prospective drug targets (Perlman et al. 2004) and to evaluate subcellular protein localization (Roques and Murphy 2002; Chen et al. 2007; Glory and Murphy 2007). Automated subcellular localization assignments have proven more sensitive than visual interpretation by human observers (Roques and Murphy 2002; Chen et al. 2007; Glory and Murphy 2007).
There are some detailed points to consider about the VCI assay. First, the brightness of our cell populations is variable, as one can see from the supplemental figures. This heterogeneity probably arises from allowing cells to settle in culture tubes before imaging; we find the most homogenous and distinct images from early to mid-logarithmic cultures that are growing actively just prior to imaging. Second, our C. albicans VCI signals do not resemble class E compartments (Kullas et al. 2004; Lee et al. 2007). We suspect that their unusual appearance arises from the overall increased expression of Vps32 in these cells; the Hog1-Vps32 and Snf4-Vps32 fusions are expressed from the HOG1 and SNF4 promoters, respectively.
The immediate value of the VCI assay is as a protein–protein interaction test for C. albicans. For some time the prevailing view was that C. albicans gene function was largely similar to S. cerevisiae gene function. For example, it appeared that processes such as filamentation (Lo et al. 1997), pH responses (Davis 2003), cell-wall integrity (Navarro-Garcia et al. 2001), and basic growth and viability (see Devasahayam et al. 2002; Michel et al. 2002) were governed by the C. albicans orthologs of known S. cerevisiae pathway participants. Such a scenario placed little importance on specific tests of C. albicans protein–protein interaction because the expectation was that they would simply recapitulate interactions among the S. cerevisiae orthologs. However, that view was driven by “sampling error”; that is, C. albicans gene function analysis rested largely upon candidate gene approaches that in turn were based on prior S. cerevisiae gene discovery. More recently, the C. albicans community has embraced new gene discovery strategies, including the screening of heterozygous, homozygous, or conditional expression C. albicans mutant libraries (Bruno and Mitchell 2004; Noble and Johnson 2007), as well as candidate gene selection based upon microarray expression profiling (Garaizar et al. 2006; Brown et al. 2007) or proteomic analysis (de Groot et al. 2004; Kusch et al. 2007). These strategies have fueled the reexamination of processes conserved between S. cerevisiae and C. albicans and have supported direct inquiry into distinct biological features of C. albicans, such as its ability to interact with host cells, invade tissues, and form biofilms. Such studies reveal that C. albicans indeed uses unique genes, pathways, and networks to meet its biological needs (see, for example, Roemer et al. 2003; Nobile and Mitchell 2005; Bruno et al. 2006; Huang et al. 2006; Srikantha et al. 2006; Zordan et al. 2006; Martchenko et al. 2007; Hogues et al. 2008). We are now poised for mechanistic studies that will yield a basic understanding of functional relationships and, ultimately, insight into the choice of therapeutic targets.
Acknowledgments
We thank members of our labs for advice and discussion. We are grateful to Sam Lee for providing the C. albicans vps4Δ/vps4Δ mutant.This work was supported by National Institutes of Health grant 5R01AI070272 (A.P.M.), National Science Foundation ITR grant EF-0331657 (R.F.M.), and a National University of Ireland Travelling Studentship (S.F.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101162/DC1.
References
- Babst, M., T. K. Sato, L. M. Banta and S. D. Emr, 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., B. Wendland, E. J. Estepa and S. D. Emr, 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggard, T., S. Linse and P. James, 2007. Methods for the detection and analysis of protein-protein interactions. Proteomics 7 2833–2842. [DOI] [PubMed] [Google Scholar]
- Besteiro, S., R. A. Williams, L. S. Morrison, G. H. Coombs and J. C. Mottram, 2006. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281 11384–11396. [DOI] [PubMed] [Google Scholar]
- Boysen, J. H., and A. P. Mitchell, 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A. J., F. C. Odds and N. A. Gow, 2007. Infection-related gene expression in Candida albicans. Curr. Opin. Microbiol. 10 307–313. [DOI] [PubMed] [Google Scholar]
- Bruno, V. M., and A. P. Mitchell, 2004. Large-scale gene function analysis in Candida albicans. Trends Microbiol. 12 157–161. [DOI] [PubMed] [Google Scholar]
- Bruno, V. M., S. Kalachikov, R. Subaran, C. J. Nobile, C. Kyratsous et al., 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byvatov, E., and G. Schneider, 2003. Support vector machine applications in bioinformatics. Appl. Bioinformatics 2 67–77. [PubMed] [Google Scholar]
- Chebira, A., Y. Barbotin, C. Jackson, T. Merryman, G. Srinivasa et al., 2007. A multiresolution approach to automated classification of protein subcellular location images. BMC Bioinformatics 8 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.-C., T. Zhao, G. J. Gordon and R. F. Murphy, 2006. A novel graphical model approach to segmenting cell images. Proceedings of the 2006 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, pp. 1–8. http://www.ieee.org.
- Chen, S. C., T. Zhao, G. J. Gordon and R. F. Murphy, 2007. Automated image analysis of protein localization in budding yeast. Bioinformatics 23 i66–i71. [DOI] [PubMed] [Google Scholar]
- Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow et al., 1997. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology 143(Pt. 2): 303–311. [DOI] [PubMed] [Google Scholar]
- Cusick, M. E., N. Klitgord, M. Vidal and D. E. Hill, 2005. Interactome: gateway into systems biology. Hum. Mol. Genet. 14(Spec. No. 2): R171–R181. [DOI] [PubMed] [Google Scholar]
- Davis, D., 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44 1–7. [DOI] [PubMed] [Google Scholar]
- de Groot, P. W., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong et al., 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasahayam, G., V. Chaturvedi and S. D. Hanes, 2002. The Ess1 prolyl isomerase is required for growth and morphogenetic switching in Candida albicans. Genetics 160 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Garaizar, J., S. Brena, J. Bikandi, A. Rementeria and J. Ponton, 2006. Use of DNA microarray technology and gene expression profiles to investigate the pathogenesis, cell biology, antifungal susceptibility and diagnosis of Candida albicans. FEMS Yeast Res. 6 987–998. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad, M., J. Berman and C. A. Gale, 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18 859–864. [DOI] [PubMed] [Google Scholar]
- Glory, E., and R. F. Murphy, 2007. Automated subcellular location determination and high-throughput microscopy. Dev. Cell 12 7–16. [DOI] [PubMed] [Google Scholar]
- Ha, D. S., J. K. Schwarz, S. J. Turco and S. M. Beverley, 1996. Use of the green fluorescent protein as a marker in transfected Leishmania. Mol. Biochem. Parasitol. 77 57–64. [DOI] [PubMed] [Google Scholar]
- Hislop, J. N., A. Marley and M. Von Zastrow, 2004. Role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J. Biol. Chem. 279 22522–22531. [DOI] [PubMed] [Google Scholar]
- Hogues, H., H. Lavoie, A. Sellam, M. Mangos, T. Roemer et al., 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosein, R. E., S. A. Williams, K. Haye and R. H. Gavin, 2003. Expression of GFP-actin leads to failure of nuclear elongation and cytokinesis in Tetrahymena thermophila. J. Eukaryot. Microbiol. 50 403–408. [DOI] [PubMed] [Google Scholar]
- Huang, G., H. Wang, S. Chou, X. Nie, J. Chen et al., 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. USA 103 12813–12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425 686–691. [DOI] [PubMed] [Google Scholar]
- Hurley, J. H., and S. D. Emr, 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis and A. Mitchell, 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Knight, R. D., S. J. Freeland and L. F. Landweber, 2001. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2 49–58. [DOI] [PubMed] [Google Scholar]
- Kranz, A., A. Kinner and R. Kolling, 2001. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullas, A. L., M. Li and D. A. Davis, 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser and M. Hecker, 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7 686–697. [DOI] [PubMed] [Google Scholar]
- Lee, S. A., J. Jones, Z. Khalique, J. Kot, M. Alba et al., 2007. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. FEMS Yeast Res. 7 973–985. [DOI] [PubMed] [Google Scholar]
- Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti et al., 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90 939–949. [DOI] [PubMed] [Google Scholar]
- Luhtala, N., and G. Odorizzi, 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., S. Kunes, P. J. Schatz and D. Botstein, 1987. Plasmid construction by homologous recombination in yeast. Gene 58 201–216. [DOI] [PubMed] [Google Scholar]
- Martchenko, M., A. Levitin, H. Hogues, A. Nantel and M. Whiteway, 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, S., S. Ushinsky, B. Klebl, E. Leberer, D. Thomas et al., 2002. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol. Microbiol. 46 269–280. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia, F., B. Eisman, E. Roman, C. Nombela and J. Pla, 2001. Signal transduction pathways and cell-wall construction in Candida albicans. Med. Mycol. 39(Suppl. 1): 87–100. [PubMed] [Google Scholar]
- Nobile, C. J., and A. P. Mitchell, 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15 1150–1155. [DOI] [PubMed] [Google Scholar]
- Noble, S. M., and A. D. Johnson, 2007. Genetics of Candida albicans, a diploid human fungal pathogen. Annu. Rev. Genet. 41 193–211. [DOI] [PubMed] [Google Scholar]
- Obita, T., S. Saksena, S. Ghazi-Tabatabai, D. J. Gill, O. Perisic et al., 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449 735–739. [DOI] [PubMed] [Google Scholar]
- Parrish, J. R., K. D. Gulyas and R. L. Finley, Jr., 2006. Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17 387–393. [DOI] [PubMed] [Google Scholar]
- Perlman, Z. E., M. D. Slack, Y. Feng, T. J. Mitchison, L. F. Wu et al., 2004. Multidimensional drug profiling by automated microscopy. Science 306 1194–1198. [DOI] [PubMed] [Google Scholar]
- Posas, F., and H. Saito, 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276 1702–1705. [DOI] [PubMed] [Google Scholar]
- Raymond, C. K., T. A. Pownder and S. L. Sexson, 1999. General method for plasmid construction using homologous recombination. Biotechniques 26 134–138, 140–131. [DOI] [PubMed] [Google Scholar]
- Remy, I., and S. W. Michnick, 2004. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol. Biol. 261 411–426. [DOI] [PubMed] [Google Scholar]
- Roemer, T., B. Jiang, J. Davison, T. Ketela, K. Veillette et al., 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50 167–181. [DOI] [PubMed] [Google Scholar]
- Roques, E. J., and R. F. Murphy, 2002. Objective evaluation of differences in protein subcellular distribution. Traffic 3 61–65. [DOI] [PubMed] [Google Scholar]
- Russell, M. R., D. P. Nickerson and G. Odorizzi, 2006. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 18 422–428. [DOI] [PubMed] [Google Scholar]
- Schuller, H. J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43 139–160. [DOI] [PubMed] [Google Scholar]
- Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu et al., 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov, K., V. Messier, C. R. Landry, S. Radinovic, M. M. Serna Molina et al., 2008. An in vivo map of the yeast protein interactome. Science 320 1465–1470. [DOI] [PubMed] [Google Scholar]
- Taylor, G. M., P. I. Hanson and M. Kielian, 2007. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 81 13631–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. B., D. Davis and A. P. Mitchell, 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan, R. E., D. J. Galgoczy and A. D. Johnson, 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 103 12807–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]