Abstract
Cichlid fishes compose an astonishingly large number of species and formed species flocks in record-breaking time. To facilitate efficient genome scans and comparisons of cichlid genomes, we constructed a medium-density genetic linkage map of microsatellite markers of Astatotilapia burtoni. The mapping cross was derived from two inbred laboratory lines to obtain F2 progeny by intercrossing. The map revealed 25 linkage groups spanning 1249.3 cM of the genome (size ∼950 Mb) with an average marker spacing of 6.12 cM. The seven Hox clusters, ParaHox C1, and two paralogs of Pdgfrβ were mapped to different linkage groups, thus supporting the hypothesis of a teleost-specific genome duplication. The A. burtoni linkage map was compared to the other two available maps for cichlids using shared markers that showed conservation and synteny among East African cichlid genomes. Interesting candidate genes for cichlid speciation were mapped using SNP markers.
THE adaptive radiations of cichlid fishes of East Africa are well-known examples for rapid diversification and explosive speciation. More than 2000 species are phylogenetically very closely related since they originated within extremely short evolutionary time spans (Meyer et al. 1990; Meyer 1993; Kornfield and Smith 2000; Verheyen et al. 2003; Kocher 2004; Salzburger and Meyer 2004; Salzburger et al. 2005). Astonishingly, large numbers of species make up the three species flocks, each composed of hundreds of species, in lakes Victoria, Malawi, and Tanganyika (Fryer and Iles 1972). Despite the huge phenotypic diversity displayed by each of the species flocks, molecular phylogenetic studies on this problem revealed that many of the species evolved similar morphologies convergently in each of these three adaptive radiations (Meyer et al. 1990; Kocher et al. 1993; Meyer 1993; Stiassny and Meyer 1999). These striking phenotypic similarities among cichlid fishes from different species flocks that have evolved in parallel make the study of the underlying genetic architecture of cichlids particularly interesting.
The observed redundant patterns in the evolutionary diversification of cichlid fishes support the view that the three large East African lakes are a natural experiment of evolution, in which this parallel evolution might ultimately help to better understand the processes that led to the repeatedly evolved patterns of diversification. Particularly in the species of cichlids in lakes Victoria and Malawi, adaptive radiations are very young and genetically extremely similar. Comparative studies on the genomic organization of these closely related yet morphologically diverse fishes will help to unravel the genetics of speciation (Kocher 2004; Albertson and Kocher 2006; Hoegg et al. 2007). Investigation of the molecular basis of those different phenotypes, i.e., the genetic and transcriptional changes that underlie differences among organisms, can be achieved through detailed comparisons of genome and transcriptome scans also including candidate gene approaches (Streelman and Kocher 2000; Braasch et al. 2006; Salzburger et al. 2007, 2008; Gerrard and Meyer 2007). For example, the gene for long wavelength-sensitive opsin (Lws) has been reported to be involved in sympatric speciation through ecological adaptation and mate choice of cichlids (Carleton et al. 2005; Maan et al. 2006; Terai et al. 2006; Seehausen et al. 2008), while the microfibril-associated glycoprotein (Mfap4) is a good candidate for examining species differences with regard to jaw development (Kobayashi et al. 2006).
Species-specific linkage maps have recently become established as important genetic tools in an effort to gain more detailed knowledge of genotype–phenotype relationships (Albertson et al. 2003; Erickson et al. 2004; Albertson and Kocher 2006). The latter approach is known as quantitative trait locus (QTL) scan, which makes use of the linkage disequilibrium created through experimental crosses between different species or laboratory strains (Falconer and Mackay 1996; Lynch and Walsh 1998). The ability to produce fertile interspecific crosses among some of these species of cichlids (Crapon de Caprona and Fritzsch 1984) and the general popularity of captive breeding support the establishment of cichlid fishes as a model system in comparative evolutionary genomic research.
The cichlid species Astatotilapia burtoni that occurs in Lake Tanganyika and in the surrounding river systems exhibits a rather generalist life style and is likely to represent a relatively ancestral type of cichlid (Meyer et al. 1991; Salzburger et al. 2005). Its phylogenetic placement between the species flock of ±500 endemic species in Lake Victoria and ≤1000 endemic species in Lake Malawi makes it an interesting species to study in this regard (Meyer et al. 1991). Since A. burtoni occupies a crucial phylogenetic position at the base of the extremely species-rich tribe of cichlids, the Haplochromini (Salzburger et al. 2002), which make up the large radiations of lakes Victoria and Malawi, its genome can serve as a sort of baseline from which comparisons to the endemic cichids of these lakes will be particularly insightful. Given these close genetic affinities, most of the genomic resources developed for A. burtoni will also be applicable to the large haplochromine cichlid species flocks from lakes Victoria and Malawi.
For A. burtoni, a BAC library (Lang et al. 2006) as well as expressed sequence tags (ESTs) have been generated (Salzburger et al. 2008), and cDNA microarrays are available as well (Renn et al. 2004; W. Salzburger, H. A. Hofmann and A. Meyer, unpublished results). In addition, there is detailed knowledge on Hox genes (Hoegg and Meyer 2005; Hoegg et al. 2007), Para-Hox genes (Siegel et al. 2007), and several other genes related to coloration (Braasch et al. 2006; Salzburger et al. 2007) and fertilization (Gerrard and Meyer 2007) for this key species. Genomic resources available from other cichlids include the tilapia Oreochromis niloticus (BAC library: Katagiri et al. 2001; genetic maps: Kocher et al. 1998; Katagiri et al. 2005; Lee et al. 2005), Haplochromis chilotes (BAC library: Watanabe et al. 2003; ESTs: Watanabe et al. 2004; cDNA microarrays: Kobayashi et al. 2006), and Metriaclima zebra (Di Palma et al. 2007; ESTs: Salzburger et al. 2008; genetic map: Albertson et al. 2003). Recently, the National Institutes of Health has committed to sequencing four cichlid genomes. A detailed description of genomic resources developed for cichlid fishes can be found at http://www.cichlidgenome.org/.
AFLPs and microsatellite loci (also termed SSR) are the most common markers used in the development of linkage maps and QTL studies. Microsatellites are preferable because of their codominant nature and extremely high degrees of intraspecific allele polymorphism, which makes them most effective. On the other hand, their generation requires high costs and is time consuming (reviewed in Erickson et al. 2004). Most linkage maps based on microsatellites have been constructed for economically important fish species, such as the Atlantic salmon (Gilbey et al. 2004), rainbow trout (Sakamoto et al. 2000), European sea bass (Chistiakov et al. 2005), and Nile tilapia (Kocher et al. 1998; Lee et al. 2005), to search for loci that affect commercially important traits. Research in the fields of ecology and evolution has recently begun to focus on identifying the genetic basis of adaptive trait evolution especially in natural populations of nonmodel organisms. The past decade has seen a proliferation of studies that employ linkage maps together with QTL approaches to shed light on evolutionary processes, for example, the parallel evolution of benthic and limnetic forms in threespine sticklebacks (Peichel et al. 2001; Colosimo et al. 2004; Miller et al. 2007) and reduction of eyes and pigmentation in the Mexican cavefish, Astyanax mexicanus (Protas et al. 2007).
Here we report on the construction of a microsatellite linkage map of the cichlid fish A. burtoni based on an F2 intercross derived from two inbred laboratory strains. We identified 25 linkage groups (LGs). The map also incorporates some EST-based markers and nuclear genes from sequenced BAC clones, e.g., the seven reported Hox genes and the two paralogs of Pdgfrβ, a gene involved in coloration (Braasch et al. 2006). This linkage map will thus provide a useful future tool for studying the genetic basis of adaptive traits that played a major role in the rapid diversification of cichlid fishes.
MATERIALS AND METHODS
Experimental crosses:
We crossed an A. burtoni female derived from our University of Konstanz stock with a male stemming from a laboratory stock that originated in the laboratory of Russell D. Fernald at Stanford University. It is now also held at the laboratory of Hans A. Hofmann (University of Texas at Austin). The stocks are originally from the Tanzania and Zambia regions of Lake Tanganyika, respectively. The resulting F1 generation was raised to sexual maturity, and groups of several females with one or two males were established for the F1 intercross. Young fry of the F2 generation, usually consisting of 10–50 individuals, were taken from the mouths of F1 females. Genotyping at 10 microsatellite markers revealed family relationships within each group. The final mapping population included 167 F2 offspring through intercrossing one male with five different females, thus constituting a half-sib family. However, we first established linkage groups by genotyping a subset of the first 90 F2 individuals that were born and then by adding the remaining 77 individuals for those markers with low LOD scores and/or those with some missing data. Loci were genotyped for an average of 106 individuals.
Microsatellite markers:
A microsatellite-enriched library was prepared from A. burtoni DNA using a magnetic bead enrichment protocol and (CA)15 and (CT)15 probes (for a detailed description, see Sanetra and Meyer 2005). A total of 1156 clones were sequenced, and 683 clones (enrichment rate ∼60%) contained repeat motifs (including ∼10% duplicate clones). Primer sets were designed for 278 putative loci using the Primer3 software (Rozen and Skaletsky 2000). Markers were considered informative when at least one F1 parent was heterozygous, which was the case for 147 of these loci.
Additional 191 microsatellite primer sequences were collected from available genomic resources for other cichlids and their usefulness for mapping the A. burtoni genome was tested. Finally, we were able to employ a total of 60 informative markers derived from the tilapia O. niloticus (Kocher et al. 1998; UNH106, UNH152, UNH130, UNH192), Copadichromis cyclicos (Kellogg et al. 1995; UNH002), Astatoreochromis allaudi (Wu et al. 1999; OSU9D, OSU13D, OSU19T, OSU20D), Tropheus moorii (Zardoya et al. 1996; TmoM5, TmoM7, TmoM27), Pundamilia pundamilia (Taylor et al. 2002; Ppun1, Ppun5-7, Ppun12, Ppun16, Ppun18-20, Ppun24, Ppun34-35, Ppun41), and M. zebra (Albertson et al. 2003; UNH2004, 2005, 2008, 2032, 2037, 2044, 2046, 2056, 2058, 2059, 2069, 2071, 2075, 2080, 2084, 2094, 2100, 2104, 2112, 2116, 2117, 2125, 2134, 2139, 2141, 2149, 2150, 2153, 2163, 2166, 2169, 2181, 2185, 2191, 2204). A comprehensive list of the markers used is provided in the supporting information, Table S1.
We also searched for microsatellite repeat motifs in 9375 nonredundant cDNA clones derived from a library using A. burtoni brains and mixed tissue including both sexes and all stages of development (Salzburger et al. 2008). For 21 microsatellite-containing cDNA clones, we developed PCR primers, 13 of which were polymorphic and gave reproducible results. GenBank accession numbers for these markers are as follows: Abur221, CN470695; Abur223, CN469772; Abur228, CN470764; Abur224, DY626128; Abur225, DY630453; Abur226, DY630491; Abur227, DY626763; Abur230, DY626468; Abur233, DY629660; Abur234, DY630828; Abur235, DY627273; Abur239, DY629088; and Abur240, DY630681. We compared these sequences to the cDNA and peptide database of medaka (Oryzias latipes), as the most closely related model organism to cichlids, using Blastview at http://www.ensembl.org/Multi/blastview.
Genotyping procedures:
Microsatellites were amplified in 10-μl PCR reactions containing 10 mm Tris–HCl, 50 mm KCl, 1.5 mm MgCl2, 0.2 mm dNTPs, 4 pmol of each locus specific primer, 0.8 units Taq polymerase (Genaxxon), and 10–30 ng genomic DNA. Forward primers were labeled with the following fluorescent dyes: 6-carboxylfluoresceine (6-FAM), hexachloro-6-carboxyfluorescein (HEX) and 2,7′,8′-benzo-5′-fluoro-2′,4,7-trichloro-5-carboxyfluorescein (NED). In addition, for 212 of the initially designed primer pairs, the M13 method for fluorescent labeling of PCR products (Schülke 2000) was used for economic reasons (see Table S1). With the latter method, 1 pmol of forward primer, 4 pmol of reverse primer, and 4 pmol of M13 6-FAM- or HEX-labeled primer were used. For the second round of genotyping, we used a PCR multiplexing kit (Qiagen) to amplify three to five loci in a single PCR reaction in a 12-μl volume containing 6.25 μl of 2× Qiagen Multiplex PCR Master Mix and 1.25 μl of a mix of primers. Final concentration of primers was 0.2 μm. All PCRs were run on a Perkin Elmer (Norwalk, CT) GeneAmp PCR 9700.
Three basic temperature protocols, depending on labeling method and multiplex scheme, were used: for forward primer labeling, one cycle of 3 min at 94°, 35 cycles at 94° for 30 sec, 48°–58° for 30 sec, 72° for 90 sec, and a final extension step at 72° for 10 min; for M13 primer labeling, one cycle of 3 min at 96°, 5 cycles at 96° for 30 sec, 62°–56° for 30 sec, 72° for 30 sec, 35 cycles at 96° for 30 sec, 58°–53° for 30 sec, 72° for 30 sec, and a final extension step at 72° for 10 min; and for multiplex PCR, 95° for 15 min, 35 cycles at 94° for 30 sec, 50°–60° for 90 sec, 72° for 60 sec, and a final extension period of 30 min at 72° (for details see Table S1). PCR products for four to six loci were combined with a mixture of ABI Genescan-500 ROX size standard and were analyzed with an ABI 3100 Automated Sequencer (Applied Biosystems). More details on fluorescent dye labeling, multiplexing schemes, and annealing temperatures (Ta) for each locus are given in Table S1. Allele sizes were scored with the Genotyper 3.7 (Applied Biosystems) software package and transferred to an electronic spreadsheet.
Type I markers:
Several clones containing interesting candidate genes have been sequenced from the BAC library of A. burtoni (Lang et al. 2006). These clones incorporate the homologous sequences of Pdgfrβ (Braasch et al. 2006) and all Hox genes reported in A. burtoni (Hoegg et al. 2007). Putative microsatellites were derived from the clone sequences with the Tandem Repeat Finder v. 3.2.1 software, so that two informative markers were obtained from BAC clone 26M7 containing Pdgfrβa (Abur209, -212) and one for 20D21 containing Pdgfrβb (Abur218). The corresponding BAC clones (clone number, accession number) for HoxAa (116M8, EF594313), HoxAb (150O18, EF594311), HoxBa (170E12, EF594310), HoxBb (34I18, EF594314), HoxCa (103K21, EF594312), HoxDa (32B18, EF594315), and HoxDb (19E16, EF594316) each yielded one to three polymorphic microsatellite markers. A tetranucleotide ATCT repeat was used for marker development of clone 99M12 (Siegel et al. 2007), which includes ParaHoxC1, representing a dispersed Hox-like gene cluster.
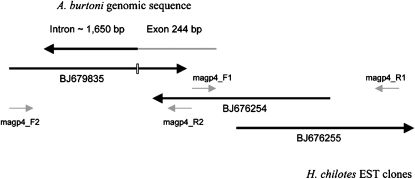
A homolog of the human microfibril-associated glycoprotein 4 (Mfap4) has been reported from cichlid EST clones derived from H. chilotes (Kobayashi et al. 2006). We used three of these clones (accession nos. BJ679835, BJ676254, and BJ680594) to form 1214 bp of continuous sequence (Figure 1). We initially designed two pairs of primers to amplify from genomic DNA of A. burtoni: Magp4_1F (5′-TCAGACCTCCACCAAACAGTC) and Magp4_1R (5′-TCCCTGAAGACCATCAGCAT), spanning 501 bp of clones BJ676254 and BJ680594, and Magp4_2F (5′-CGGTGCAGGTGTACTGTGAC) and Magp_2R (5′-ACTGCACAGGACGGATCTTC) covering 544 bp of clone BJ679835. After identifying a SNP in an intronic region, two additional primers were designed from the A. burtoni sequence for genotyping: SNP_5F (5′-GGCTTGTCTCATGTGCCTTC) and SNP_6R (5′-ACCAGCTGTCCTGGTCTTTTG), yielding a 339-bp fragment.
Figure 1.—
Schematic of the amplification of the Mfap4 gene from A. burtoni. Position of primers, alignment of EST clones of H. chilotes, and the location of an intron in A. burtoni derived from genomic DNA are shown. The clones and primers are not drawn to scale.
Lws has been characterized from a BAC library of H. chilotes. Numerous primers to amplify upstream and downstream regions of the gene are available (Terai et al. 2006). We used the following primers to amplify this region from genomic DNA in A. burtoni, all resulting in ∼1-kb fragments: LWSB_LF and LWSB_R1, LWSB_F2 and LWSB_R3, LWSB_F3 and LWSB_R4, LWSB_F4 and LWSB_R5, LWSB_F8 and LWSB_R9, and LWSB_F9 and LWSB_R10.
The primers AroI_Ex7F (5′-GGTGATCGCAGCTCCGGACACTCTCTCC), AroI_Ex8R (5′-CCTGTGTTCAGAATGATGTTTGTGC), and AroI_1600R (5′-GTACAGCTAAAGGTTCGGGTC), for amplification of 600–1200 bp of ovarian cytochrome P450 aromatase (Cyp19a1), were designed using a genomic sequence from tilapia deposited in GenBank (AF472620) and using sequences from Lake Malawi cichlids at http://www.ncbi.nlm.nih.gov/blast/mmtrace.shtml (D. T. Gerrard and A. Meyer, unpublished results). Following SNP identification, the nested primers AroI_Fw (5′-ATGGCTGCATTCCACCAC) and AroI_Rv (5′-TTCTTCATGCTTCTGCTCCTC) were designed for A. burtoni, producing 447 bp of intron sequence located between exon 7 and exon 8.
The primary sequences of the three gene regions, Mfap4, Lws, and Cyp19a1, were amplified using the annealing temperatures of 55°, 58°, and 54°, respectively, and the following PCR conditions: one cycle of 3 min at 94°, 35 cycles at 94° for 30 sec, 55°–58° for 30 sec, 72° for 90 sec, and a final extension step at 72° for 10 min. Resulting sequences of A. burtoni were screened for SNPs in the parents of the mapping cross, and identified SNPs were then sequenced and scored using nested primers (with annealing of 55°) in 167 F2 individuals.
Linkage mapping:
Linkage groups, distances, and maker orderings were determined with LocusMap 1.1 (Garbe and Da 2003), JoinMap 4 (Van Ooijen 2006), and Map Manager QTX (Manly et al. 2001). Non-inheritance errors provided by Locusmap were checked by reevaluating the original chromatograms and either corrected or omitted from the data set. Most of these errors were due to rounding errors of a 1-base difference in allele size; others were classified as possible allele drop-outs or allele mutations. The assignment of markers to linkage groups was carried out by using a grouping LOD threshold of ≥4.0. This value was increased from the commonly used LOD score of 3.0 to minimize the risk of false linkage due to the large number of two-way tests being performed (Ott 1991). Maps of each of the linkage groups were obtained using a pairwise LOD threshold of 3 and a maximum recombination threshold of 0.4. The Kosambi mapping function, which accounts for double recombinations, was used to convert recombination frequencies to centimorgans for all analyses. The order of the markers was manually optimized to decrease the number of double recombinations. Markers that present shared alleles in the grandparents have reduced statistical power, since paternal/maternal homozygotes and heterozygotes cannot be discriminated in some F2 families (e.g., when one F1 parent is homozygous for the shared allele). These markers could assume equally probable positions, and therefore their positions are represented as ranges in the map. The final map distances were calculated using Map Manager QTX. Graphics of the linkage groups were produced with the MapChart 2.1. software (Vorrips 2002).
RESULTS
Polymorphic microsatellites:
We have characterized 278 new microsatellite sequences from genomic DNA of A. burtoni, 225 of which could be used to amplify PCR products of the expected size. One hundred forty-seven markers were informative (at least one grandparent heterozygous), while 49 were not variable in this mapping cross. The remaining 28 loci showed banding patterns that were difficult to interpret and thus could not be reliably scored. An additional 60 microsatellite markers were derived from prior studies that employed microsatellite markers in cichlids (Kellogg et al. 1995; Zardoya et al. 1996; Kocher et al. 1998; Taylor et al. 2002; Albertson et al. 2003). Most of the microsatellites used consist of pure and compound dinucleotide tandem repeats, composing mainly CA and. to a lesser extent, CT repeat motifs. In addition, there were 13 tetranucleotide loci developed from the genus Pundamilia (Taylor et al. 2002) and one from a BAC clone containing ParaHoxC1. An investigation of the relationship between repeat length and rate of polymorphism showed no significant correlation; however, no repeats shorter than eight times were used initially.
A. burtoni linkage map:
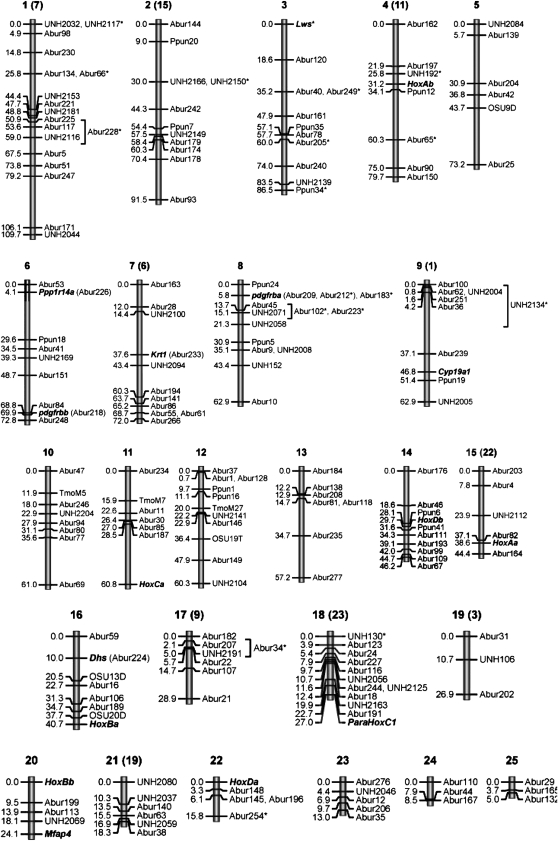
Significant linkages were identified for 204 genetic markers, including 191 microsatellite loci and 13 type I (gene) markers (Figure 2). Only six markers (Abur58, Abur147, Abur190, UNH002, UNH2075, and UNH2185) could not be linked to any other marker (97% of markers could be linked). We found 25 linkage groups with the number of markers per group ranging from 3 (LG19, -24, -25) to 18 (LG1). The largest linkage groups were LG1 and LG2 with 109.7 and 91.5 cM, respectively. The A. burtoni karyotype (2n = 40) shows at least two large subtelocentric-telocentric chromosomes, although karyotypic size variation among chromosomes was not as pronounced when compared to Tilapia and Sarotherodon (Thompson 1981).
Figure 2.—
Linkage map of A. burtoni comprising 25 linkage groups. Corresponding linkage groups for tilapia are in parentheses as inferred from shared markers among the three African cichlid maps. Distances in Kosambi centimorgans are indicated at the left of each linkage group. Asterisks indicate loci at which the crossed F0 grandparents share a common allele and paternity/maternity of the particular allele could not be identified unambiguously for all six half-sib F2 families.
The total sex-averaged length of the map was 1249.3 cM. Marker spacing was on average 6.12 cM, with the largest distance being 33 cM. Markers that had a common allele in both grandparents had reduced mapping power (since the origin of the allele cannot be identified) and presented a range of equally likely positions, which are indicated in Figure 2. Since the genome size of this species has been reported to be 0.97 pg (Lang et al. 2006), which equals ∼950 Mb, we estimated the physical-to-map distance as 760 kb/cM.
Hox gene clusters:
In A. burtoni, seven Hox gene clusters have been detected by means of BAC library screening and sequencing of positive clones (Hoegg et al. 2007). The mapping results using microsatellite flanking regions revealed a distinctive distribution of Hox clusters throughout the genome. HoxAa, HoxAb, HoxBa, and HoxBb were mapped to LG15, LG4, LG16, and LG20, respectively. While HoxCa was assigned to LG11, HoxDa and HoxDb mapped to LG22 and LG14, respectively. Overall, this pattern supports the hypothesis of the fish-specific genome duplication (FSGD) because all Hox clusters are found on different chromosomes (Hoegg and Meyer 2005; Meyer and van de Peer 2005). ParaHoxC1 is on LG18 with no association with one of the other conventional Hox clusters, corroborating the idea that these Hox-like genes constitute dispersed homeobox genes forming novel clusters somewhere else in the genome (Siegel et al. 2007).
Platelet-derived growth factor receptor β (Pdgfrβ):
Two paralogons of this gene are known to be present in cichlids due to the FSGD (Braasch et al. 2006). The mapping of adjacent microsatellite markers of type (GT/CA)12–15 from corresponding BAC clones showed the location of Pdgfrβa on LG8, while Pdgfrβb was mapped to LG6. In the same BAC clone, the two markers Abur209 and Abur212 flank the Pdgfrβa–Csf1r tandem, spanning ∼50 kb between them. It is thus not surprising that they are regarded as identical in the map. The observation of Pdgfrα (see the above treatment on ParaHoxC1) and the A- and B-copies of Pdgfrβ occurring on different linkage groups, as in Hox-genes, favors the origin of this gene family by whole-genome duplication and not by tandem duplication.
Long wavelength-sensitive opsin (Lws):
In the sequence upstream of the Lws gene using the primers LWSB_F2 and LWSB_R3, we found a SNP at position 231 (using nested primers) in the F0 grandparents of the mapping cross. The male parent was CT heterozygous and the female parent was TT homozygous, thereby permitting linkage analyses (yet with reduced statistical power) through genotyping of F2 individuals. The analysis revealed that this gene, which is believed to be important for color vision and probably speciation by female mate choice (Terai et al. 2006), is located on LG3 in the linkage map of A. burtoni. Lws therefore does not appear to be linked to any other candidate gene investigated in this study.
Microfibril-associated glycoprotein 4 (Mfap4):
Amplification from genomic DNA with the two primer pairs directly derived from EST clones of H. chilotes led to the discovery of an intronic region in sequence BJ679835 (Figure 1). Since the expected size of the product from cDNA was 544 bp and the observed size was 2.2 kb, the size of the intron could be estimated to be ∼1.65 kb in length. Within this intron we identified a SNP at position 392 in the sequence given by primer Magp4_2R when aligned to clone BJ679835. At this site the male parent was TT homozygote and the female parent was CC homozygote, while all F1 individuals were CT heterozygotes, as expected. We also compared the amplified coding regions between H. chilotes and A. burtoni and discovered a surprisingly large number of 15 substitutions in only 195 bp. For SNP genotyping of the F2, we used nested primers in the intron region yielding a shorter fragment of 339 bp, subsequent linkage analyses of which showed the Mfap4 locus to be positioned on the small LG20 (five markers, 24.1 cM). The latter also comprises the HoxBb cluster and the marker UNH2069, which shows an associated QTL for jaw morphology in the Malawi cichlid Metriaclima (Albertson et al. 2003).
Ovarian cytochrome P450 aromatase (Cyp19a1):
We were able to identify two SNPs (AF472620:g.4167A>C and AF472620:g.4440G>T) in the F0 parental DNA of the A. burtoni mapping cross: one in the intron bridging exons 7 and 8 and the second one in exon 8. The male parent was homozygous for cytosine and guanine at positions g.4167 and g.4440, respectively, while the female parent was homozygous for adenine and thymine in these same positions. Linkage analysis with microsatellite markers using an F2 intercross revealed the map location of Cyp19a1 on LG9 in A. burtoni. In the Nile tilapia, Cyp19a1 was found in the vicinity of the presumed SEX locus on LG1 (Lee and Kocher 2007). This linkage group corresponds to the original LG6 reported by Albertson et al. (2003) for Metriaclima, but this has now been renamed in accordance with the tilapia map as LG1 (Albertson et al. 2005).
EST-linked microsatellites:
Thirteen informative microsatellite markers (Abur221, 223–228, 230, 233–235, 239–240; Table S1) were produced from EST clones of A. burtoni. These markers were widely distributed on several different linkage groups, such as LG1, -3, 6–9, -11, -13, -16, and -18, making them especially suitable for genome scans to discover functional polymorphisms. Comparisons of A. burtoni EST clones with known sequences of medaka revealed a few homologies with protein-coding genes. The sequence adjoining marker Abur224 on LG16 was indicated as part of the transcript of deoxyhypusine synthase (Dhs), which occurs in a single copy in medaka and is essential for cell viability. Despite the short overlap of 42 bp and therefore the high e-value [2.2e−11; percentage identity (PID): 79%), alignment structure and orientation to the poly(A) tail with the microsatellite in the 3′-UTR strongly support this assumption. Abur226 on LG6 (6.4e−42; PID 78%) corresponds to protein phosphatase 1 regulatory subunit 14A (Ppp1r14a), a cytosolic inhibitory protein of PP1 with a molecular weight of 17 kDa. Abur233 on LG7 (5.6e−50; PID 77%) was found adjacent to a member of the protein family keratin, type I (cytoskeletal cytokeratin), accounting for the keratin filaments in epithelia. Teleost fish show an excess of keratin type I over type II genes; thus as many as 17 type I gene members are present in medaka. For more annotations of EST sequences from this library, see Salzburger et al. (2008).
Comparison of the A. burtoni and the M. zebra/ Labeotropheus fueleborni maps:
A genetic linkage map is available from a hybrid cross of two closely related Lake Malawi cichlids, L. fueleborni and M. zebra (Albertson et al. 2003). It contains 127 microsatellite markers, 33 of which we were able to use as informative markers in the linkage map of A. burtoni. A comparison of the two maps revealed good concordance in some parts, in that all markers located in a single linkage group in Lake Malawi cichlids, e.g., LG2 with UNH2037, -2059, and -2080, were found on LG21 of A. burtoni as well. In addition, numerous markers appeared jointly—i.e., in synteny—in the same order (with the exception of UNH2116 and UNH2181) on differently named linkage groups in the two maps, suggesting conservation of genomic regions in those linkage groups across different species of cichlids. It appears that A. burtoni linkage groups LG1, -2, -7, -8, -9, -12, -18, and -21 are homologous to LG1, -16, -5, -4, -6, -3, -10, and -2, respectively, in the Lake Malawi cichlid map (Albertson et al. 2003) when using the criterion of at least two shared markers per LG. The observed correspondences of microsatellite markers are reported in the Oxford plot in Table 1.
TABLE 1.
Oxford plot comparing the linkage maps of A. burtoni and M. zebra/L. fueleborni
|
M. zebra/L. fueleborni
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. burtoni | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| 1 | 6 | |||||||||||||||||||||||
| 2 | 3 | |||||||||||||||||||||||
| 3 | 1 | |||||||||||||||||||||||
| 4 | ||||||||||||||||||||||||
| 5 | 1 | |||||||||||||||||||||||
| 6 | 1 | |||||||||||||||||||||||
| 7 | 2 | |||||||||||||||||||||||
| 8 | 3 | |||||||||||||||||||||||
| 9 | 3 | |||||||||||||||||||||||
| 10 | 1 | |||||||||||||||||||||||
| 11 | ||||||||||||||||||||||||
| 12 | 2 | |||||||||||||||||||||||
| 13 | ||||||||||||||||||||||||
| 14 | ||||||||||||||||||||||||
| 15 | 1 | |||||||||||||||||||||||
| 16 | ||||||||||||||||||||||||
| 17 | 1 | |||||||||||||||||||||||
| 18 | 3 | |||||||||||||||||||||||
| 19 | ||||||||||||||||||||||||
| 20 | 1 | |||||||||||||||||||||||
| 21 | 3 | |||||||||||||||||||||||
| 22 | ||||||||||||||||||||||||
| 23 | 1 | |||||||||||||||||||||||
| 24 | ||||||||||||||||||||||||
| 25 | ||||||||||||||||||||||||
Comparison between the A. burtoni and the tilapia (Oreochromis spp.) map:
We initially screened 51 microsatellite markers from the available linkage maps of tilapia (Kocher et al. 1998; Lee et al. 2005) for their use in linkage mapping of A. burtoni and found rather low levels of polymorphism in our testcross, although amplification success was ∼50%. Thus, only a small number of markers are shared between the two maps, namely UNH106, UNH2191, UNH192, UNH2150, UNH2166, and UNH130, that could be used for comparison. In the tilapia map containing 24 linkage groups, markers on LG3, -9, -11, -15, and -23 indicated correspondence of these linkage groups to LG19, -17, -4, -2, and -18, respectively, in the A. burtoni map. Due to the small number of tilapia microsatellites that were successfully mapped, the correspondence of LGs was determined with the sharing of a single marker, with the exception of LG2, which shares two markers with LG15 of tilapia.
SNP mapping of Cyp19a1 in A. burtoni mapped this gene to LG9, which therefore might be homologous to LG1 in tilapia, which contains the genes SEX and Cyp19a1 (Lee and Kocher 2007). In accordance with that, Albertson et al. (2005) have renamed LG6 from an earlier article on Metriaclima (Albertson et al. 2003) to LG1 as in tilapia. This might suggest that similarities among all three East African cichlid genetic maps could be used to find probable synonymies of linkage groups. We attempt to reconcile the nomenclature of the present genetic linkage maps of cichlids (supporting information, Table S2). Also, using the tilapia map as a standard, some of these relationships were included in Figure 2.
DISCUSSION
Here we present the third genetic map of a cichlid fish. A. burtoni occurs in Lake Tanganyika and its surrounding rivers. It significantly adds to the knowledge of previous linkage maps for the generalist and geographically widespread tilapia, Oreochromis spp. (Kocher et al. 1998; Lee et al. 2005), and the specialist Lake Malawi endemic M. zebra/L. fueleborni (Albertson et al. 2003). In the A. burtoni map, we identified linkages among 204 genetic markers, mainly microsatellites and 13 type I (gene) markers, which were assigned to 25 linkage groups. The difference in size of the linkage groups corresponds quite well with chromosome morphology reported from investigations of the karyotype. While most chromosomes are relatively small and of metacentric-submetacentric or metacentric type, there are also four large subtelocentric-telocentric chromosomes in the karyotype (Thompson 1981). The A. burtoni karyotype consists of 20 chromosomes; it is therefore expected that some of the linkage groups will coalesce with the addition of more markers. Lake Malawi cichlids, on the other hand, have a slightly higher chromosome number (N = 23) (Thompson 1981) in very good agreement with the 24 linkage groups found by Albertson et al. (2003) for an interspecific cross between M. zebra and L. fueleborni. The current map of tilapia, although one of the most detailed fish linkage maps, still contains two linkage groups more than is expected from the 22 chromosomes of the karyotype (Lee et al. 2005).
There is a quite good correspondence between the A. burtoni and the Lake Malawi genetic maps (Table 1). Synteny between microsatellite markers was common while only a few unexpected syntenies among shared markers were found, such as UNH2084 that was mapped to LG5 instead of LG12 in A. burtoni. Those marker locations might indicate genomic rearrangements such as translocations of chromosomal sections. The synteny of markers used for the map of tilapia could not be directly explored in A. burtoni possibly because of the considerable evolutionary distance (∼15 MY) between these species. This is the most probable explanation for the observation that a large portion of the PCR reactions for particular markers did not work. Many of the markers that did amplify did not show polymorphism in the A. burtoni mapping cross. Hence, the hope of a wide applicability for the tilapia map for the >2000 species of cichlid fishes in the East African lakes (Lee et al. 2005) might have been somewhat optimistic. Nevertheless, by using the combined information from the three maps of East African cichlids, many linkage groups could be interpreted as being orthologous with some linkage groups of the tilapia map on the basis of shared markers (see Table S2 and Figure 2).
Comparison of the map location of the gene for ovarian cytochrome P450 aromatase (Cyp19a1) on LG1 in tilapia (Lee and Kocher 2007), together with the overlapping markers used in M. zebra and A. burtoni (Table S2), suggests that the orthology of LG6 in M. zebra and LG9 in A. burtoni to that tilapia LG1 is highly likely (see also Albertson et al. 2005). Cyp19a1 is involved in sex differentiation of mammals and could also be important in determining sex in vertebrate species that lack sex chromosomes. However, in the Nile tilapia, this gene was found 27 cM away from the presumed sex-determining locus on LG1, calling into question its function as a master control gene for sex determination. In general, the sex locus in tilapia (O. niloticus) behaves like an XY male heterogametic system (Lee et al. 2003; Lee and Kocher 2007). On the other hand, Lee et al. (2004) found microsatellite markers consistent with a WZ (female heterogametic) system on LG3 in O. aureus. Thus, the mechanism of sex determination appears highly variable among species of African cichlids—as appears to be the case in fishes more generally (Volff et al. 2007). The occurrence of Cyp19a1 on LG9 in A. burtoni raises the possibility that a sex-determining factor is located on this chromosome. It is known, however, that alternative chromosomal sex-determining mechanisms have evolved independently in closely related fish species (Takehana et al. 2007; Henning et al. 2008).
Using microsatellites from BAC clone sequences, we were able to map all seven clusters of Hox genes that have been reported from A. burtoni (Hoegg et al. 2007) as well as the ParaHoxC1 cluster (Siegel et al. 2007). The surprising variation in Hox cluster numbers among vertebrates has been widely used to study the evolution of vertebrate genomic organization (Hoegg and Meyer 2005). While the phylogenetic timing of the first two rounds of genome duplications (that also resulted in the Hox cluster duplications) during the chordate-tetrapod evolution is still somewhat debated, the evidence is solid that the eight Hox clusters in ray-finned fish originated through a whole-genome duplication (the FSGD or 3R hypothesis) (Malaga-Trillo and Meyer 2001; Hoegg et al. 2004; Meyer and van de Peer 2005). In accordance with the FSGD hypothesis, all Hox clusters were found to be scattered throughout the genome of A. burtoni. Similarly, zebrafish have seven Hox clusters on seven different chromosomes (Postlethwait et al. 1998), but their Db cluster was lost instead of the Cb cluster in A. burtoni (Hoegg et al. 2007). The ParaHoxC1 paralogon mapped to LG18, which does not carry any other genes of the Hox complex. Sequence comparisons showed that ParaHoxC1 and its 3′ adjoining genes of Danio rerio are located on chromosome 20 (Siegel et al. 2007). In general, synteny of the duplicated genes in teleosts seems to be less conserved in the ParaHox genes compared to the Hox genes.
One important cause for the vast amount of cichlid diversification might be related to the abundance of color morphs in different populations that through mate choice and male–male competition might lead to speciation (e.g., Turner and Burrows 1995; Seehausen and Schluter 2004). Therefore, the mapping of candidate genes involved in fish coloration would seem to aid in the study speciation processes in cichlids. For example, the orange blotch color pattern in M. zebra, which is expressed mainly in females, has been mapped and candidate genes for this pigmentation phenotype have been identified (Streelman et al. 2003). We mapped two paralogons of the tandems Pdgfrβ-Csf1r and one of Pdgfrα-Kita, a family of receptor tyrosine kinase genes that have been shown to influence coloration in teleosts (Braasch et al. 2006; Salzburger et al. 2007). While the kit gene is essential for the development of neural-crest-derived dark melanocytes in mammals and zebrafish (Parichy et al. 1999), Csf1r promotes the development of yellow xanthophores in zebrafish (Parichy and Turner 2003), and there is evidence for its role in the development of cichlid egg spots (Salzburger et al. 2007). The location of the Pdgfrß paralogons on two different linkage groups of A. burtoni (Pdgfrßa on LG8 and Pdgfrßb on LG6) lends further support to the hypothesis of the fish-specific genome duplication. In general, the teleost A-paralogon has retained a longer stretch of synteny with the single copy of the tetrapod locus compared to the B-paralogon (see Braasch et al. 2006). It has therefore been suggested that the B-paralogon underwent functional divergence (possibly neofunctionalization) of the cell-surface receptors. These duplicated receptor genes would be important targets for future QTL studies of cichlid coloration patterns.
Divergent evolution of the visual system is a likely mechanism to explain incipient speciation and diverse patterns in the male breeding coloration in cichlids (Terai et al. 2006). A. burtoni is a close relative of the sibling species pairs in the genus Pundamilia, for which the sensory drive hypothesis (differences in male coloration evolving as a consequence of divergent visual sensitivities) has been suggested to involve the gene for long wavelength-sensitive opsin (Lws) (Seehausen et al. 2008). This gene shows the highest variability among cichlid opsins and appears to be under strong divergent selection at least in the Lake Victoria species flock (Carleton et al. 2005). This gene mapped in A. burtoni to LG3, which corresponds to LG5 in tilapia that carries the genes for the Blue Opsin and for c-ski I (Lee et al. 2005). Remarkably, the latter marker has been found to be in close association with the orange blotch color polymorphism in M. zebra (Streelman et al. 2003). It will thus be interesting to examine whether other linked genes for coloration co-occur on that chromosome.
Apart from color, evolutionary diversification in cichlids is also believed to be driven by trophic specialization and associated altered jaw morphologies and tooth shape in different species (e.g., Kocher 2004; Albertson and Kocher 2006; Streelman and Albertson 2006). A QTL study by Albertson et al. (2003) focused on these feeding adaptations and suggested that the oral jaws of Lake Malawi cichlids evolved in response to strong divergent selection. Closely related species of Lake Victoria cichlids such as H. chilotes and sp. “Rock Kribensis” can exhibit quite divergent types of jaws. A conspicuous difference in expression of the Mfap4 gene (encoding a microfibril-associated glycoprotein) was recently discovered in these species (Kobayashi et al. 2006). Mfap4 therefore may be partly responsible for the morphological differences among cichlid species. Magp4 is also known to be involved in a human heritable disease, the Smith–Magenis–Syndrome, which results in a characteristic phenotype with a flattened mid-face and striking jaw and forehead (Zhao et al. 1995). The ci-Mfap4 mapped to the small LG20 in A. burtoni. It also contains the HoxBb cluster and three anonymous microsatellite markers. The corresponding (shared marker UNH2069) LG20 in M. zebra also carries a QTL for differences in lower-jaw width (Albertson et al. 2003). Another candidate gene for craniofacial diversity, bone morphogenetic protein 4 (bmp4), is located on LG2 (renamed LG19 in Albertson et al. 2005) in M. zebra (which corresponds to LG21 in A. burtoni) and explains >30% of the phenotypic variation in the opening and closing levers of the cichlid lower jaw (Albertson et al. 2005).
Thirteen EST-based microsatellite markers derived from brain and mixed tissue libraries of A. burtoni (Salzburger et al. 2008) mapped to its entire genome. Homology searches using the medaka database resulted in three reliable hits for deoxyhypusine synthase (Dhs), protein phosphatase 1 (Ppp1r14a), and a member of the keratin, type I protein family. The remaining EST-linked A. burtoni markers came from anonymous clones, for which no homology with known sequences could be established. Nevertheless, they might prove to be useful gene-associated markers for detecting signatures of divergent selection (Vasemägi et al. 2005). In particular, the broad taxonomic application range of at least some microsatellite markers in East African cichlids provides a rare opportunity to use such markers with a known chromosomal location for comparative studies on polymorphism and to examine the footprints of selection.
Genetic linkage maps are valuable genomic resources that have been widely used for a number of different applications, but particularly interesting may be their use in QTL approaches for evolutionary questions (Erickson et al. 2004; Slate 2005). One of the most debated questions in evolutionary biology is whether major genes play a key role in species differences or whether a large number of small changes bring about phenotypic diversification during speciation (Orr 2001). QTL studies have made significant contributions to this issue (e.g., Hawthorne and Via 2001; Peichel et al. 2001; Albertson et al. 2003). In threespine sticklebacks, for example, it was shown that a major QTL causes large shifts in phenotyp such as the morphological differences seen between the sympatric benthic and limnetic forms (Colosimo et al. 2004). These first studies support the hypothesis that large effects in a small number of genes, more often than small effects in large numbers of loci, tend to bring about large phenotypic changes and might even cause evolutionary novelties. However, whether these first results will turn out to be typical is still unclear.
Studies in East African cichlids found new relationships between genomic regions involved in feeding adaptations and jaw types (Albertson et al. 2003, 2005), including the discovery of a region explaining ∼40% of the phenotypic variance in cichlid tooth patterning (cusp number) (Streelman and Albertson 2006). It is thus promising that whole-genome sequencing has been initiated for several species of cichlid fishes, including draft sequencing (5×) of tilapia (O. niloticus) and 2× coverage each of three haplochromine species (A. burtoni, Pundamilia, and M. zebra will be among these haplochrome cichlids whose genomes will be sequenced (http://www.cichlidgenome.org/) so that the medium-density genetic map of A. burtoni can be used with QTL analyses to ultimately identify evolutionarily important genes (those that are presumably selectively adaptive). Comparisons of the sequenced cichlid genomes will then give insights into the similarity or differences in the underlying molecular changes that caused their phenotypic divergence among closely related species or caused convergent similarities among more distantly related cichlid species.
Acknowledgments
We thank W. Salzburger for assistance with the mapping cross and T. D. Kocher for comments on a previous version of this manuscript. C. Chang-Rudolf, I. Eichstätter, U. Ernst, E. Hespeler, D. Leo, C. Michel, K. Nübling, M. Pehr, N. Siegel, and H. Singer helped with the lab work; C. Stemshorn provided advice on the LocusMap software; and S. Kuraku provided advice on the medaka database. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to A.M., a Ph.D. grant from the Deutscher Akademischer Austausch Dienst/Brazilian National Counsel of Technological and Scientific Development to F.H., and the Long-Term Fellowship of the International Human Frontier Science Program to S.F. (00059/2005-L).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.089367/DC1.
References
- Albertson, R. C., and T. D. Kocher, 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity 97 211–221. [DOI] [PubMed] [Google Scholar]
- Albertson, R. C., J. T. Streelman and T. D. Kocher, 2003. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci. USA 100 5252–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson, R. C., J. T. Streelman, T. D. Kocher and P. C. Yelick, 2005. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. USA 102 16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch, I., W. Salzburger and A. Meyer, 2006. Asymmetric evolution in two fish-specifically duplicated receptor tyrosin kinase paralogons involved in teleost coloration. Mol. Biol. Evol. 23 1192–1202. [DOI] [PubMed] [Google Scholar]
- Carleton, K. L., J. W. Parry, J. K. Bowmaker, D. M. Hunt and O. Seehausen, 2005. Colour vision and speciation in Lake Victoria cichlids of the genus Pundamilia. Mol. Ecol. 14 4341–4353. [DOI] [PubMed] [Google Scholar]
- Chistiakov, D. A., B. Hellemans, C. S. Haley, A. S. Law, C. S. Tsigenopoulos et al., 2005. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics 170 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo, P. F., C. L. Peichel, K. Nereng, B. K. Blackman, M. D. Shapiro et al., 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. Plos Biol. 2 e109. [DOI] [PMC free article] [PubMed]
- Crapon de Caprona, M. D., and B. Fritzsch, 1984. Interspecific fertile hybrids of haplochromine Cichlidae (Teleostei) and their possible importance for speciation. Neth. J. Zool. 34 503–538. [Google Scholar]
- DiPalma, F., C. Kidd, R. Borowsky and T.D. Kocher, 2007. Construction of bacterial artificial chromosome libraries for the Lake Malawi cichlid (Metriaclima zebra), and the Blind Cavefish (Astyanax mexicanus). Zebrafish 4 41–47. [DOI] [PubMed] [Google Scholar]
- Erickson, D. L., C. B. Fenster, K. Stenøien and D. Price, 2004. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13 2505–2522. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, New York.
- Fryer, G., and T. D. Iles, 1972. The Cichlid Fishes of the African Great Lakes. Oliver & Boyd, Edinburgh.
- Garbe, J., and Y. Da, 2003. Locusmap User Manual, Version 1.1. Department of Animal Science, University of Minnesota. http://animalgene.umn.edu/locusmap/index.html.
- Gerrard, D. T., and A. Meyer, 2007. Positive selection and gene conversion in SPP120, a fertilization-related gene, during East African cichlid fish radiation. Mol. Biol. Evol. 24 2286–2297. [DOI] [PubMed] [Google Scholar]
- Gilbey, J., E. Verspoor, A. McLay and D. Houlihan, 2004. A microsatellite linkage map for Atlantic salmon (Salmo salar). Anim. Genet. 35 98–105. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D. J., and S. Via, 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412 904–907. [DOI] [PubMed] [Google Scholar]
- Henning, F., V. Trifonov, M. A. Ferguson-Smith and L. F. Almeida-Toledo, 2008. Non-homologous sex chromosomes in two species of the genus Eigenmannia (Teleostei: Gymnotiformes). Cytogenet. Genome Res. 121 55–58. [DOI] [PubMed] [Google Scholar]
- Hoegg, S., and A. Meyer, 2005. Hox clusters as models for vertebrate genome evolution. Trends Genet. 21 421–424. [DOI] [PubMed] [Google Scholar]
- Hoegg, S., H. Brinkmann, J. S. Taylor and A. Meyer, 2004. Phylogenetic timing of the fish-specific genome duplication correlates with phenotypic and taxonomic diversification in fishes. J. Mol. Evol. 59 190–203. [DOI] [PubMed] [Google Scholar]
- Hoegg, S., J. L. Boore, J. V. Kuehl and A. Meyer, 2007. Comparative phylogenomic analyses of teleost fish Hox gene clusters: lessons from the cichlid fish Astatotilapia burtoni. BMC Genomics 8 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, T., S. Asakawa, S. Minagawa, N. Shimizu, I. Hirono et al., 2001. Construction and characterization of BAC libraries for three fish species: rainbow trout, carp and tilapia. Anim. Genet. 32 200–204. [DOI] [PubMed] [Google Scholar]
- Katagiri, T., C. Kidd, E. Tomasino, J. T. Davis, C. Wishon et al., 2005. A BAC-based physical map of the Nile tilapia genome. BMC Genomics 6 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, K. A., J. A. Markert, J. R. Stauffer, Jr. and T. D. Kocher, 1995. Microsatellite variation demonstrates multiple paternity in lekking cichlid fishes from Lake Malawi, Africa. Proc. R. Soc. Lond. B 260 79–84. [Google Scholar]
- Kobayashi, N., T., M. Watanabe, T. Kijimoto, K. Fujimura, M. Nakazawa et al., 2006. magp4 gene may contribute to the diversification of cichlid morphs and their speciation. Gene 373 126–133. [DOI] [PubMed] [Google Scholar]
- Kocher, T. D., 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 4 288–298. [DOI] [PubMed] [Google Scholar]
- Kocher, T. D., J. A. Conroy, K. R. McKaye and J. R. Stauffer, 1993. Similar morphologies of cichlids in lakes Tanganyika and Malawi are due to convergence. Mol. Phylogenet. Evol. 2 158–165. [DOI] [PubMed] [Google Scholar]
- Kocher, T. D., W-J. Lee, H. Sobolewska, D. Penman and B. McAndrew, 1998. A genetic linkage map of a cichlid fish, the tilapia (Oreochromis niloticus). Genetics 148 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfield, I., and P. F. Smith, 2000. African cichlid fishes: model systems for evolutionary biology. Annu. Rev. Ecol. Syst. 31 163–196. [Google Scholar]
- Lang, M., T. Miyake, I. Braasch, D. Tinnemore, N. Siegel et al., 2006. A BAC library of the East African haplochromine cichlid fish Astatotilapia burtoni. J. Exp. Zoolog. B Mol. Dev. Evol. 306B 35–44. [DOI] [PubMed] [Google Scholar]
- Lee, B-Y., and T. D. Kocher, 2007. Exclusion of Wilms tumour (WT1b) and ovarian cytochrome P450 aromatase (Cyp19a1) as candidates for sex determination genes in Nile tilapia (Oreochromis niloticus). Anim. Genet. 38 85–86. [DOI] [PubMed] [Google Scholar]
- Lee, B-Y., D. J. Penman and T. D. Kocher, 2003. Identification of a sex-determining region in Nile tilapia (Oreochromis aureus). Heredity 92 543–549. [DOI] [PubMed] [Google Scholar]
- Lee, B-Y., G. Hulata and T. D. Kocher, 2004. Two unlinked loci controlling sex of blue tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim. Genet. 34 379–383. [DOI] [PubMed] [Google Scholar]
- Lee, B-Y., W-J. Lee, J. T. Streelman, K. L. Carleton, A. E. Howe et al., 2005. A second-generation linkage map of Tilapia (Oreochromis spp.). Genetics 170 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Maan, M. E., K. D. Hofker, J. J. van Alphen and O. Seehausen, 2006. Sensory drive in cichlid speciation. Am. Nat. 167 947–954. [DOI] [PubMed] [Google Scholar]
- Malaga-Trillo, E., and A. Meyer, 2001. Genome duplications and accelerated evolution of Hox genes and cluster architecture in teleost fishes. Am. Zool. 41 676–686. [Google Scholar]
- Manly, K. F., J. R. H. Cudmore and J. M. Meer, 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12 930–932. [DOI] [PubMed] [Google Scholar]
- Meyer, A., 1993. Phylogenetic relationships and evolutionary processes in East African cichlids. Trends Ecol. Evol. 8 279–284. [DOI] [PubMed] [Google Scholar]
- Meyer, A., and Y. van de Peer, 2005. From 2R to 3R: evidence for a fish specific genome duplication (FSGD). BioEssays 27 937–945. [DOI] [PubMed] [Google Scholar]
- Meyer, A., T. D. Kocher, P. Basasibwaki and A. C. Wilson, 1990. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 347 550–553. [DOI] [PubMed] [Google Scholar]
- Meyer, A., T. D. Kocher and A. C. Wilson, 1991. African fishes. Nature 350 467–468. [Google Scholar]
- Miller, C. T., S. Beleza, A. A. Pollen, D. Schluter, R. A. Kittles et al., 2007. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2001. The genetics of species differences. Trends Ecol. Evol. 16 343–350. [DOI] [PubMed] [Google Scholar]
- Ott, J., 1991. The Analysis of Human Genetic Linkage, Ed. 2. Johns Hopkins Press, Baltimore.
- Parichy, D. M., and J. M. Turner, 2003. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development 130 817–833. [DOI] [PubMed] [Google Scholar]
- Parichy, D. M., J. F. Rawls, S. J. Pratt, T. T. Whitfield and S. L. Johnson, 1999. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 126 3425–3436. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., K. S. Nereng, K. A. Ohgi, B. L. Cole, P. F. Colosimo et al., 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414 901–905. [DOI] [PubMed] [Google Scholar]
- Postlethwait, J. H., Y.-L. Yan, M. A. Gates, S. Horne, A. Amores et al., 1998. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18 345–349. [DOI] [PubMed] [Google Scholar]
- Protas, M., M. Conrad, J. B. Gross, C. Tabin and R. Borowsky, 2007. Regressive evolution in the Mexican Cave Tetra, Astyanax mexicanus. Curr. Biol. 17 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn, S. C., N. Aubin-Horth and H. A. Hofmann, 2004. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics 5: 42. [DOI] [PMC free article] [PubMed]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger, W., and A. Meyer, 2004. The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften 91 277–290. [DOI] [PubMed] [Google Scholar]
- Salzburger, W., A. Meyer, S. Baric, E. Verheyen and C. Sturmbauer, 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst. Biol. 51 113–135. [DOI] [PubMed] [Google Scholar]
- Salzburger, W., T. Mack, E. Verheyen and A. Meyer, 2005. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 5 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger, W., I. Braasch and A. Meyer, 2007. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger, W., D. Steinke, S. C. Renn, H. A. Hofmann, R. D. Fernald et al., 2008. Annotation of expressed sequence tags for the East African cichlid fish species Astatotilapia burtoni and evolutionary analyses of cichlid ORFs. BMC Genomics 9 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanetra, M., and A. Meyer, 2005. Microsatellites from the burbot (Lota lota), a freshwater gadoid fish (Teleostei). Mol. Ecol. Notes 5 390–392. [Google Scholar]
- Schülke, M., 2000. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18 233–234. [DOI] [PubMed] [Google Scholar]
- Seehausen, O., and D. Schluter, 2004. Male-male competition and nuptial color displacement as diversifying force in Lake Victoria cichlid fishes. Proc. R. Soc. Lond. B 271 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen, O., Y. Terai, I. S. Magalhaes, K. L. Carleton, H. D. Mrosso et al., 2008. Speciation through sensory drive in cichlid fish. Nature 455 620–626. [DOI] [PubMed] [Google Scholar]
- Siegel, N., S. Hoegg, W. Salzburger, I. Braasch and A. Meyer, 2007. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics 8 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate, J., 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14 363–379. [DOI] [PubMed] [Google Scholar]
- Stiassny, M. L. J., and A. Meyer, 1999. Buntbarsche Meister der Anpassung. Spektrum der Wissenschaft 6 36–43. [Google Scholar]
- Streelman, J. T., and R. C. Albertson, 2006. Evolution of novelty in the cichlid dentition. J. Exp. Zool. 306B 216–226. [DOI] [PubMed] [Google Scholar]
- Streelman, J. T., and T. D. Kocher, 2000. From phenotype to genotype. Evol. Dev. 2 166–173. [DOI] [PubMed] [Google Scholar]
- Streelman, J. T., R. C. Albertson and T. D. Kocher, 2003. Genome mapping of the orange blotch color pattern in cichlid fishes. Mol. Ecol. 12 2465–2471. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., K. Naruse, S. Hamaguchi and M. Sakaizumi, 2007. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116 463–470. [DOI] [PubMed] [Google Scholar]
- Taylor, M.I., F. Meardon, G. Turner, O. Seehausen, H. D. J. Mrosso et al., 2002. Characterization of tetranucleotide microsatellite loci in a Lake Victorian, haplochromine cichlid fish: a Pundamilia pundamilia × Pundamilia nyererei hybrid. Mol. Ecol. Notes 2 443–445. [Google Scholar]
- Terai, Y., O. Seehausen, T. Sasaki, K. Takahashi, S. Mizoiri et al., 2006. Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PloS Biol. 12 e433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, K. W., 1981. Karyotypes of six species of African Cichlidae (Pisces: Perciformes). Experientia 37 351–352. [DOI] [PubMed] [Google Scholar]
- Turner, G. F., and M. T. Burrows, 1995. A model of sympatric speciation by sexual selection. Proc. R. Soc. Lond. B 260 287–292. [Google Scholar]
- Van Ooijen, J. W., 2006. JoinMap 4. Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen, The Netherlands.
- Vasemägi, A., J. Nilson and C. R. Primmer, 2005. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent evolution in Atlantic salmon (Salmo salmar L.). Mol. Biol. Evol. 22 1067–1076. [DOI] [PubMed] [Google Scholar]
- Verheyen, E., W. Salzburger, J. Snocks and A. Meyer, 2003. The origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300 325–329. [DOI] [PubMed] [Google Scholar]
- Volff, J-N., I. Nanda, M. Schmid and M. Schartl, 2007. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex. Dev. 1 85–99. [DOI] [PubMed] [Google Scholar]
- Vorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., N. Kobayashi, A. Fujiyama and N. Okada, 2003. Construction of a BAC library for Haplochromis chilotes, a cichlid fish from Lake Victoria. Genes Genet. Syst. 78 103–105. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., N. Kobayashi, T. Shin-i, T. Horiike, Y. Tateno et al., 2004. Extensive analysis of ORF sequences from two different cichlid species in Lake Victoria provides molecular evidence for a recent radiation event of the Victoria species flock: identity of EST sequences between Haplochromis chilotes and Haplochromis sp. “Redtailsheller.” Gene 343 263–269. [DOI] [PubMed] [Google Scholar]
- Wu, L., L. Kaufman and P. A. Fuerst, 1999. Isolation of microsatellite markers in Astatoreochromis alluaudi and their cross-species amplifications in other African cichlids. Mol. Ecol. 8 895–897. [DOI] [PubMed] [Google Scholar]
- Zardoya, R., D. M. Vollmer, C. Craddock, J. T. Streelman, S. Karl et al., 1996. Evolutionary conservation of microsatellite flanking regions and their use in resolving the phylogeny of cichlid fishes (Pisces: Perciformes). Proc. R. Soc. Lond. B 263 1589–1598. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., C. C. Lee, S. Jiralerspong, R. C. Juyal, F. Lu et al., 1995. The gene for a human microfibril-associated glycoprotein is commonly deleted in Smith-Magenis syndrome patients. Hum. Mol. Genet. 4 589–597. [DOI] [PubMed] [Google Scholar]