Abstract
Meiotic recombination enhances genetic diversity as well as ensures proper segregation of homologous chromosomes, requiring Spo11-initiated double-strand breaks (DSBs). DNA deaminases act on regions of single-stranded DNA and deaminate cytosine to uracil (dU). In the immunoglobulin locus, this lesion will initiate point mutations, gene conversion, and DNA recombination. To begin to delineate the effect of induced base lesions on meiosis, we analyzed the effect of expressing DNA deaminases (activation-induced deaminase, AID, and APOBEC3C) in germ cells. We show that meiotic dU:dG lesions can partially rescue a spo11Δ phenotype in yeast and worm. In rec12 Schizosaccharomyces pombe, AID expression increased proper chromosome segregation, thereby enhancing spore viability, and induced low-frequency meiotic crossovers. Expression of AID in the germ cells of Caenorhabditis elegans spo-11 induced meiotic RAD-51 foci formation and chromosomal bivalency and segregation, as well as an increase in viability. RNAi experiments showed that this rescue was dependent on uracil DNA-glycosylase (Ung). Furthermore, unlike ionizing radiation-induced spo-11 rescue, AID expression did not induce large numbers of DSBs during the rescue. This suggests that the products of DNA deamination and base excision repair, such as uracil, an abasic site, or a single-stranded nick, are sufficient to initiate and alter meiotic recombination in uni- and multicellular organisms.
MEIOSIS is a specialized form of cell division in sexually reproducing organisms, which can be divided into different stages. Following premeiotic DNA replication, homologous chromosomes are aligned and crossovers between homologous chromosomes are generated by meiotic recombination during meiosis I. This permits accurate segregation of homologous chromosomes into daughter cells at the first meiotic division. During the second meiotic division, the sister chromatids are segregated and haploid gametes are formed. Most meiotic recombination is thought to initiate via a double-strand break (DSB) in DNA, induced by the evolutionary conserved protein Spo11 (or Rec12 in Schizosaccharomyces pombe) (Klapholz et al. 1985; Lin and Smith 1994; Keeney et al. 1997; Whitby 2005). In all organisms studied, loss of Spo11 has a partial or full detrimental effect on gamete formation. This deficiency can be partially rescued by the artificial introduction of double-strand breaks, such as treatment with gamma radiation (Thorne and Byers 1993; Dernburg et al. 1998) or inactivation of proper DNA processing (Farah et al. 2005). In fission yeast, loss of Rec12 has a 3-fold effect: tetrad formation in the asci is decreased almost 50% with a concomitant increase in dyad formation (a consequence of failed meiosis I), spore viability is decreased almost 4-fold, and meiotic recombination is reduced by at least 100-fold (De Veaux et al. 1992; Sharif et al. 2002). In Caenorhabditis elegans, Spo11 deficiency has a comparable effect on the organism, manifested in reduced viability and increased frequency of male offspring due to the lack of crossovers and impaired segregation of chromosomes. The importance of Spo11-induced DSBs for proper germ cell formation is clear, yet there has been little progress in understanding how DNA lesions other than DSBs can influence meiosis or how these lesions alter meiotic recombination in the absence of Spo11—although recent data may imply a role for them (Cromie et al. 2006).
In somatic cells most DNA damage is derived from chemical (oxygen, radicals, or alkylating molecules) or physical (e.g., UV or gamma radiation) agents and is repaired by numerous evolutionary conserved pathways. Recently though, protein-induced DNA deamination, almost exclusively found in vertebrates (Conticello et al. 2005), has been shown to provide physiological benefits and has been implicated in systems as diverse as the development of acquired immunity (Di Noia and Neuberger 2002; Petersen-Mahrt et al. 2002; Rada et al. 2002; Bransteitter et al. 2003; Chaudhuri et al. 2003; Neuberger et al. 2003; Chaudhuri and Alt 2004; Maizels 2005), active viral inhibition in innate immunity (Sheehy et al. 2002; Harris et al. 2003; Huthoff and Malim 2005; Cullen 2006), and the reprogramming of epigenetic markers (Morgan et al. 2004). Acting on single-stranded DNA, DNA deaminases recognize cytosine and catalyze a hydrolytic deamination, producing uracil. This foreign DNA base can be a substrate for a number of different DNA repair pathways. In the acquired immune system a dU:dG mismatch induced by activation-induced deaminase (AID) can lead to somatic hypermutation (SHM) in the variable region, presented as point mutations at the lesion and the surrounding bases (Chaudhuri and Alt 2004; Neuberger et al. 2003, 2005; Maizels 2005). Furthermore, dU:dG lesions near the constant region, if recognized by mismatch repair (MMR) proteins or base excision repair (BER) proteins and the nonhomologous end-joining pathway (NHEJ), lead to intrachromosomal DNA recombination and class switch recombination (CSR) (Manis et al. 1998; Rada et al. 2002, 2004; Imai et al. 2003). In some vertebrates, the postrecognition and processing of dU:dG can be mediated through the RAD-51c paralogs (XRCC2 and XRCC3), leading to immunoglobulin gene conversion (iGC) in the variable region (Sale et al. 2001). Targeting of AID outside the immunoglobulin locus can induce mutations, but at a much reduced frequency (Pasqualucci et al. 2001). Importantly, dU lesions do not generally lead to DSBs, as DSBs have been implicated only during CSR and only if AID is recruiting specific cofactors (Barreto et al. 2003). It is therefore the targeting region (variable vs. constant vs. nonimmunoglobulin), chromatin configuration, AID recruited cofactors, and cell state that will determine how the dU:dG will be processed leading to mutation, recombination, or repair.
Because the catalytic activity of DNA deaminases is independent of its host and has been demonstrated by exogenous addition in Escherichia coli (Harris et al. 2002; Petersen-Mahrt et al. 2002) and yeast (Poltoratsky et al. 2004; Mayorov et al. 2005; Schumacher et al. 2005; Krause et al. 2006; Rogozin and Pavlov 2006), we wanted to know if a DNA deaminase-induced dU:dG mismatch could be processed by the meiotic nuclei to trigger meiotic recombination. Our data indicate that expression of AID and APOBEC3C during meiosis of S. pombe can partially rescue the rec12Δ phenotype, as seen by an increase in tetrad formation, spore viability, and meiotic recombination. Furthermore, AID expression in the germline of C. elegans spo-11 revealed that dU lesions induce postreplicative meiotic RAD-51 foci, which was dependent on the BER protein Ung. These lesions are also processed to give rise to crossovers and accurate chromosome segregation at the first meiotic division. Importantly, AID's induced rescue does not appear to be due to excessive DSB formation. These data lead us to conclude, that the products of DNA deaminases and/or BER, a dU, an abasic site, or a single-stranded DNA nick, are sufficient to initiate meiotic recombination.
MATERIALS AND METHODS
Strains and media:
The genotypes of the S. pombe strains used in this study were JCF108 (h− ade6-M210 his3-D1 leu1-32 ura4-D18), JCF109 (h+ wt ade6-M216 his 3-D1 leu1-32 ura4-D18), FO1693 (h− rec12-171∷ura4+ ura4-D18 leu1-32 arg3-D4 ade6+), FO1694 (h+ rec12-171∷ura4+ ura4-D18 leu1-32 his3-D1 ade6-M216), FO1695 (h− rec12-167 leu1-32 his3-D1 arg3-D4 ade6-M216), and FO1696 (h+ rec12-169∷HA6HIS-KanMX ura4-D18 leu1-32 ade6+). S. pombe strains were grown vegetatively in liquid/solid yeast extract with supplements (YE5S) or liquid/solid Edinburgh minimal medium (EMM) with supplements at 30° as described previously (Moreno et al. 1991). Diploids were distinguished from haploid cells by adding 2.5 mg/liter phloxin B (Sigma) to the YE5S media or EMM plus supplements. In the presence of phloxin B, diploid colonies stain dark pink whereas haploid colonies stain light pink. Strains were crossed on EMM low nitrogen lacking leucine ± thiamine at 25°.
Constructs for fission yeast experiments:
Exogenous expression of the human AID was achieved by using the pREP1 and pREP41 expression vectors described previously (Maundrell 1990, 1993). These vectors contain the nmt (no message in thiamine) promoter, allowing efficient repression of gene expression by thiamine. In this study, a final concentration of 15 μm thiamine was used to repress the nmt promoter. pREP41 has a weaker promoter compared to pREP1 and therefore less protein is produced by this expression vector. The protocol for lithium acetate transformation of fission yeast was used as described previously (Bahler et al. 1993).
Tetrad analysis:
Tetrad analysis was performed as described (Moreno et al. 1991), with slight modifications. Asci from 3-day-old matings were suspended in 0.5 ml of dH2O. A drop of water with asci was run down the side of YE5S plates and left to dry at room temperature. Thereafter plates were incubated at 37° for 4–5 hr until the asci walls were nearly disrupted. During this process, the lid of the plate was kept partially open to dry the agar surface to facilitate the picking of the spores. Spores from tetrads, triads, or dyads were individually picked and sequestered with a micromanipulator. YE5S plates were then incubated at 30° until colonies were visible. The colonies were counted to estimate the viabilities of spores from tetrads, triads, and dyads. Missegregation was assessed in mature asci by the presence of more than four spores, less than two spores, or asci in which there were clearly detectable aberrances in the DNA content according to DAPI staining and in the relative sizes of the spores.
Random spore analysis:
Approximately 1 × 105 cells from 3-day-old matings were suspended in 250 μl of water containing 1500 units of β-glucuronidase (Sigma) and were incubated overnight at 30° to digest vegetative cells. Spores were centrifuged for 20 sec at 13,000 rpm, the supernatant was aspirated, and cells were resuspended in 1 ml of dH2O. After spores were counted with a hemocytometer, 1000 spores were grown on YE5S or YE5S with 15 μm of thiamine plates for 2–3 days at 30° and the corresponding total number of colonies was counted.
Recombination assay:
A loop-full of 3-day-old matings was treated with β-glucuronidase to digest asci and kill vegetative cells. The liberated spores were then counted and plated (50,000 spores per plate) on selective media EMM −Leu −Ade −Ura +Phloxin B with or without thiamine and nonselective media EMM −Leu +Ade +Ura +Phloxin B with thiamine. Plates were incubated at 30° for 5 days. Light pink colonies representing haploid cells were counted, and dark pink colonies representing diploid cells were excluded from the counting. For detection of the reciprocal recombinatorial phenotype (ade6-M216 ura4-D18), spores were plated on low adenine (10 mg/liter) and 5-FOA (0.1%) EMM −Leu plates.
Analysis of GFP-AID expression during nmt promoter activation or repression:
To analyze the activation of nmt promoter, the following procedure was carried out. Cells were vegetatively grown in 4 ml EMM −Leu +thiamine at 30° to an OD of ∼1.0 and then centrifuged at 3000 rpm for 5 min. The supernatant was removed and the cell pellet washed with 1 ml of dH2O. Cells were centrifuged again and strains were mixed to start the cross. The cross was carried out on EMM low nitrogen plates in the absence of thiamine at 25°. To analyze the repression of nmt promoter by thiamine, cells were first vegetatively grown in the absence of thiamine and strains mixed together in water containing 15 μm thiamine to promote efficient nmt repression. Thereafter, the cross was performed on EMM low nitrogen plates containing 15 μm thiamine. Every hour, a loop-full of cross was fixed in 1 ml of cold 70% ethanol to track the changes of GFP signal strength. Cells were vortexed and stored at 4° until the last time point. Cells were spun at 13,000 rpm for 30 sec, the supernatant was removed, and the cell pellet was washed in 1 ml 50 mm sodium citrate buffer pH 7.0 and then harvested in 300 μl 50 mm sodium citrate buffer pH 7.0. Samples were analyzed using a Beckton-Dickinson FACScan, counting a total of 50,000 cells per sample.
Nocodazole-induced block during meiosis:
Cells were grown vegetatively and crossed in the presence of thiamine for 24 hr to promote the fusion of the cells from opposite mating types. Thereafter cells were treated with nocodazole (final concentration 300 μm) for 24 hr to finish DNA replication of the fused cells and to block other cells from entering meiosis. Cells were then washed extensively to remove thiamine, but continuously treated with nocodazole, while inducing AID expression for 24 hr. GFP-AID induction was confirmed by flow cytometry (data not shown). Nocodazole was then removed to induce spore formation while inhibiting AID expression with the addition of thiamine. Spores were liberated by β-glucuronidase treatment and plated on EMM media with thiamine lacking adenine, uracil, and leucine to assess recombinants or EMM with thiamine lacking leucine to assess the overall numbers of live spores.
Worm strains and culture conditions:
C. elegans strains were cultured as described previously (Brenner 1974). The strains wild-type Bristol N2 and spo-11 (ok79) (Dernburg et al. 1998) were kindly provided by the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN). RNAi for ung-1 (Y56A3A.29a) was performed for 24 hr by feeding, as described previously (Fire et al. 1998; Boulton et al. 2002; Dengg et al. 2006). The ung-1 strain was kindly provided by Hilde Nilsen (University of Oslo) (Dengg et al. 2006).
Worm viability analysis and hermaphrodite/male identification:
Worm viability was determined by counting viable progeny generated by individual hermaphrodites. The percentage of male progeny was determined by sex-specific counting. Statistical analysis of worm viability was done using chi-square analysis, with the following modification: in the wild-type worm we arbitrarily set the count of dead eggs to two, even though we did not observe any. This alteration was to facilitate statistical analysis (in effect being more conservative than the observed data).
Irradiation of worms:
Irradiation of nematodes was performed in a 137Cs Irradiator (IBL437, Cis Bio International) with low-dose (15 Gy) irradiation. Animals were analyzed for TUNEL staining 30 min postirradiation and for RAD-51 staining 4 hr postirradiation.
Generation of human AID-expressing worm strain:
Human AID sequence was cloned into the Gateway System (Invitrogen, Carlsbad, CA) entry vector p221, followed by cloning into pID3 vector, generating an AID∷GFP fusion protein under the control of the pie-1 promoter (Tenenhaus et al. 1998). The AID∷GFP transgenic worm line was generated by microparticle bombardment of unc-119 (ed3) animals (Praitis et al. 2001) with pID3_AID∷GFP. spo-11 AID∷GFP worms were generated by crossing unc-119 AID∷GFP worms with a GFP balanced spo-11 strain. The genotype of the worms was verified by single-worm PCR, using primers for AID and spo-11 as described below.
Single-worm PCR:
Five microliters of lysis buffer (50 mm KCl, 10 mm Tris pH 8.3, 2.5 mm MgCl2, 0.45% IGEPAL CA 630, 0.45% Tween-20, 0.01% gelatin) with proteinase K (1 mg/ml) were placed into 0.2 ml DNase- and RNase-free PCR tubes or a 96-well PCR plate. Single worms were washed in a drop of PBS and then transferred into lysis buffer and frozen at −80° or on dry ice for at least 20 min. Tubes were then incubated at 65° for 90 min to lyse the worms. Proteinase K was inactivated by heating the tubes to 95° for 15 min. Thereafter, a nested PCR reaction was performed. For the first PCR reaction, 1 μl of worm lysate was added to a 12.5-μl PCR reaction that included the primers at 0.5 μm, 1× buffer, 1 mm MgCl2, 0.2 mm dNTPs and 1.25 unit of Platinum Taq DNA polymerase. The second PCR reaction was performed using 1 μl of the first PCR reaction in a 12.5-μl PCR reaction mixture as indicated for the first reaction. AID-GFP fusion construct was detected by nested PCR, using external primer sequences 5′ TAGACCCTGGCCGCTGCTACC 3′ and 5′ CAAAAGGATGCGCCGAAGCTGTCTGGAG 3′ and nested primers 5′ GAGGCAAGAAGACACTCTGG 3′ and 5′ GTGACATTCCTGGAAGTTGC 3′. spo-11 deletion was detected with nested PCR using external primers 5′ CGTGTTTCCCAAGATGCTC 3′ and 5′ CGGAATGCGTGCAAGTG 3′ and nested primers 5′ CCGAACAGCATATTGAAGAGG 3′ and 5′ GCGCATATAAAACACGGAGAC 3′, as previously described (Dernburg et al. 1998).
Visualization of GFP in C. elegans:
Molten 4% agarose was placed on prewarmed (55°) slides with a final thickness of ∼1 mm. The agarose was left to solidify before five worms were placed on the agar surface in 10 μl of PBS. Slides were left to dry for 5 min before covering with a coverslip and analyzed using a Deltavision microscope (Applied Precision).
Cytological preparation and immunostaining of C. elegans:
Gravid hermaphrodites were transferred to 30 μl of phosphate-buffered saline (PBS) on poly-l-lysine-coated slides to remove bacteria from worm bodies. Thereafter, worms were transferred to 50 μl of 10 mm levamisole. Germlines were extruded by removing the head and tail with a fine needle (27 gauge). Levamisole was replaced with 1% paraformaldehyde in PBS for 10 min. After fixation, the germlines were permeabilized for 5 min in Tris-buffered saline, 0.5% bovine serum albumin, and 0.1% Triton X-100 (TBSBT) and washed with TBSBT at least three times for 5 min each time, followed by blocking in TBSBT for 30 min. Primary antibodies were diluted in TBSB [1:50 for both 4.18.1 and 4.26.1 (Aoufouchi et al. 2008) anti-AID, 1:200 for anti-ung (ab13668; Abcam, Cambridge, UK), 1:200 for anti-RAD-51 antibody (Martin et al. 2005)] and incubated overnight at 4° in a humid chamber. Germlines were subsequently washed at least three times for 5 min each time with TBSB before incubation with secondary antibodies conjugated with Cy3 [anti-rabbit, 1:10,000 (Sigma, United Kingdom); anti-mouse, 1:10,000 (Sigma)] for 1–2 hr at room temperature. Finally, the germlines were washed at least three times with TBSB for 5 min before adding ProLong Gold Antifade with DAPI (Molecular Probes) and mounted with coverslips.
TUNEL assay:
The terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) assay was carried out as described before (Parusel et al. 2006), with minor modifications. For the assay, gravid nematodes were washed in PBS on poly-l-lysine-covered slides and dissected in PBS, followed by fixation in 4% formaldehyde in PBS at room temperature (RT) for 20 min. Gonads were rinsed in PBS, 0.4% Triton X-100 (PTX) three times, incubated in 100 mm sodium citrate, 0.1% Triton X-100 at 65° for 20 min, and then washed twice in PTX. Gonads were incubated for 30 min at RT in 0.1 m Tris-HCl (pH 7.5) containing 3% BSA and 20% bovine serum and washed twice with PBS at RT, and excess fluid was drained off. Thereafter, the gonads were incubated in TUNEL reaction mixture (Roche, Indianapolis) at 37° for 1.5 hr in the dark. The reaction was stopped by rinsing three times in PTX, before adding ProLong Gold Antifade with DAPI (Molecular Probes, United Kingdom) and mounting the samples with coverslips.
Fluorescence microscopy:
Delta-vision microscopy was used to examine germlines with a ×40 or a ×63 1.4-NA Planapochromat lens on an Olympus inverted microscope (IX71), and images were captured using SoftWorx computer software (Applied Precision). Three-dimensional data sets were computationally deconvolved if needed, and regions of interest were then projected into one dimension. Merged or single-color images were recorded using GIMP software.
RESULTS
To determine how dU:dG mismatches can influence recombination and chromosome segregation during meiosis, we expressed DNA deaminases in two genetically tractable organisms: S. pombe and C. elegans. In both organisms, DNA deaminase activity was analyzed on a Spo11 (Rec12)-deficient background, which produced low but sufficient offspring for analysis. Since the AID/APOBEC3 DNA deaminase family has been identified only in vertebrates (Conticello et al. 2005), it is highly unlikely that either organism used in this study possesses such enzymes in its germline, providing precisely controlled systems to delineate the effect of cytosine to uracil deamination in meiotic nuclei.
Induction of base mismatches rescues tetrad formation in S. pombe rec12Δ:
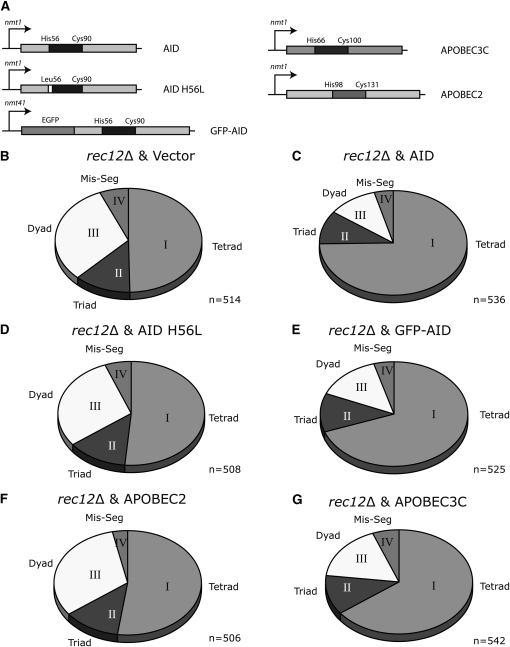
As previously published (De Veaux et al. 1992), rec12Δ mutants are deficient in tetrad formation (Figure 1B); asci analysis showed that only 50% formed tetrads (94% in wild type), with an increase to 30% dyads (1–2% dyads wild type, supporting information, Table S1), indicative of impaired meiotic recombination. Expressing AID under the thiamine repressor promoter nmt (Figure 1A) during meiosis in the rec12Δ strain significantly rescued tetrad formation with a concomitant decrease in dyad formation (Figure 1C). Tetrad formation was increased from 50 to 73%, while dyad formation decreased from 30 to 12%. Strains repressed for AID expression by addition of thiamine did not show any rescue (Table S1). To determine if the catalytic activity of AID is required for rec12Δ rescue, we generated a catalytically inactive mutant of AID, by mutating the zinc coordinating amino acid histidine 56 (H56L) (Papavasiliou and Schatz 2002; Chaudhuri et al. 2003; Pham et al. 2003; Morgan et al. 2004) (Figure 1A). Expression of AID H56L in the rec12Δ background failed to rescue the deficiency in tetrad formation (Figure 1D), even though similar levels of H56L and wild-type AID protein were produced (data not shown). To visually follow protein expression, we also generated a GFP-AID fusion protein (Figure 1A), which induced a similar phenotype to the untagged AID (Figure 1E and Table S1).
Figure 1.—
DNA deaminases partially restore tetrad formation in rec12Δ S. pombe. (A) Schematic representation of the constructs used in this study. Proteins were expressed under the control of thiamine-repressible nmt1 or nmt41 promoter. Expression is driven in the absence of thiamine. Amino acids essential for the formation of the catalytic domain (blue) are indicated as well as the substitution of His56 with Leu56 in human AID (AID), which renders the protein catalytically inactive. (B–G) Relative distribution of tetrads (I), triads (II), dyads (III), and missegregations (IV) that are formed during meiosis in a rec12Δ background transfected with an empty vector (B), with human AID (C), with a catalytically mutant human AID (D), with GFP-human AID (E), with human APOBEC2 (F), and with human APOBEC3C (G). Cells were transfected, grown in the presence of thiamine for 24 hr, washed, grown in the absence of thiamine for 6 hr, and crossed for 2 days on thiamine (−) plates. Tetrads, triads, dyads, and missegregations were quantitated by microscopy.
AID expression in rec12Δ increases spore viability:
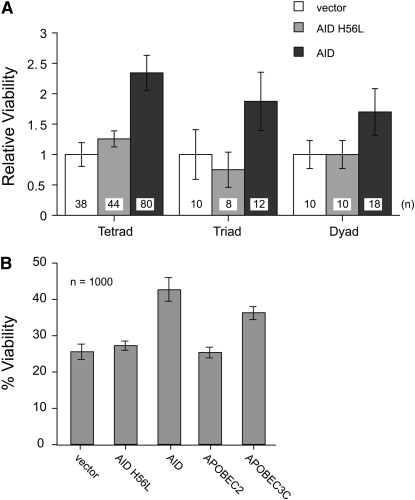
The rescue of spo11 mutant phenotypes by gamma radiation can be at the expense of overall cell viability (Thorne and Byers 1993). Therefore to confirm that improved tetrad formation is not due to excessive amounts of AID-induced DNA damage, which could impair survival, we directly analyzed spore viability by dissecting tetrads, triads, and dyads into individual spores and plated them for growth. The average spore viability of a tetrad of rec12Δ was raised from 22 to 51% after AID expression (Figure S1). This is shown as a more than twofold increase in Figure 2A. We also observed a significant increase in viability when spores were picked from dyads or triads (Figure 2A) or when we performed random spore analysis from cultures (Figure 2B). As in the tetrad formation assay, the catalytic inactive AID mutant was unable to rescue the viability. We noted a slight effect on cell numbers during vegetative growth after AID expression, but the rate of doubling was not markedly changed (data not shown).
Figure 2.—
Deaminases increase spore viability after meiosis. (A) Ascus dissection analysis: relative viabilities of rec12Δ single spores from tetrads, triads, and dyads in control-transfected cells (white), in AID catalytic mutant expressing cells (orange), and in catalytically active AID expressing cells (red). The numbers of the spores that were isolated are indicated at the bottom of each column, and the viability of the vector-only rec12Δ strain was set to one. (B) Percentage of viability of random spore analysis, from rec12Δ cells expressing different deaminases during meiosis.
AID induces meiotic recombination:
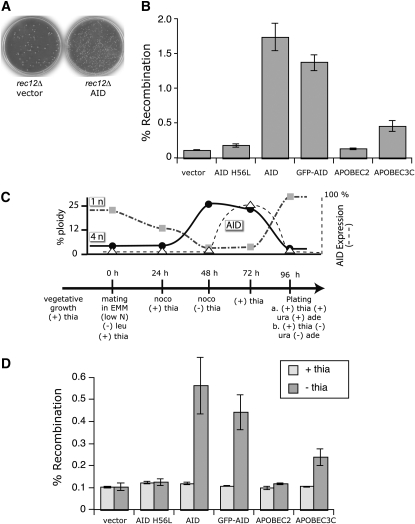
Double-strand breaks generated by Spo11/Rec12 induce interhomolog recombination, which can promote proper chromosome segregation through the establishment of chiasmata (Bahler et al. 1993; Gerton and Hawley 2005; Whitby 2005). To see whether upon AID expression the formation of dU could lead to meiotic crossovers, we analyzed the recombination frequency between ade6-M216 and ura4-D18, two auxotrophic markers (>50 cM apart) on chromosome III. Meiotic recombination frequency between these two marker loci was enhanced 15-fold after AID expression, from 0.11 to 1.7% (Figure 3 and Table S2). Again, this was dependent on the expression of AID (no recombination was detected in the presence of thiamine; Figure 3D) and its catalytic activity (AID H56L did not significantly increase recombination; Figure 3, B and D). The effect was also unlikely due to an enhanced number of diploid cells after meiotic recombination, as rec12Δ cells with vector alone had ∼2.1% diploid cells (13 red/615 total) and rec12Δ cells with AID produced 0.32% diploid cells (18 red/5640 total). At the same time, screening recombinant cells on low adenine and 5-fluoroorotic acid (5-FOA), we could determine that reciprocal crossovers (double mutant of ade6-M216 and ura4-D18) were also generated. Again, AID expression enhanced crossovers by >10-fold, since vector alone produced 0.20% and AID 2.3% recombinants.
Figure 3.—
Meiotic recombination initiated by AID. (A) Representative plates showing the increase in recombinants after meiosis of rec12Δ cells transfected with empty vector (left plate) or with AID (right plate). Cells auxotrophic for either adenine (ade6) or uracil (ura4) were crossed on EMM low N and spores were plated on EMM media supplemented with phloxin B and in the absence or the presence of both adenine and uracil, to assess the relative number of recombinants compared to the total number of viable spores. Shown are the EMM plates lacking adenine and uracil. (B) Fold increase in ade6 ura4 recombinants after the expression of AID catalytic mutant, functional AID, GFP-AID, APOBEC2, or APOBEC3C during meiosis compared to the empty vector (set to one) control. (C) Time line of postreplicative induction of AID-induced recombination. Time points represent the relative timing of changing the media or the addition of the indicated compounds nocodazole (noco), leucine (leu), and thiamine (thia); + and − indicate the presence or the absence of the compounds starting from the corresponding time point. Cell ploidy, visualized with propidium iodide in FACS, is shown as a solid line (solid circles, 4n) and a shaded dashed-dotted line (shaded squares, 1n), and AID expression, visualized by GFP in FACS, is shown as a dashed line (based on Figure S2, open triangle). (D) DNA deaminase-mediated recombination events can be initiated after DNA replication. Cells were treated as indicated in the time line (C). The graph shows the recombinants obtained when AID, mutant (H56L) AID, and the APOBEC proteins were expressed after removal of thiamine (− thia) at a 48-hr time point compared to the results when deaminase expression is repressed during meiosis by thiamine (+ thia). For details see materials and methods.
It was also possible that AID-induced lesions that conferred meiotic rescue were DSBs resulting only from replication fork collapse in premeiotic S phases. To exclude this possibility, we used nocodazole (known to depolymerize microtubules) to stall cells after DNA replication and inhibit them from going through meiosis. Treatment was monitored by FACS analysis (Figure S2) for propidium-iodide (DNA content) and GFP staining (AID expression). Yeast were grown in the presence of thiamine (see time line in Figure 3C) and induced to mate at time 0 hr; after 24 hr nocodazole was added, and a further 24 hr later AID was expressed by removing thiamine (expression was monitored via GFP-AID; see FACS data in Figure S2) while the cells were still in nocodazole; by 72 hr postmating initiation cells were released from the nocodazole block and AID expression was inhibited with thiamine, and a further 24 hr later they were plated to score for meiotic recombination. As shown in Figure 3D, we were still able to identify a significant increase in the frequency of meiotic recombination between the two loci if AID was induced after the nocodazole block. Again, this effect required both AID expression and AID catalysis (Figure 3D). Although we cannot fully exclude the possibility that the AID-induced rescue was solely due to collapsed replication forks, we think this is unlikely on the basis of the significant proportion of postreplication arrested cells that underwent meiotic division after AID induction (see discussion for details).
APOBEC3C can rescue rec12Δ meiosis and initiate recombination:
In its physiological environment, cytoplasmic AID is known to enter and function in the nucleus, whereas the APOBEC3 family members (except for APOBEC3B) are usually confined to the cytoplasm (Mangeat et al. 2003; Marin et al. 2003). This subcellular restriction may be a means of regulating the function of DNA deaminases (Coker and Petersen-Mahrt 2007). On the other hand, expression of cytoplasmic APOBEC3G in yeast was able to inhibit the movement of retrotransposable elements, by a mechanism involving APOBEC3G possibly entering the nucleus (Dutko et al. 2005; Schumacher et al. 2005; Esnault et al. 2006). We therefore wanted to know if other DNA deaminases could be used to induce dU:dG mismatches during meiosis, leading to rescue of the rec12Δ phenotype as well as homologous recombination (HR).
We constructed expression vectors of APOBEC3C [a DNA deaminase known to inhibit SIV and Line1 retrotransposons (Harris et al. 2002; Yu et al. 2004; Dutko et al. 2005; Muckenfuss et al. 2006)] and APOBEC2 [a DNA deaminase family member with no apparent catalytic activity toward polynucleotides (Harris et al. 2002; Jarmuz et al. 2002; Conticello et al. 2005; Mikl et al. 2005)]. Expression of APOBEC2 failed to rescue tetrad formation (Figure 1F), increase viability (Figure 2B), or increase meiotic recombination (Table S2). Analogous to AID, expression of APOBEC3C did increase tetrad formation (Figure 1G) and viability (Figure 2B), as well as meiotic recombination (Table S2), indicating that dU from other DNA deaminases can also induce meiotic recombination events.
AID expression in C. elegans spo-11 rescues RAD-51 focus formation and bivalency:
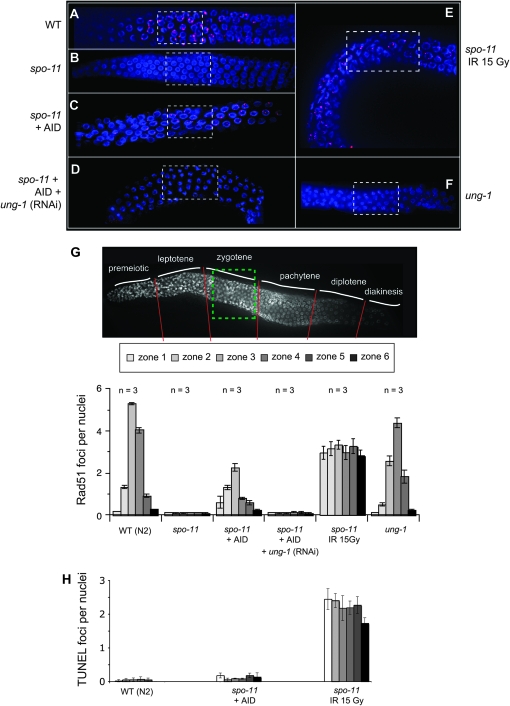
Genetic analysis in S. pombe has revealed that ectopic AID expression during meiosis is capable of inducing meiotic recombination in a single-cell organism. To extend this analysis to a multicellular organism we employed C. elegans as a model system. Here, the spatial and temporal arrangement of the worm's germline allows for a visual determination of the various stages of meiosis [e.g., DNA replication is limited to the distal tips: the premeiotic zone (Wood 1988)]. Furthermore, in C. elegans, meiotic recombination is limited to a single crossover event per homologous chromosome pair (Villeneuve 1994). At the molecular level, spo-11 mutants are devoid of meiotic RAD-51 foci and fail to generate the obligate crossover per homolog pair; cytologically, this manifests as chromosome univalency at diakinesis (Dernburg et al. 1998). We generated worms expressing AID via the pie-1 promoter, a gene that is expressed throughout meiotic prophase (Tenenhaus et al. 1998). We used RAD-51 focus formation as a measure for meiotic recombination initiation (Figure 4). Wild-type worms exhibit the normal distribution of Spo11-induced meiotic RAD-51 foci (Figure 4, A and G), with the majority of foci present in late leptotene to pachytene. As expected, the spo-11 mutants are devoid of RAD-51 foci (Figure 4B). Analogous to our observations in fission yeast, ectopic germline expression of AID partially rescues the spo-11 deficiency, as seen by the presence of meiotic RAD-51 foci (Figure 4C). The distribution of foci was broader and less numerous than in the wild-type organism, but interestingly was most abundant near the leptotene/zygotene zone (Figure 4G). The absence of AID-induced RAD51 lesions in the premeiotic zone would indicate that the dU lesions are induced postreplication.
Figure 4.—
AID induces RAD-51 foci during meiosis in the spo-11 worm. Representative pictures of germlines visualized for RAD-51 foci, using anti-RAD-51 antibodies (red) as well as DAPI staining to visualize the nuclei, are shown: (A) wild-type animals, (B) spo-11, (C) spo-11 worm expressing AID, (D) spo-11 worm expressing AID with RNAi treatment for ung-1, (E) spo-11 worm irradiated with 15 Gy and analyzed after 4 hr, and (F) ung-1 defective nematode. Dashed squares represent the proposed location of zygotene stage in the germline. The germlines were divided into six zones according to the developmental stage of the germline. (G) The number of RAD-51 foci per nucleus was counted in worm germlines with respective genotypes (A–F) in the six developmental zones (30 nuclei per zone per worm, analyzing three worms per genotype). (H) The number of TUNEL stains per nucleus in the six developmental zones was counted from the germline of the indicated worms (for details see Figure S3). The fold difference between spo-11 (IR 15 Gy) and spo-11 + AID was >20-fold and >50-fold for the wt.
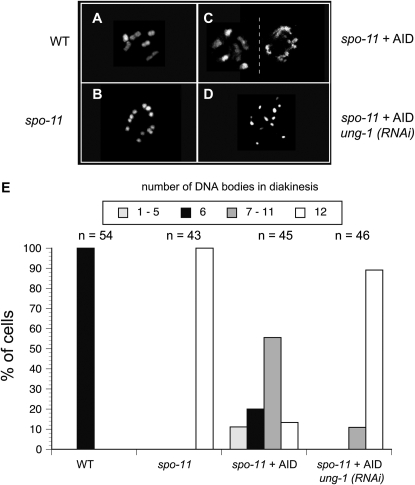
In wild-type animals, 6 bivalents are detectable by DAPI staining in oocyte nuclei arrested at diakinesis (Figure 5A). Each bivalent represents a condensed homologous chromosome pair held together by a single chiasma (the physical manifestation of crossing over). In a spo-11 worm, there was no manifestation of bivalency, leading to the formation of 12 univalent chromosomes (Figure 5B). In contrast, AID-induced mismatches in the spo-11 mutant were correctly processed to induce meiotic recombination leading to a substantial recovery of bivalency (Figure 5, C and E).
Figure 5.—
AID alters the chromosome morphology during the diakinesis stage. Representative pictures are shown of DAPI-stained chromosomes during diakinesis from (A) wild type, (B) spo-11, (C) two nuclei of an AID-expressing spo-11 worm, and (D) an AID-expressing spo-11 worm after treatment with ung-1 (RNAi). (E) The numbers of DAPI-staining bodies per nucleus were counted for each of the corresponding genotypes from the indicated number of cells. Different shadings for the columns represent the numbers of DAPI-stained bodies as indicated on the top of the graph.
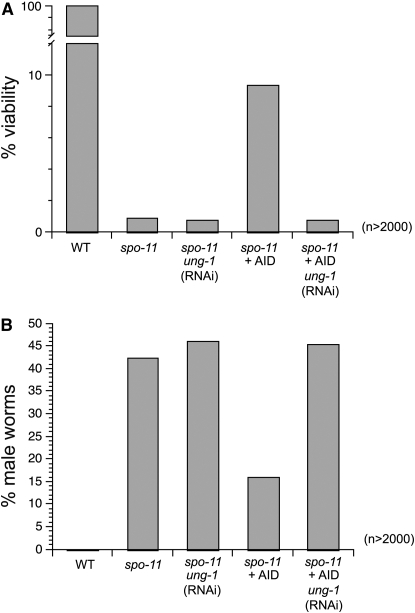
AID increases viability and decreases males in spo-11 worms:
While RAD-51 focus formation and bivalency are a good measure of some of the molecular events taking place during meiosis, we wanted to know if AID could also increase the viability of the spo-11 mutant worms. Due to aberrant chromosome segregation, spo-11 worms usually produce <1% viable offspring when compared with wild-type worms (Figure 6A and Table 1). The elevation in chromosome nondysjunction also manifests as an increased frequency of male offspring. The frequency of males is significantly increased from ∼0.1% in wild type to >40% in spo-11 mutants (Figure 6B and Table 1) (Dernburg et al. 1998). As seen with S. pombe, where AID increased tetrad formation as well as the viability, C. elegans spo-11 expressing AID exhibit a >10-fold increase in viable brood size (Figure 6A and Table 1) and a 2.5-fold reduction in males (Figure 6B and Table 1). This suggests that the induction of meiotic recombination in spo-11 mutants by ectopic expression of AID is sufficient to confer increased viability to progeny.
Figure 6.—
AID increases viability and hermaphrodite frequency in spo-11 worms. (A) Worm viabilities for the indicated genotypes were assessed by counting viable offspring and dead eggs in a brood from young adults. (B) The frequencies of males among viable progeny from the sample in A were assessed by microscopic analysis of tail morphology and movement. A total of at least 2000 worms were counted per genotype (for extended data see Table 1).
TABLE 1.
Brood viability and male frequency
| Strain | Dead eggs | Live progeny | Total counts | % lethality | % viability | Male | Hermaph. | % male | % hermaph. |
|---|---|---|---|---|---|---|---|---|---|
| wt | 0 | 4333 | 4333 | 0.0 | 100.0 | 0 | 4333 | 0.0 | 100.0 |
| spo-11 | 4797 | 42 | 4839 | 99.1 | 0.9 | 151 | 208 | 42.1 | 57.9 |
| wt + AID | 17* | 2043 | 2060 | 0.8 | 99.2 | 2 | 2041 | 0.1 | 99.9 |
| spo-11 + AID | 4401 | 440 | 4841 | 90.9 | 9.1*** | 64 | 376 | 14.5*** | 85.5 |
| spo-11 + AID si-UNG | 2267 | 13 | 2280 | 99.4 | 0.6***** | 80 | 97 | 45.2***** | 54.8 |
| wt si-UNG | 5 | 2027 | 2032 | 0.3 | 99.8 | 2 | 2025 | 0.1 | 99.9 |
| spo-11 si-UNG | 4853 | 25** | 4878 | 99.5 | 0.5 | 159 | 184 | 46.4 | 53.6 |
| wt + AID si-UNG | 59 | 1962 | 2021 | 2.9**** | 97.1 | 37 | 1925 | 1.9**** | 98.1 |
| ung-1 | 12* | 2134 | 2146 | 0.6 | 99.4 | 9* | 2125 | 0.4 | 99.6 |
| wt IR 15 Gy | 134 | 2119 | 2253 | 6.0 | 94.1 | 5 | 2114 | 0.2 | 99.8 |
| spo-11 IR 15 Gy | 2291 | 25 | 2316 | 98.9 | 1.1 | 96 | 163 | 37.1 | 63.9 |
Viability (% viability) is based on the number of viable offspring/(dead eggs + viable offspring); % lethality = 1/viability; male frequency (% male) is based on the numbers of animals with a male phenotype (hook, slim torso, more active movement) among at least 170 viable offspring; hermaphrodite frequency (% hermaph.) = 1/% male; wt, N2 or unc119; si-UNG, siRNA UNG-treated worms; IR 15 Gy, 15 Gy of ionizing radiation; *P < 0.01 (to wt); **P < 0.05 (to spo-11); ***P < 0.001 (to spo-11); ****P < 0.01 (to wt si-UNG); *****P < 0.01 (to spo-11 + AID).
AID-induced rescue of spo-11 is dependent on Ung:
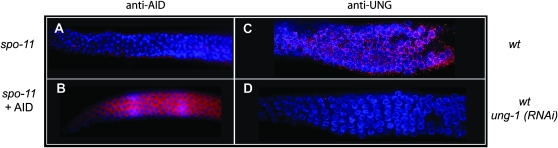
To begin to determine which of the above mentioned DNA repair pathways may be involved in processing the AID-induced dU:dG lesion, we used RNAi to inactivate UNG-1, the single uracil DNA glycosylase encoded in the C. elegans genome (Dengg et al. 2006), in AID-expressing spo-11 worms. RNAi depletion of UNG-1 completely abolished AID-induced RAD-51 focus formation in spo-11 mutants (Figure 4, D and G). This indicates that AID-induced dU lesions are predominantly processed through the BER pathway to produce abasic sites, and that these lesions are sufficient to generate HR substrates in meiosis. As indicated in Figure 4C, the RAD-51 foci in AID-expressing worms peak at the zygotene stage. The density of RAD-51 foci in this region could be due to limited expression of AID, limited expression of BER proteins such as UNG, or other novel aspects of the meiotic recombination pathway. Immunohistochemical analysis of AID expression indicated that AID was expressed during the mitotic stage and throughout meiosis (Figure 7, A and B), as would be expected from the pie-1 promoter. Using an UNG-specific antibody, we were able to demonstrate expression of endogenous UNG during oogenesis in a multicellular organism (Figure 7, C and D), highlighting the potential role for BER in meiosis after DNA replication. Although the distribution of UNG appeared partially overlapping but different from AID expression, there is no clear indication that the AID-induced RAD-51 foci are formed solely due to a limited localized expression of UNG.
Figure 7.—
Expression of AID and Ung during oogenesis. AID was detected with anti-AID antibodies (red) in (A) a spo-11 worm and (B) an AID-expressing spo-11 worm. UNG was detected with anti-Ung antibody (red) in (C) a wild-type worm germline and (D) a wild-type worm treated with ung-1 (RNAi). Nuclei were counterstained with DAPI.
As with the RAD-51 focus formation, AID-induced bivalency was dependent on the BER protein UNG (Figure 5, D and E), since RNAi of ung-1 reverted the AID-expressing nematode back to univalent chromosomes. Furthermore, the AID-induced increase in viability is fully dependent on the expression of UNG-1 during meiosis, as is the decrease in male formation (Figure 6 and Table 1).
To what extent BER proteins play a role in meiosis is still to be determined, but analysis of ung-1 (RNAi)-treated wild-type (wt) worms showed a minor but significant increase in dead eggs and male formation (Table 1). Extending this analysis to the ung-1 worm (Dengg et al. 2006), we observed an even larger and statistically significant increase in dead eggs and male formation. Furthermore, in a SPO-11-deficient background siRNA of ung-1 decreased the viability even further (Table 1). Together this indicates that BER proteins can play a role in meiosis.
AID- and gamma radiation-induced rescue proceed differently:
In previous studies, ionizing radiation has been used to artificially induce DNA damage in spo-11, mimicking some of the functions of Spo11 (Dernburg et al. 1998). Gamma radiation-induced RAD-51 focus formation in spo-11 worms is distributed randomly throughout meiotic prophase (Figure 4E), which is in contrast to the AID-induced lesions, which peaked around zygotene (Figure 4G). As stated above, only under certain circumstances can AID-induced lesions become DSBs. To directly observe the formation of DSBs as well as accessible DNA ends, we employed a previously published TUNEL assay (Parusel et al. 2006). Although we could observe a large number of TUNEL positive cells after gamma radiation treatment (sublethal dose), we did not detect foci in the AID-expressing worms (Figure 4H and Figure S3). This demonstrated that AID did not induce a large number of DSBs or accessible DNA ends, unlike the ionizing radiation treatment (see discussion).
DISCUSSION
DNA lesions derived from chemical or physical damage need to be removed by DNA repair mechanisms to maintain the integrity of the genome. One of the most common single-base-altering lesions in nonreplicative DNA is deaminated cytosine, resulting in the formation of a dU:dG mismatch. It has been hypothesized that DNA lesions other than Spo11-induced DSBs may initiate meiotic recombination (Farah et al. 2005; Cromie et al. 2006). What, therefore, is the effect of a dU:dG mismatch during meiosis, or, more precisely, (1) Can DNA deaminases such as AID function in a meiotic nucleus?, (2) Is the dU:dG product recognized by DNA repair machinery and processed in a meiotic nucleus?, (3) Can processing of the lesion affect meiosis?, and (4) Do the involved processing pathways lead to meiotic recombination? Expression of DNA deaminases in the rec12Δ strains of S. pombe leads to a rescue of tetrad formation (Figure 1), increased spore viability (Figure 2), and increased meiotic recombination (Figure 3). More importantly, the partial rescue of C. elegans spo-11 by AID demonstrates that in multicellular organisms the meiotic nucleus is also capable of processing a dU lesion for recombination. The formation of RAD-51 foci near the leptotene/zygotene zone in the worm clearly indicates that AID-induced dU lesions are processed by the postreplicative meiotic nucleus to HR substrates (Figure 4). Furthermore, the increased formation of bivalents (Figure 5), viable offspring (Figure 6A), and decrease in males (Figure 6B) demonstrate that AID-induced base mismatches can give rise to crossovers that are capable of guiding chromosomal segregation at the first meiotic division.
Similar to the AID-induced diversification of the acquired immune system (via mutation and recombination), meiosis does induce DNA recombination and potentially mutations. Importantly, in both instances, external selection forces drive the survival of the altered genome. How these alterations are achieved is dependent on the way DNA lesions are processed by the different DNA repair pathways. In the immunoglobulin locus different DNA repair pathways can process the dU:dG lesion, leading to various phases of SHM, iGC, and CSR (Petersen-Mahrt et al. 2002; Neuberger et al. 2003, 2005; Chaudhuri and Alt 2004; Maizels 2005). The outcome of a dU:dG mismatch leading to class switching relies on BER (UNG) as well as MMR (Msh2/Msh6, MLH1-PMS2, and MSH5) proteins to initiate the recombination (Rada et al. 2004; Larson et al. 2005; Sekine et al. 2007) and NHEJ (Ku70/Ku80 DNA PKs) to complete the process (Manis et al. 1998; Rooney et al. 2004). Both mouse knockout and yeast genetic studies have implicated MMR proteins to play a critical role in meiosis (Hunter et al. 1996), which include the Msh4–Msh5, Mlh1–Pms2, and Mlh1–Mlh3 complexes (Prolla et al. 1998; Lipkin et al. 2002; Kunz and Schar 2004; Snowden et al. 2004). Regardless of which of the DNA repair pathways is initiated by the dU lesion, the potential of using AID as a tool will allow us to readdress if and how HR can proceed from a single-stranded nick (see references in Smith 2004). This could provide analogous analysis of how the RAG proteins have been used to study single-stranded nicks as HR substrates in mammalian cells (Lee et al. 2004).
There are of course other possible pathways that could induce the formation of HR substrates (RAD-51 foci) from a dU lesion. In worms BER seems to be an early necessary step. This also seems to be the case in S. pombe, as the work by the group of Primo Schär indicates, where removal of the BER protein TDG inhibited the rescue by AID (P. Schär, personal communication). It still needs to be determined how the abasic site and subsequent processing steps can lead to the formation of HR substrates. In line with this, we observed that missegregations did not decrease significantly upon AID induction whereas dyads did. Improper dyad formation is possibly due to a defect in meiosis I, whereas missegregation can be derived from defects in meiosis I and II. Therefore, where AID is expressed and during which cell stage will determine if the lesion can “relieve” the chromosomal abnormality.
In both organisms AID induction led to significant rescue of the SPO-11 deficiency, which is, in light of the different lesions generated (AID, a dU, and SPO-11, a DSB), quite remarkable. That we did not detect a complete rescue is perhaps not surprising, as SPO-11 may have auxiliary functions during meiosis, and the different lesions may have been processed to different extents. In the worm, we detected RAD-51 foci after AID induction, albeit at a slightly reduced level (Figure 4G), yet the viability and male rescue were not complete nor to the same extent to which the RAD-51 foci would have indicated (Figures 4 and 6). As each chromosome pair has to acquire at least one RAD-51 focus, AID-induced lesions may not have allowed for sufficient HR substrate formation; this in turn may also have contributed to the formation of irregular bivalency numbers (Figure 5E). Future work will be able to delineate if this difference is due to a lack of auxiliary function of AID or a difference in lesion processing. It also remains to be determined if the zygotene-limited AID-induced RAD-51 foci are due to a lack of ssDNA access for AID in the premeiotic phase, lack of processing of the dU in the premeiotic phase, or a difference in DNA repair between the premeiotic and meiotic phases, even though UNG-1 appears to be present in both stages (Figure 7). It is possible that the dU lesions are repaired during DNA replication without leading to DSBs, whereas postreplicative dU lesions are preferentially processed into HR substrates. The latter possibility is analogous to recent work on B cells, where inactivation of error-free repair is required for proper SHM to proceed (Arakawa et al. 2006; Simpson et al. 2006).
It is important to note that ectopic germline expression of AID did not induce RAD-51 foci in the premeiotic zone of the germline (Figure 4), which contains the only actively proliferating cells in the germline. This would suggest that the majority of RAD-51 foci induced by AID in spo-11 mutants occur in postreplicative cells. Our work in S. pombe (Figure 3D) is possibly in agreement with an AID-induced meiotic recombination rescue independent of DNA replication. This is based on (Figure 3, C and D, and Figure S2) the following: (1) the total number of 1n (ploidy) cells after nocodazole release (96 hr) was 60% of the 4n cells during AID expression, suggesting that the majority of the spores were derived from the 4n cells; (2) there is a reciprocal relationship of simultaneous loss of 4n cells and gain of 1n cells; (3) after 48 hr there are <5% of 1n cells remaining (cells that could have undergone fusion after nocodazole release and therefore could be a source of replication-dependent AID damage); and (4) a significant portion of the 2n peak is derived from vegetative cells, which were removed with glucuronidase prior to plating.
It is interesting to note that in the yeast rescue experiments, there appears to be a numerical discrepancy between the rescue of tetrad formation/viability and meiotic crossovers. Spore viability in a rec12 mutant is ∼22%, which is better than expected from a solely random process of chromosome segregation, and has been attributed to achiasmata segregation (Davis and Smith 2005). With AID, spore viability increases to ∼50%. However, the frequency of crossing over in the ade6–ura4 interval, while increasing 10-fold, is still only 1.7%. The ade6–ura4 interval is ∼10% of the overall genome size, and therefore if we assume that this interval is representative of crossing over induced by AID genomewide, then one might speculate that AID-induced lesions result in a crossover event in only 17% of meioses. This would seem to be insufficient to account for the improvement in spore viability. Again a possible explanation for this is that AID might induce distinct types of lesions (e.g., single-strand breaks and double-strand breaks), which could be differently disposed to give rise to crossover recombination, but are both capable of promoting correct chromosome segregation. Perhaps recombination acting at single-strand breaks aids homolog pairing and alignment without generating crossovers. Future studies using AID will allow us to investigate this possibility.
In B cells, the formation of DSBs from AID-induced lesions seems to be dependent on the genetic loci, the cell state, AID cofactors, and the chromatin status, as DSBs are limited to the highly repetitive switch regions during class switch recombination, but are less likely to occur during somatic hypermutation of the variable regions (Wuerffel et al. 1997; Petersen et al. 2001; Stavnezer et al. 2008). Because gamma radiation can be an alternative source of DSB, we could compare the potential AID-induced DSB with other DSBs. Using low dose (15 Gy) gamma-irradiated worms, we observed extensive TUNEL staining throughout the germline. Importantly, although gamma radiation has been shown to induce apoptosis, due to the cellular physiology of the nematode, apoptosis and genome fragmentation do not happen until late stages of meiosis (cellularization; zone 6 in Figure 4) (Lettre and Hengartner 2006). Although the TUNEL assay has a detection limit, it is not clear if this lack of signal is due to the infrequency of breaks or their transient nature, and therefore below the threshold of detection, or if the Spo11-induced DSBs are inaccessible. The latter is plausible, since not only has it been shown that Spo11 is temporarily covalently bound to the DSB during meiosis (Keeney et al. 1997), but also Spo11 can colocalize with the lesion to recruit downstream processing pathways, thereby possibly “hiding” the available DSB until other repair factors are present. From the TUNEL staining, there did not appear to be an excessive number of AID-induced DSBs or free ends during meiosis. This implies that, although base mismatches are processed into recombination events (indicated by RAD-51 foci and chromosome bivalency), the lesions seem to be different in their accessibility to TUNEL staining. Importantly, the lack of TUNEL staining and the enhanced efficiency of the rescued viability of AID-induced base mismatches over irradiated (IR) ones (Table 1) indicated that random DNA DSB generation is not an efficient or a viable means of rescue, and AID is unlikely to have induced such lesions in large numbers.
This study has for the first time demonstrated that DNA lesions other than DSBs (as has been suggested by Cromie et al. 2006), such as dU, abasic site, or ssDNA nicks, can initiate meiotic recombination in yeast and the multicellular organism C. elegans. This suggests an evolutionary conserved mechanism of DNA repair for dU lesions in the meiotic nucleus of organisms as diverse as fission yeast and the nematode. Neither of these organisms expresses members of the DNA deaminase family physiologically, indicating that the simple expression of such an enzyme in meiotic nuclei can lead to proper processing and initiation of meiotic recombination, paralleling the notion that the immune system's acquisition of SHM may have been a “simple” addition of DNA deaminases to already existing DNA repair processing pathways. In extension, because of AID's expression (as well as other DNA deaminases) in oocytes (Morgan et al. 2004) and during spermatogenesis (Schreck et al. 2006), one should not fully disregard the potential that protein-induced DNA deamination can affect meiotic recombination in vertebrates. This warrants a reevaluation of meiosis in AID-deficient mice, with respect to recombination, crossover, and chromosome segregation analysis.
Acknowledgments
We thank Hilde Nilsen for kindly providing us with the ung-1 C. elegans. We thank Heather Coker for helpful discussions and critical reading of the manuscript and Julie Cooper for advice and support. We thank Primo Schär for sharing unpublished data with us. S.P. was supported in part by SA Archimedes, Estonian Foundation of European Union Education and Research. M.C.W. and F.O. were supported by a Wellcome Trust Senior Research Fellowship to M.C.W. This work was supported by Cancer Research United Kingdom. The authors declare no conflict of interest.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101683/DC3.
References
- Aoufouchi, S., A. Faili, C. Zober, O. D'Orlando, S. Weller et al., 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa, H., G. L. Moldovan, H. Saribasak, N. N. Saribasak, S. Jentsch et al., 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4 e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., T. Wyler, J. Loidl and J. Kohli, 1993. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto, V., B. Reina-San-Martin, A. R. Ramiro, K. M. McBride and M. C. Nussenzweig, 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell 12 501–508. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson et al., 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295 127–131. [DOI] [PubMed] [Google Scholar]
- Bransteitter, R., P. Pham, M. D. Scharff and M. F. Goodman, 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, J., and F. W. Alt, 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4 541–552. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud et al., 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422 726–730. [DOI] [PubMed] [Google Scholar]
- Coker, H. A., and S. K. Petersen-Mahrt, 2007. The nuclear DNA deaminase AID functions distributively whereas cytoplasmic APOBEC3G has a processive mode of action. DNA Repair 6 235–243. [DOI] [PubMed] [Google Scholar]
- Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt and M. S. Neuberger, 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22 367–377. [DOI] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter et al., 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B. R., 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2005. Dynein promotes achiasmate segregation in Schizosaccharomyces pombe. Genetics 170 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengg, M., T. Garcia-Muse, S. G. Gill, N. Ashcroft, S. J. Boulton et al., 2006. Abrogation of the CLK-2 checkpoint leads to tolerance to base-excision repair intermediates. EMBO Rep. 7 1046–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94 387–398. [DOI] [PubMed] [Google Scholar]
- Di Noia, J., and M. S. Neuberger, 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419 43–48. [DOI] [PubMed] [Google Scholar]
- Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen and M. J. Curcio, 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault, C., J. Millet, O. Schwartz and T. Heidmann, 2006. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 34 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah, J. A., G. Cromie, L. Davis, W. W. Steiner and G. R. Smith, 2005. Activation of an alternative, rec12 (spo11)-independent pathway of fission yeast meiotic recombination in the absence of a DNA flap endonuclease. Genetics 171 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Gerton, J. L., and R. S. Hawley, 2005. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6 477–487. [DOI] [PubMed] [Google Scholar]
- Harris, R. S., S. K. Petersen-Mahrt and M. S. Neuberger, 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10 1247–1253. [DOI] [PubMed] [Google Scholar]
- Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt et al., 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113 803–809. [DOI] [PubMed] [Google Scholar]
- Hunter, N., S. R. Chambers, E. J. Louis and R. H. Borts, 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Huthoff, H., and M. H. Malim, 2005. Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins. Virology 334 147–153. [DOI] [PubMed] [Google Scholar]
- Imai, K., G. Slupphaug, W. I. Lee, P. Revy, S. Nonoyama et al., 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4 1023–1028. [DOI] [PubMed] [Google Scholar]
- Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham et al., 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79 285–296. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Klapholz, S., C. S. Waddell and R. E. Esposito, 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, K., K. B. Marcu and J. Greeve, 2006. The cytidine deaminases AID and APOBEC-1 exhibit distinct functional properties in a novel yeast selectable system. Mol. Immunol. 43 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, C., and P. Schar, 2004. Meiotic recombination: sealing the partnership at the junction. Curr. Biol. 14 R962–R964. [DOI] [PubMed] [Google Scholar]
- Larson, E. D., M. L. Duquette, W. J. Cummings, R. J. Streiff and N. Maizels, 2005. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 15 470–474. [DOI] [PubMed] [Google Scholar]
- Lee, G. S., M. B. Neiditch, S. S. Salus and D. B. Roth, 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117 171–184. [DOI] [PubMed] [Google Scholar]
- Lettre, G., and M. O. Hengartner, 2006. Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell. Biol. 7 97–108. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and G. R. Smith, 1994. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin, S. M., P. B. Moens, V. Wang, M. Lenzi, D. Shanmugarajah et al., 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31 385–390. [DOI] [PubMed] [Google Scholar]
- Maizels, N., 2005. Immunoglobulin gene diversification. Annu. Rev. Genet. 39 23–46. [DOI] [PubMed] [Google Scholar]
- Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin et al., 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424 99–103. [DOI] [PubMed] [Google Scholar]
- Manis, J. P., Y. Gu, R. Lansford, E. Sonoda, R. Ferrini et al., 1998. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 187 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, M., K. M. Rose, S. L. Kozak and D. Kabat, 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9 1398–1403. [DOI] [PubMed] [Google Scholar]
- Martin, J. S., N. Winkelmann, M. I. Petalcorin, M. J. McIlwraith and S. J. Boulton, 2005. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K., 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265 10857–10864. [PubMed] [Google Scholar]
- Maundrell, K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123 127–130. [DOI] [PubMed] [Google Scholar]
- Mayorov, V. I., I. B. Rogozin, L. R. Adkison, C. Frahm, T. A. Kunkel et al., 2005. Expression of human AID in yeast induces mutations in context similar to the context of somatic hypermutation at G-C pairs in immunoglobulin genes. BMC Immunol. 6 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikl, M. C., I. N. Watt, M. Lu, W. Reik, S. L. Davies et al., 2005. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 25 7270–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Morgan, H. D., W. Dean, H. A. Coker, W. Reik and S. K. Petersen-Mahrt, 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279 52353–52360. [DOI] [PubMed] [Google Scholar]
- Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower et al., 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281 22161–22172. [DOI] [PubMed] [Google Scholar]
- Neuberger, M. S., R. S. Harris, J. Di Noia and S. K. Petersen-Mahrt, 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28 305–312. [DOI] [PubMed] [Google Scholar]
- Neuberger, M. S., J. M. Di Noia, R. C. Beale, G. T. Williams, Z. Yang et al., 2005. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 5 171–178. [DOI] [PubMed] [Google Scholar]
- Papavasiliou, F. N., and D. G. Schatz, 2002. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 195 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parusel, C. T., E. A. Kritikou, M. O. Hengartner, W. Krek and M. Gotta, 2006. URI-1 is required for DNA stability in C. elegans. Development 133 621–629. [DOI] [PubMed] [Google Scholar]
- Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R. S. Chaganti et al., 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412 341–346. [DOI] [PubMed] [Google Scholar]
- Petersen, S., R. Casellas, B. Reina-San-Martin, H. T. Chen, M. J. Difilippantonio et al., 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature 414 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt, S. K., R. S. Harris and M. S. Neuberger, 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418 99–103. [DOI] [PubMed] [Google Scholar]
- Pham, P., R. Bransteitter, J. Petruska and M. F. Goodman, 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424 103–107. [DOI] [PubMed] [Google Scholar]
- Poltoratsky, V. P., S. H. Wilson, T. A. Kunkel and Y. I. Pavlov, 2004. Recombinogenic phenotype of human activation-induced cytosine deaminase. J. Immunol. 172 4308–4313. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla, T. A., S. M. Baker, A. C. Harris, J. L. Tsao, X. Yao et al., 1998. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 18 276–279. [DOI] [PubMed] [Google Scholar]
- Rada, C., G. T. Williams, H. Nilsen, D. E. Barnes, T. Lindahl et al., 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12 1748–1755. [DOI] [PubMed] [Google Scholar]
- Rada, C., J. M. Di Noia and M. S. Neuberger, 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16 163–171. [DOI] [PubMed] [Google Scholar]
- Rogozin, I. B., and Y. I. Pavlov, 2006. The cytidine deaminase AID exhibits similar functional properties in yeast and mammals. Mol. Immunol. 43 1481–1484. [DOI] [PubMed] [Google Scholar]
- Rooney, S., J. Chaudhuri and F. W. Alt, 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200 115–131. [DOI] [PubMed] [Google Scholar]
- Sale, J. E., D. M. Calandrini, M. Takata, S. Takeda and M. S. Neuberger, 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412 921–926. [DOI] [PubMed] [Google Scholar]
- Schreck, S., M. Buettner, E. Kremmer, M. Bogdan, H. Herbst et al., 2006. Activation-induced cytidine deaminase (AID) is expressed in normal spermatogenesis but only infrequently in testicular germ cell tumours. J. Pathol. 210 26–31. [DOI] [PubMed] [Google Scholar]
- Schumacher, A. J., D. V. Nissley and R. S. Harris, 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102 9854–9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, H., R. C. Ferreira, Q. Pan-Hammarstrom, R. R. Graham, B. Ziemba et al., 2007. Role for Msh5 in the regulation of Ig class switch recombination. Proc. Natl. Acad. Sci. USA 104 7193–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, W. D., G. G. Glick, M. K. Davidson and W. P. Wahls, 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy, A. M., N. C. Gaddis, J. D. Choi and M. H. Malim, 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418 646–650. [DOI] [PubMed] [Google Scholar]
- Simpson, L. J., A. L. Ross, D. Szuts, C. A. Alviani, V. H. Oestergaard et al., 2006. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 7 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., 2004. How homologous recombination is initiated: unexpected evidence for single-strand nicks from v(d)j site-specific recombination. Cell 117 146–148. [DOI] [PubMed] [Google Scholar]
- Snowden, T., S. Acharya, C. Butz, M. Berardini and R. Fishel, 2004. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15 437–451. [DOI] [PubMed] [Google Scholar]
- Stavnezer, J., J. E. Guikema and C. E. Schrader, 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus, C., C. Schubert and G. Seydoux, 1998. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev. Biol. 200 212–224. [DOI] [PubMed] [Google Scholar]
- Thorne, L. W., and B. Byers, 1993. Stage-specific effects of X-irradiation on yeast meiosis. Genetics 134 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33 1451–1455. [DOI] [PubMed] [Google Scholar]
- Wood, W. B. E., 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wuerffel, R. A., J. Du, R. J. Thompson and A. L. Kenter, 1997. Ig Sgamma3 DNA-specifc double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol. 159 4139–4144. [PubMed] [Google Scholar]
- Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz et al., 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279 53379–53386. [DOI] [PubMed] [Google Scholar]