Abstract
Expression quantitative trait loci (eQTL) mapping concerns finding genomic variation to elucidate variation of expression traits. This problem poses significant challenges due to high dimensionality of both the gene expression and the genomic marker data. We propose a multivariate response regression approach with simultaneous variable selection and dimension reduction for the eQTL mapping problem. Transcripts with similar expression are clustered into groups, and their expression profiles are viewed as a multivariate response. Then, we employ our recently developed sparse partial least-squares regression methodology to select markers associated with each cluster of genes. We demonstrate with extensive simulations that our eQTL mapping with multivariate response sparse partial least-squares regression (M-SPLS eQTL) method overcomes the issue of multiple transcript- or marker-specific analyses, thereby avoiding potential elevation of type I error. Additionally, joint analysis of multiple transcripts by multivariate response regression increases power for detecting weak linkages. We illustrate that M-SPLS eQTL compares competitively with other approaches and has a number of significant advantages, including the ability to handle highly correlated genotype data and computational efficiency. We provide an application of this methodology to a mouse data set concerning obesity and diabetes.
EXPRESSION quantitative trait loci (eQTL) mapping is a genetic mapping of genomewide gene expression. It combines traditional quantitative trait mapping and microarray technology. eQTL mapping provides an opportunity to investigate a large and unbiased set of traits that are immediately connected to DNA sequence variation; thereby it enables the study of gene networks. eQTL mapping studies have been applied in several model organisms and humans (Brem et al. 2002; Schadt et al. 2003; Morley et al. 2004; Chesler et al. 2005; Stranger et al. 2005; Wang et al. 2006). These studies have demonstrated several advantages of this line of research, from identifying candidate genes (Schadt et al. 2003) to elucidating regulatory networks (Brem et al. 2002; Schadt et al. 2003; Yvert et al. 2003).
Typical eQTL studies involve an N × G matrix of gene expression, where rows are different individuals (e.g., mice, in the order of tens) and columns are transcripts (in the order of thousands), and an N× p matrix (Xp) with genomic marker (in the order of hundreds or more) information. eQTL analysis differs from traditional quantitative trait loci (QTL) analysis in the number of traits considered. We refer to Kendziorski and Wang (2006) for a comprehensive review of general statistical issues concerning eQTL studies. Initial methods for eQTL mapping can be grouped into two (Kendziorski et al. 2006): (1) transcript-specific analysis in which mapping of a single expression trait is considered at a time and the entire analysis consists of thousands of transcript-specific analyses and (2) marker-specific analysis in which differentially expressed transcripts are identified at a single marker (by considering a marker genotype as a treatment) and the complete analysis requires scanning for all the markers. Both approaches are multiple applications of traditional methods (QTL mapping and identification of differentially expressed transcripts) and are prone to elevation of the false positive rate. Moreover, these approaches analyze data disjointly, either at the transcript or the marker level, leading to loss of power.
Mapping methods based on a notion of meta-transcript, which combine multiple similarly behaving transcripts by clustering or principal components analysis of genomewide gene expression data (Lan et al. 2003; Yvert et al. 2003), are viable approaches for reducing the number of tests and improving the power of linkage detection. However, these methods do not produce transcript-specific information because identified markers associate with a meta-transcript and not with individual transcripts.
Recent efforts for eQTL analysis focus on combined analysis of all the transcript and marker data by collapsing the aforementioned approaches 1 and 2. The mixture over markers (MOM) model of Kendziorski et al. (2006) is the first approach to facilitate information sharing across transcripts by an empirical Bayes method. It identifies transcripts that map to at least one marker (mapping transcripts) and then characterizes one or more markers per mapping transcript by utilizing transcript-specific highest posterior density regions. Recently, Gelfond et al. (2007) improved on the MOM model by utilizing genomic locations of the transcripts. To identify mapping transcripts and related eQTL simultaneously, Jia and Xu (2007) proposed a shrinkage analysis through a Bayesian hierarchical model called BAYES. This approach treats eQTL mapping in a variable selection context, where expression values of transcripts are modeled by linear functions of markers and variable selection is promoted by a special prior distribution specification, namely, the spike and slab distribution (Mitchell and Beauchamp 1988), on regression coefficients. When fitting transcript-level regression models, BAYES uses all the transcripts and markers simultaneously to achieve better power in detecting linkages. These transcript-level regression models share the same set of prior distributions. Although BAYES is flexible enough to map multiple markers per transcript, it is highly parametric, relies on prior specifications, and requires intense computations. Furthermore, properties of BAYES when markers are highly correlated are not studied. This is an important practical challenge because markers in close proximity are often highly correlated due to linkage disequilibrium (LD). These high correlations may hamper the performance of variable selection schemes that do not explicitly accommodate such a grouping structure.
In this article, we propose a multivariate response regression framework, named eQTL mapping with multivariate response sparse partial least-squares regression (M-SPLS eQTL). We utilize sparse partial least-squares (SPLS) regression (Chun and Keleş 2007), a novel statistical methodology for multivariate response regression with built-in dimension reduction and variable selection. Such a formulation is motivated by the apparent power advantages of multiple phenotype modeling observed in traditional multitrait QTL mapping (Jiang and Zeng 1995; Allison et al. 1998). It aims to capitalize on correlations between multiple transcripts while simultaneously dealing with all the markers. Recent computational models of eQTL mapping in the yeast Saccharomyces cerevisiae suggest that most eQTL have weak effects and that half of transcripts require more than five loci (markers) under additive models (Brem and Kruglyak 2005). This study further elucidated the importance of joint analysis of the multiple transcripts and markers to boost weak linkage signals. In our approach, we cluster genes into groups on the basis of their expression similarity. This helps us to view the expression values within a cluster as a multivariate response. Then, we form a cluster-level multivariate response regression and employ SPLS regression to identify markers affecting all or a subgroup of genes within the cluster. In the next two sections, we review underlying principles of the SPLS regression by focusing on aspects important to our application and describe our method in detail. In the simulation studies section, we study the operating characteristics of our approach and compare it to other approaches. We show that the proposed framework has excellent power and very small type-I error and significantly outperforms its univariate counterpart. In the case study: application to mouse data from a study of obesity and diabetes section, we illustrate our approach with a mouse data set of obesity and diabetes research (Lan et al. 2006) and then discuss potential extensions.
eQTL MAPPING WITH MULTIVARIATE SPLS REGRESSION
SPLS regression:
Partial least-squares (PLS) regression has been an alternative to ordinary least squares (OLS) regression in ill-conditioned linear regression models that arise in several disciplines such as chemistry, economics, psychology, and pharmaceutical science (De Jong 1993). At the core of PLS regression is a dimension reduction technique that operates under the assumption of a basic latent decomposition of a response matrix 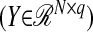 and a predictor matrix
and a predictor matrix 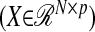 ,
,
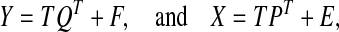 |
where 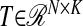 is a matrix that produces K linear combinations (scores),
is a matrix that produces K linear combinations (scores), 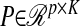 and
and 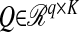 are matrices of coefficients (loadings), and
are matrices of coefficients (loadings), and 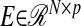 and
and 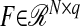 are matrices of random errors.
are matrices of random errors.
To specify the latent component matrix T such that T = XW, PLS requires finding the columns of W = (w1, w2, …, wK) from successive optimization problems. The criterion for the kth estimated direction vector 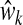 is formulated as
is formulated as
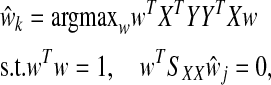 |
(1) |
for j = 1, …, k − 1, where SXX is the sample covariance matrix of X. After estimating the latent components (T), loadings (Q) are estimated via OLS for the model Y = TQT + F. βPLS is estimated by 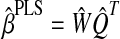 , where
, where 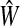 and
and 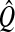 are estimates of W and Q, since Y = XWQT + F = XβPLS + F.
are estimates of W and Q, since Y = XWQT + F = XβPLS + F.
In Chun and Keleş (2007), we investigated theoretical properties of PLS regression and showed that although it had been traditionally promoted for regression problems with a large number of variables, it suffers from the curse of dimensionality in the contemporary large p, small N setting. To address this, we developed a sparse PLS regression that aims to promote sparsity by imposing an L1 penalty onto the direction vector of PLS. The SPLS objective function is given by
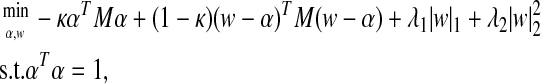 |
(2) |
where M = XTYYTX. This formulation promotes an exact zero property by imposing an L1 penalty onto a surrogate of the direction vector (w) instead of the original direction vector (α), while keeping α and w close to each other. This formulation is discussed in Chun and Keleş (2007), where we also characterized the solution of the minimization problem. The first L1 penalty encourages sparsity on w, and the second L2 penalty takes care of potential singularity in M when solving for w. The parameter κ is for reducing the concavity of the problem and avoiding locally optimal solutions. We show in Chun and Keleş (2007) that a κ-value of <0.5 performs well in practice and considering multiple κ-values has the effect of initiating the algorithm with different starting values. After obtaining α and w, we rescale the solution of w to have norm 1 and use this scaled version as the estimated direction vector.
The direction vector objective function in (2) is utilized in the course of the SPLS algorithm to select active (relevant) variables. We define 𝒜 to be an index set for active variables, K as the number of components, and 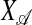 as the matrix of covariates of which indexes are contained in 𝒜. Then, the computational SPLS algorithm can be summarized as follows:
as the matrix of covariates of which indexes are contained in 𝒜. Then, the computational SPLS algorithm can be summarized as follows:
1. Set
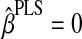 ,
, 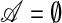 , k = 1, and Y1 = Y.
, k = 1, and Y1 = Y.2. While (k ≤ K),
2.1. Find
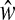 by solving the minimization problem in (2) with
by solving the minimization problem in (2) with 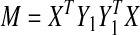 .
.2.2. Update 𝒜 as
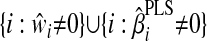 .
.2.3. Fit PLS with
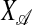 by using k numbers of latent components.
by using k numbers of latent components.2.4. Update
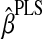 by using the new PLS estimates of the direction vectors, and update Y1 and k through
by using the new PLS estimates of the direction vectors, and update Y1 and k through 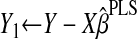 and
and 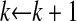 .
.
As seen in formulation (2), SPLS has tuning parameters λ1, λ2, and K. Since this formulation becomes highly singular when q = 1 or Y's are highly correlated, i.e., favorable scenarios for SPLS regression, we set λ2 to ∞ with an elastic net penalty (Zou and Hastie 2005). This leads to the form of a soft thresholded estimator (Chun and Keleş 2007). As a result, step 2.1 of the SPLS algorithm takes the form of simple soft thresholding driven only by λ1. In principle, each direction vector requires its own soft thresholding parameter. However, tuning K numbers of parameters is computationally prohibitive. Thus, we utilize the following adaptive form of a soft thresholded estimator where we need only to tune η, 0 ≤ η ≤ 1:
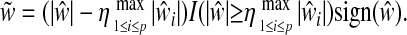 |
This form of soft thresholding retains components that are greater than some fraction of the maximum component. As a result, SPLS has two tuning parameters, η and K, and these are tuned by cross-validation (CV).
SPLS regression can select a higher number of relevant variables than the available sample size since the number of variables that contribute to each direction vector is not limited by the sample size. This property is shared by recent variable selection methods such as elastic net (Zou and Hastie 2005) and supervised principal components (Bair et al. 2006). Additionally, as apparent from the formulation in (2), SPLS regression is able to handle multivariate 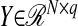 , q ≥ 1, without additional computational complexity. This property motivates the use of SPLS regression within the context of eQTL mapping where the goal is to utilize transcript and marker information simultaneously.
, q ≥ 1, without additional computational complexity. This property motivates the use of SPLS regression within the context of eQTL mapping where the goal is to utilize transcript and marker information simultaneously.
M-SPLS eQTL:
Our approach consists of two steps.
Step 1. Clustering of the G × N expression matrix: Current eQTL studies typically have a total of N experimental units from two or more distinct populations. There is a vast literature on clustering of gene expression data. Among simple methods are nonparametric clustering methods such as k-means, partitioning around medoids (Kaufman and Rousseeuw 1990), and hierarchical clustering (Eisen et al. 1998) or parametric clustering methods such as a mixture of Gaussian distributions (Fraley and Raftery 2002). We view the choice of the clustering method as a design-dependent decision and present an example within our case study. The thrust of the clustering step is to provide a transition from transcript-level regression models to module/cluster-level regression models.
Step 2. Cluster-specific multivariate response SPLS regression with bootstrap confidence intervals: After the clustering/grouping step, at each cluster k, we define a Gk-dimensional response vector
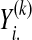 to denote the expressions of all the Gk genes, measured on the ith subject. We then consider a cluster-specific marker model
to denote the expressions of all the Gk genes, measured on the ith subject. We then consider a cluster-specific marker model
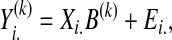 |
where Ei. denotes the random error matrix and B(k) is a p × Gk matrix representing the contribution of each marker m ∈ {1, …, p} to the expression variation of each transcript g ∈ {1, …, Gk} of cluster k. Such a model is fitted for every cluster using the SPLS regression.
Two apparent gains are expected from this approach. First, we expect it to be more powerful than both the individual transcript- and marker-specific analyses because transcripts with similar patterns are considered simultaneously and correlations among the transcripts are taken into account. Thus, it will be able to detect weak linkages. Second, it is expected to avoid type-I error inflation by eliminating multiple model fittings. We illustrate these points with simulations. SPLS regression tends to select a set of highly correlated markers rather than a single one among them when the covariates that are collectively associated with a phenotype have a grouping structure. This group selection property is easily realized in the SPLS algorithm. The minimization problem in (2), with an updated M in step 2.1, can allow a set of variables to be admitted to the active set 𝒜 simultaneously in step 2.2. This property is especially attractive in the following two cases. First, when a region of the genome, covered by a set of markers (e.g., in the form of haplotypes), is associated with a phenotype, SPLS regression can localize the region rather than select a single marker from the set. Selecting the set of highly correlated markers is more desirable when the data do not discriminate among these due to small sample size. Second, when quantitative traits are linked to several physically linked loci with small effects, suggested by several QTL mapping studies (reviewed in Flint et al. 2005), SPLS regression can capture these linked loci.
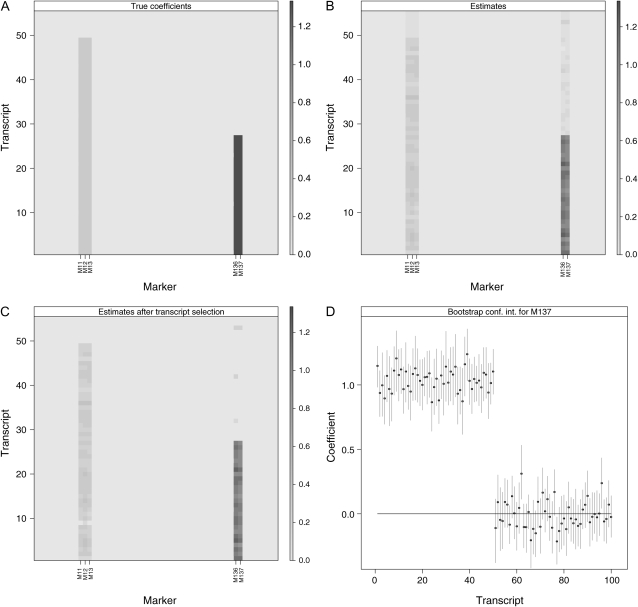
The final stage of cluster-specific SPLS regression is constructing bootstrap confidence intervals for transcript selection. The outcome of multivariate SPLS regression is a set of selected markers that significantly associate with one or more transcripts in the cluster and their estimated regression coefficients. We provide an example of such an outcome for a data set from our simulation study (simulation C-1) in Figure 1. Figure 1A depicts true linkages simulated for a cluster of 100 genes over 145 markers. Figure 1B displays linkages estimated by the M-SPLS regression. As evident in this plot, M-SPLS is able to select the true set of markers, but several false linkages, albeit with very small sizes, are also revealed for the selected markers. This is not realistic because, generally, a given marker or a set of markers is likely to associate with a subset of the genes within a cluster since cluster analysis is also prone to errors. To circumvent this, we construct bootstrap confidence intervals for transcript selection. After the initial application of M-SPLS regression, subjects are randomly selected with replacement and multivariate response PLS regression is fitted using only the selected markers from the original fit. An empirical distribution of estimated regression coefficients is obtained for each marker/transcript combination after a large number of bootstrap iterations. Using these empirical distributions, a 95% confidence interval is constructed for each marker/transcript combination. The final summary of linkages contains marker/transcript combinations for which the confidence intervals exclude zero. Figure 1C summarizes the linkages after the bootstrap confidence intervals are taken into account. Here, only the relevant transcripts have nonzero coefficients at the selected markers. For illustration purposes, we provide bootstrap confidence intervals for marker 137 (D18Mit123) across all 100 transcripts in Figure 1D.
Figure 1.—
(A) Set of true linkages. (B) Absolute values of the linkages estimated by M-SPLS regression. (C) Absolute values of the estimated linkages after considering bootstrap confidence intervals. In A–C, the x-axis represents markers, and the y-axis (on the left) represents transcripts. The shading of each pixel represents the strength of linkage signal. (D) Ninety-five percent C.I.'s for marker 137 across all the transcripts in the cluster. The y-axis depicts the size of the coefficients.
SIMULATION STUDIES
We performed simulation studies to investigate the operating characteristics of M-SPLS eQTL by comparing it to available methods under various eQTL architectures (simulations A and B). We paid attention to having both simple single-marker and more complex multiple-marker eQTL architectures. In addition, we allowed a large number of transcripts to be affected by a single architecture following some of the recent eQTL mapping findings (Wu et al. 2008). We also examined the advantages of multivariate response SPLS regression by comparing it to its univariate counterpart (simulation C). In these simulations, we intentionally skipped the clustering step and treated all the transcripts as a group. However, the performance of multivariate response SPLS regression might depend on the composition of a given cluster. In simulation C, we investigated the robustness of M-SPLS regression to different inaccuracies in cluster assignments and evaluated its ability to identify regions of the genome with a large number of mapping transcripts, i.e., hotspots (Schadt et al. 2003).
Simulation A—comparison of M-SPLS eQTL to BAYES and MOM in the absence of a strong LD structure among markers:
We first compared M-SPLS eQTL to BAYES (Jia and Xu 2007) and MOM (Kendziorski et al. 2006) by adopting the simulation experiments of Jia and Xu (2007). Ten markers (p = 10) are generated on a 360-cM genome by using the Haldane map function (Haldane 1919) and four eQTL are located at markers 1, 3, 6, and 10. A total of G = 1000 transcripts and N = 50 samples are generated following the Bayesian regression model that forms the backbone of Jia and Xu's (2007) BAYES method. In the first scenario (A-1), each subgroup of transcripts is affected by only a single marker. Transcripts 1–50 are under the influence of marker 10, transcripts 601–604 of marker 3, transcripts 605–610 of marker 1, and transcripts 961–1000 of marker 6. The remaining transcripts do not map to any markers and their expression values are determined by the error terms. eQTL control sizes, which are essentially coefficients of the relevant markers in the BAYES regression model, are generated from N(0, 32), and error terms are generated from N(0, 0.12). In the second scenario (A-2), multiple-marker sets affect the expression of subgroups of transcripts as follows: transcripts 1–16 are controlled by markers 1 and 10, transcripts 17–20 by markers 1, 3, and 10, and transcripts 971–990 by markers 1 and 6. Data for the remaining transcripts as well as eQTL effects and error terms are generated as in the first scenario.
We generated 100 replicates of each simulation scenario and applied SPLS regression. We then compared the operating characteristics with the results reported in Jia and Xu (2007). We note that Jia and Xu (2007) use only 20 replicates, which is presumably due to the computational complexity of the BAYES method. However, the results are overall comparable because our results for 20 vs. 100 simulation replicates are very similar. We used 99% bootstrap confidence intervals based on 1000 bootstrap samples for transcript selection whereas Jia and Xu (2007) use some unspecified false discovery rate (FDR), which is ≪1%, for linkage thresholding.
The simulation averages of power and type-I error are reported in Table 1. Here, U-SPLS refers to univariate SPLS regression where we fit an SPLS regression per transcript. U-SPLS is expected to produce many false positives due to multiple fitting of the regression model. As indicated in Table 1, indeed this approach has highly inflated type-I error. It is possible to argue that the performance of U-SPLS can be improved by implementing a bootstrap confidence interval step similar to that of M-SPLS. However, this increases computation time considerably; i.e., if M-SPLS replicates 1000 bootstrap samples, U-SPLS would replicate 1000 for each Gk transcript. We observe that M-SPLS has quite small type-I error and performs comparably to BAYES in terms of power despite the fact that the underlying data generating model precisely follows the assumptions of BAYES. Additionally, we observe that M-SPLS has the ability to accommodate the case where multiple transcripts do not form a homogeneous group. This is a desired property since different groups of transcripts within a cluster could easily be associated with multiple-marker sets. We revisit this point in simulation C-2.
TABLE 1.
Type-I error and power results based on the simulation setup of Jia and Xu (2007)
| A-1: single marker, multiple transcripts
|
A-2: multiple markers, multiple transcripts
|
|||
|---|---|---|---|---|
| Method | Type-I error | Power | Type-I error | Power |
| MOM | 0 | 0.9800 | 0.0004 | 0.642 |
| BAYES | 0 | 0.9800 | 0 | 0.993 |
| M-SPLS | 0.007 | 0.9870 | 0.007 | 0.986 |
| U-SPLS | 0.126 | 0.9910 | 0.1430 | 0.928 |
Simulation B—comparison of M-SPLS eQTL and BAYES with a strong LD structure among markers:
The current literature on eQTL mapping utilizes a small number of markers that typically lack a strong LD structure when investigating operating characteristics of methods by simulations (e.g., simulation A). In this next set of simulations, we use all of the 145 markers from 60 mice (p = 145 and N = 60) (Lan et al. 2006) to increase the number of markers and to reflect the ranges of LD structure that might exist among markers. We consider two types of eQTL architectures. In the first scenario (B-1), we select 6 markers (D2Mit17, D3Mit22, D4Mit190, D10Mit42, D12Mit217, and D13Mit66) genomewide. This represents a case where the markers in the eQTL architecture do not necessarily have a grouping structure since these markers have a relatively low LD structure. In the second scenario (B-2), we select three chromosomes (2, 5, and 15) randomly, and 2 highly correlated adjacent markers per chromosome, depicting three eQTL covered by 6 markers (D2Mit274 at 69.6 cM, D2Mit17 at 73.9 cM, D5Mit259 at 43 cM, D5Mit9 at 46 cM, D15Mit193 at 58.4 cM, and D15Mit16 at 70.1 cM). In both B-1 and B-2, transcripts 1–5 are directly regulated by an architecture due to these 6 markers, components of which are described in supporting information, Table S1. Transcripts 6–30 are regulated by transcript 3, and transcripts 31–50 are regulated by transcript 5 to allow within-group correlation that is not due to markers. Remaining transcripts 51–60 are determined by a Gaussian error term. Average heritability across these 60 transcripts is 0.75. M-SPLS is tuned by 10-fold CV and we use 10,000 bootstrap samples for constructing confidence intervals. We use a cutoff of 0.2 for FDR control for BAYES.
As seen in Table 2, M-SPLS eQTL has significantly higher power than BAYES at the cost of a small increase in the type-I error. Power gain of M-SPLS eQTL is more noticeable in simulation B-2 with the strong LD structure among the markers of the eQTL architecture. This suggests that the grouping property of M-SPLS regression could be beneficial in genetic mapping studies in the presence of linkages with strongly correlated markers. Although this grouping property slightly increases the type-I error by including extra markers that have high correlations with the true set of relevant markers, SPLS starts to select a smaller set among the set of correlated markers as the sample size increases and the data start to discriminate these markers (simulation data not shown).
TABLE 2.
Type-I error and power results for simulation B
| B-1: weak LDa
|
B-2: strong LDa
|
|||
|---|---|---|---|---|
| Method | Type-I error (SD) | Power (SD) | Type-I error (SD) | Power (SD) |
| BAYES | 0.009 (0.002) | 0.832 (0.023) | 0.004 (0.001) | 0.612 (0.02) |
| M-SPLS eQTL | 0.014 (0.002) | 0.915 (0.021) | 0.008 (0.001) | 0.942 (0.03) |
LD among the markers in the eQTL architecture.
Simulation C—sensitivity of M-SPLS eQTL to the quality of cluster assignments:
In simulations A-1 and A-2, we observed that M-SPLS regression overcomes the elevation of type-I error compared to U-SPLS by avoiding multiple model fits. However, the gain due to M-SPLS might depend on the quality of cluster assignments. Thus, we next compare performances of U-SPLS and M-SPLS regression by imposing different types of inaccuracies on cluster assignments. We consider two cases: (C-1) expression values of some of the cluster transcripts are determined by noise, i.e., presence of nonmapping transcripts, and (C-2) subgroups of transcripts within a cluster are controlled by different combinations of architectures. In addition, in simulation C-3, we investigate the hotspot detection property of M-SPLS eQTL that is likely to be affected by the quality of cluster assignments.
Simulation C-1—noisy clusters with nonmapping transcripts:
We assume that there is only one eQTL architecture involving several markers and affecting a percentage of the genes in the cluster; i.e., the observed correlation mechanism among the genes is a result of a single eQTL architecture. This corresponds to considering three factors in the data-generating scheme: r, number of relevant markers in the eQTL architecture (3, 10); ρ, proportion of cluster genes affected by the eQTL architecture (10, 30, 60, and 90%); and c, control size of the eQTL architecture (weak vs. strong).
As in simulation B, we use the full set of markers from 60 mice. For each combination of r, ρ, and c, we simulate 100 transcripts, treated as a group for M-SPLS regression, as follows. We first generate a norm 1 eQTL architecture direction vector with r nonzero components. The sizes of the coefficients are controlled to a constant multiplied by this direction vector. We consider the constants c = 1 and c = 2 for weak and strong effects, ρ proportion of transcripts are controlled by the eQTL architecture, and random error terms are generated from N(0, 1). We use fivefold CV for marker selection and 95% bootstrap confidence intervals based on 1000 bootstrap samples for transcript selection. The simulations are replicated 100 times. More details on data generation of these simulations are provided in Table S2.
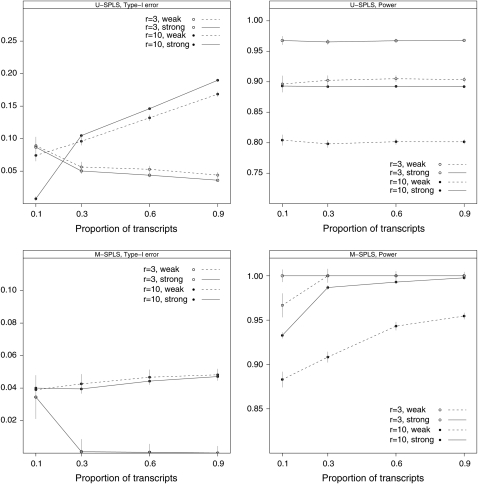
Results are presented in Figure 2 in terms of power and type-I error. U-SPLS regression exhibits inflated type-I error as expected on the basis of the earlier simulations following Jia and Xu's (2007) design. Additionally, the power of U-SPLS does not change as the proportion of transcripts associated with the eQTL mechanism increases. This is also expected since U-SPLS considers separate regression fits for each transcript. On the other hand, M-SPLS regression has very small type-I error and, overall, has significantly higher power than U-SPLS. We note that in our data-generating scheme, the sizes of the coefficients are inherently decreasing as the number of markers r in the eQTL mechanism increases. This is because the sizes are proportional to the elements of the direction vector and the norm of the direction vector is by definition 1. As a result, the r = 10 markers and weak control configuration have the highest noise level among the 16 configurations considered. Despite the decreasing overall signal at the transcript level, the power of M-SPLS increases as the proportion of transcripts affected by the eQTL mechanism increases. This provides evidence that M-SPLS successfully utilizes information across multiple transcripts; therefore, low signal linkages that might be missed by examining individual markers separately become detectable. Additionally, M-SPLS has more power than U-SPLS even when only 10% of the transcripts in the cluster are affected by the same set of markers (at both control sizes when r = 10).
Figure 2.—
Results for simulation C-1 (noisy cluster with many nonmapping transcripts). Symbols represent different numbers of markers (○, r = 3; •, r = 10) associated with ρ ∈ {0.1, 0.3, 0.6, 0.9} proportion of transcripts in the cluster. Different line types indicate weak (dashed line) or strong (solid line) control by a single eQTL architecture.
Simulation C-2—heterogeneous clusters with subgroups of transcripts controlled by different eQTL architectures:
We next study the case where two hidden components, therefore two different eQTL architectures, are present. These two components are (1) eQTL mechanism 1, linear combinations of markers 11, 12, and 13; and (2) eQTL mechanism 2, linear combinations of markers 136 and 137. Mechanism 1 is set to have a weaker control size than mechanism 2. We consider two cases for the multiple-eQTL architectures simulation:
C-2.1: Transcripts 1–50 are under the influence of mechanisms 1 and 2, and transcripts 51–90 are affected only by mechanism 1. Expression values of the rest of the transcripts are set by the error terms.
C-2.2: The same as C-2.1 but with a larger control size.
Details on the parameter settings are provided in Table S3.
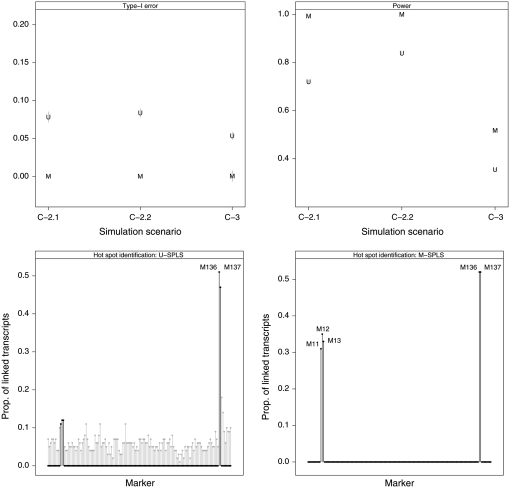
Results of these multiple-eQTL architecture simulations are provided in Figure 3. M-SPLS has greater power than U-SPLS with a smaller type-I error. This observation is consistent with our earlier simulation experiments, suggesting that M-SPLS has the ability to accommodate cases where different groups of transcripts within a cluster are associated with multiple-marker sets.
Figure 3.—
Results for simulations C-2 (heterogeneous cluster: C-2.1 represents weaker control by the two eQTL architectures compared to C-2.2) and C-3 (cluster with weak linkages). Top panels represent type-I error and power for U-SPLS (U) and M-SPLS (M) with vertical lines representing simulation standard errors. Bottom panels report the proportion of linked transcripts for each marker by U-SPLS (bottom left panel) and M-SPLS (bottom right panel) in simulation C-3. Hotspot markers are indicated with solid lines.
Simulation C-3—clusters with weak eQTL effects:
Hotspot regions are defined as loci of a genome that are mapped by a large number of genes (Schadt et al. 2003). They lead to widespread changes in the expression of distant genes. Hotspots that exhibit strong control of their target transcripts are often easily identified with transcript-specific approaches. However, if the hotspot locus exerts weak control over its targets, except maybe for a few directly related transcripts (e.g., cis-regulation), univariate approaches tend to miss these linkages and thus fail to identify the hotspot. In contrast, a multivariate approach might capture these weak linkages by utilizing correlations among transcripts.
We assume that two subgroups of transcripts form a cluster and are controlled by different combinations of two architectures. Each of the architectures can be interpreted as hotspots as they control 50 and 80% of the transcripts. One of the architectures exerts weak control except for one transcript, but the other exhibits strong control of all linked transcripts. This setting is similar to that of simulation C-2.1 and more details are provided in Table S3.
Figure 3 summarizes the results from this simulation. Linkages with the first eQTL mechanism cannot be detected by U-SPLS because the control size is very small, resulting in poor power for U-SPLS. In contrast, M-SPLS has at least twice the power of U-SPLS, although M-SPLS misses some of the linkages with this architecture. This result is also reflected in the hotspot selection performance of the methods. The first eQTL mechanism cannot be revealed as a hotspot from individual regression analyses by U-SPLS (Figure 3, bottom left). However, M-SPLS is able to identify markers involved in this mechanism as hotspots (Figure 3, bottom right).
CASE STUDY: APPLICATION TO MOUSE DATA FROM A STUDY OF OBESITY AND DIABETES
We present an application of our method to a mouse data set published in Lan et al. (2006). This data set contains expression measurements of 45,265 transcripts from liver tissues of 60 mice. Mice were collected from a (B6 × BTBR) F2-ob/ob cross where animals lacked a functional leptin protein hormone, known to be important for reproduction and regulation of body weight and metabolism (Zhang et al. 1994), and segregated for obesity- and diabetes-related phenotypes. We utilized the preprocessed data that are publicly available at GEO (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE3330). The marker map for these data consists of 145 microsatellite markers from 19 nonsex mouse chromosomes. Following Jia and Xu (2007), we performed an initial screening of the transcripts on the basis of their variability across 60 mice and excluded transcripts with sample variances <0.12 from our analysis. This left a total of G = 1573 transcripts.
Next, we clustered these remaining transcripts. As discussed earlier, the clustering method in an application is highly design dependent. For a time-course experiment, methods that utilize dependencies among different time points (Yuan and Kendziorski 2006) or methods specifically parameterizing cluster profiles (Jörnsten and Keleş 2008) might be more desirable. For the mouse data, we considered the following approach motivated by the successful use of the topological overlap measure (TOM) (Ravasz et al. 2002) in clustering analysis (Zhang and Horvath 2005). First, we constructed undirected, unweighted gene networks on the basis of the expression data, using the Gaussian graphical model (GGM) approach of Schäfer and Strimmer (2005). The constructed network is then used to compute TOM for each pair of transcripts. Dissimilarity measure 1-TOM between 2 transcripts represents a lack of closeness based on the number of shared neighbors in the expression network. Since 95 transcripts did not share any neighbors with other transcripts, they were analyzed by U-SPLS regression. Hierarchical clustering on the remaining transcripts using this dissimilarity measure resulted in 47 clusters based on the average silhouette measure (Kaufman and Rousseeuw 1990). The within-cluster Pearson correlations ranged from 0.027 to 0.948 with a mean of 0.226 across 47 clusters.
We present the results for one of the clusters in more detail. This cluster contains 3 lipid metabolism transcripts, namely, Scd1, Elovl6, and Fasn, that were investigated by a different analysis of the same data set (Lan et al. 2006; Jia and Xu 2007). There are a total of 83 transcripts in this cluster with a median within-cluster correlation of 0.12. An application of our approach with M-SPLS yields 27 markers, presented in Table 3, that are associated with one or more transcripts. The total number of linkages identified for this cluster is 487 and there are 62 transcripts that do not map to any marker. An image plot of the estimated effects of this cluster across markers and transcripts is provided in Figure 4. The entire M-SPLS eQTL analysis, including both the tuning and the bootstrap steps, for this cluster of 83 transcripts took only 3 min on a 64-bit machine with 2.66-Ghz CPU.
TABLE 3.
Markers identified for a cluster of size 83 including three lipid metabolism transcripts: Scd1, Elovl6, and Fasn
| Marker | Map (cM) | No. of mapping transcripts | Reference |
|---|---|---|---|
| D2Mit274 | 69.6 | 1 | Close to obesity modifier locus D2Mit9 (Stoehr et al. 2004) |
| D2Mit17 | 73.9 | 16 | Obesity/diabetes syndrome (Stoehr et al. 2000) |
| D2Mit106 | 77.9 | 19 | Liver weight (Jerez-Timaure et al. 2005) |
| Obesity/diabetes syndrome (Stoehr et al. 2000) | |||
| D2Mit194 | 85.4 | 18 | Obesity locus (Diament et al. 2004) |
| Body weight and fat (Jerez-Timaure et al. 2005) | |||
| Obesity/diabetes syndrome (Stoehr et al. 2000) | |||
| D2Mit263 | 98.7 | 21 | Lipid metabolism (Lan et al. 2006) |
| Obesity/diabetes syndrome (Stoehr et al. 2000) | |||
| D2Mit51 | 101.7 | 20 | |
| D2Mit49 | 104.2 | 21 | |
| D2Mit229 | 110.5 | 21 | |
| D2Mit148 | 121.6 | 21 | |
| D5Mit348 | 6.3 | 17 | |
| D5Mit75 | 11.8 | 21 | |
| D5Mit267 | 17.1 | 21 | Reproduction in leptin-deficient obese mice (Ewart-Toland et al. 1999) |
| D5Mit259 | 43 | 21 | |
| D5Mit9 | 46 | 21 | |
| D5Mit240 | 49.1 | 21 | Lipid metabolism (Lan et al. 2006) |
| D5Mit136 | 54.9 | 21 | |
| D8Mit249 | 58.1 | 18 | Fat gene (Naggert et al. 1995) |
| Triglyceride level (Colinayo et al. 2003) | |||
| D8Mit211 | 72 | 21 | |
| D8Mit113 | 77.6 | 21 | |
| D9Mit21 | 43.8 | 12 | |
| D9Mit207 | 45.3 | 17 | |
| D9Mit8 | 58.6 | 18 | Fat-pad mass (Mehrabian et al. 1998) |
| D9Mit15 | 93.6 | 21 | |
| D9Mit18 | 110.7 | 17 | |
| D15Mit174 | 0 | 11 | |
| D15Mit136 | 11.5 | 17 | |
| D15Mit63 | 21 | 13 | Early life body weight (Miller et al. 2002) |
| Diabetic modifier (Takeshita et al. 2006) |
Figure 4.—
M-SPLS solution for a cluster of 83 transcripts including the 3 lipid metabolism transcripts.
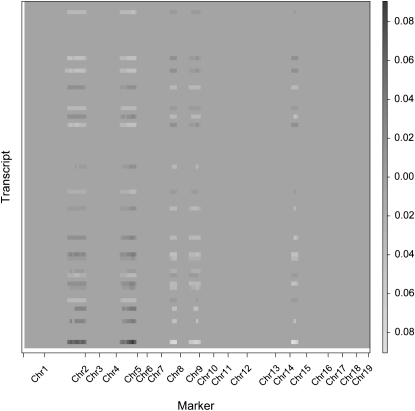
We note that many of the selected markers are in close proximity to each other on the mouse genome. These physically close markers are highly correlated (pairwise correlations on chromosomes 2, 5, and 15 are displayed in Figure S1). The fact that these highly correlated markers are identified relates to the group selection property of the SPLS regression. Since SPLS can choose more than one variable at each step of the selection process, it is able to capture all the relevant correlated variables rather than arbitrarily selecting one. One can argue that, perhaps, the selected markers cover too large of a region on each chromosome. The problem of identifying such large regions is driven by the nature of the data. Since the markers are highly correlated, it is hard to select finer areas of the genome with data from 60 mice. Flint et al. (2005) argue that 300 F2 animals are needed to map a QTL with an effect size of 5% onto a 40-cM interval with 50% power, using markers that are spaced every 20 cM across the genome. We anticipate that more mice are needed to localize finer areas of the genome in eQTL studies. In each correlated marker group in Table 3, there is at least one marker that is previously declared as an obesity- and diabetes-related locus. This result is encouraging since it provides a list of transcripts mapping to markers known to be related to obesity and diabetes.
Expression profiles of lipid metabolism transcripts Scd1, Elovl6, and Fasn are highly correlated (pairwise plots are in Figure S2; minimum pairwise correlation is 0.756). Therefore, it is reasonable to expect similar linkages for these transcripts. Indeed, M-SPLS reveals that these transcripts map to similar markers, whereas BAYES yields different linkages. This could be due to high correlation among markers. Unlike the markers generated by the Haldane map function (simulations A-1 and A-2), markers from the mouse study exhibit very high correlations. This multicollinearity problem is not explicitly addressed in BAYES, and priors for regression coefficients are assumed to be independent. In fact, similar mixture priors were used by Sha et al. (2006) in the context of a different model and a decrease in the variable selection performance was observed for the correlated variable case. It is plausible that BAYES also suffers from a similar problem and tends to select only one variable among a set of correlated variables.
Lan et al. (2006) highlighted that transcripts that were highly correlated with Scd1 mapped to the same genomic locations as Scd1, and found major QTL peaks for most of the 20 lipid metabolism traits at markers D2Mit263 and D5Mit240. These two markers are successfully identified by our approach. Among the five hotspots reported by MOM, one of them is also identified by M-SPLS eQTL with the group of transcripts we considered. This is marker D8Mit249, which is close to the “fat” gene known to affect obesity and diabetes (Naggert et al. 1995). M-SPLS eQTL identified D5Mit348, which is adjacent to D5Mit1, instead of D5Mit1, which affects triglyceride levels. Marker D15Mit63, emphasized in the findings of BAYES, is also identified by M-SPLS eQTL. Markers on chromosome 2 have been the most popular candidates for obesity and diabetes (Stoehr et al. 2000; Diament et al. 2004; Jerez-Timaure et al. 2005), but hotspots from MOM and BAYES do not have a noticeable indication of this. In particular, BAYES does not find any hotspots on chromosome 2. In contrast, M-SPLS eQTL yields strong effects for markers on chromosome 2. Furthermore, although marker D5Mit267, which is identified by M-SPLS eQTL but missed by MOM and BAYES, does not seem to be directly related to obesity and diabetes, it is associated with reproduction, which is another known function of leptin protein hormone (Ewart-Toland et al. 1999).
DISCUSSION
The advent of microarray technology is providing an unprecedented opportunity for investigating complex genetics underlying inheritance of transcript levels in segregating populations. One of the statistical challenges is the eQTL mapping problem that concerns identification of linkages between thousands of transcripts and markers. We formulated the eQTL mapping problem as a variable selection problem in a multivariate response regression. We then utilized sparse partial least squares (Chun and Keleş 2007) as a simultaneous variable selection and dimension reduction approach to identify linkages. This framework, implemented as an R package named SPLS (File S1), offers offers a computationally fast alternative for analyzing multiple transcript and marker data simultaneously to gain power and avoid multiplicities for good error control.
We demonstrated the advantages of our method with simulation experiments. These experiments included eQTL architectures with strong effects on a small fraction as well as weak effects on a large fraction of transcripts. These studies showed that as the number of mapping transcripts increases, the power of M-SPLS increases whereas its univariate analog with transcript-level regressions cannot capitalize on this phenomenon. We illustrated the utility of our approach with an example from mouse obesity and diabetes research. This case study highlighted the ability of SPLS regression to select groups of correlated markers. BAYES, an alternative variable selection approach to the eQTL problem, lacks this property and tends to select only one marker among the group of correlated markers. Our approach was able to consistently yield similar linkages for highly correlated transcripts. Furthermore, we were able to identify a marker that was missed by the previous analysis of the same data set but could potentially be important since it relates to another function of the leptin protein hormone (Ewart-Toland et al. 1999).
In this article, we allowed the markers to appear as main terms in the regression model. Identifying interactions among markers is a challenging problem. With an appropriate prescreening of markers, SPLS regression has the potential to handle a large number of interactions. In Chun and Keleş (2007), this property is illustrated with as many as 5000 variables. Another important research question in eQTL mapping is allowing for linkages with locations between markers using interval mapping (Chen and Kendziorski 2007). Our current formulation allows for mapping only at exact marker locations. However, a first pass with our approach and then a more focused traditional interval mapping (Sen and Churchill 2001) based on the selected markers might be a viable strategy.
Acknowledgments
We thank two anonymous referees for their constructive and valuable comments. This research has been supported in part by a Pharmaceutical Research and Manufactures of America Foundation Research Starter Grant in Informatics, by National Institutes of Health grant HG003747, and by National Science Foundation grant DMS 0804597 to S.K.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.100362/DC1.
References
- Allison, D. B., B. Thiel, P. S. Jean, R. C. Elston, M. C. Infante et al., 1998. Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am. J. Hum. Genet. 63 1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair, E., T. Hastie, D. Paul and R. Tibshirani, 2006. Prediction by supervised principal components. J. Am. Stat. Assoc. 101 119–137. [Google Scholar]
- Brem, R., and L. Kruglyak, 2005. The landscape of genetic complexity across 5700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296 752–755. [DOI] [PubMed] [Google Scholar]
- Chen, M., and C. Kendziorski, 2007. A statistical framework for expression quantitative trait loci (eQTL) mapping. Genetics 177 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler, E. J., L. Lu, S. Shou, Y. Qu, J. Gu et al., 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37 233–242. [DOI] [PubMed] [Google Scholar]
- Chun, H., and S. Keleş, 2007. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. http://www.stat.wisc.edu/∼keles/Papers/spls_jrssb.pdf. [DOI] [PMC free article] [PubMed]
- Colinayo, V. V., J. H. Qiao, X. P. Wang, K. L. Krass, E. Schadt et al., 2003. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm. Genome 14 464–471. [DOI] [PubMed] [Google Scholar]
- de Jong, S., 1993. SIMPLS: an alternative approach to partial least squares regression. Chemometrics Intell. Lab. Syst. 18 251–263. [Google Scholar]
- Diament, A., P. Farahani, S. Chiu, J. Fisler and C. Warden, 2004. A novel mouse chromosome 2 congenic strain with obesity phenotypes. Mamm. Genome 15(6): 452–459. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart-Toland, A., K. Mounzih, J. Qiu and F. F. Chehab, 1999. Effect of the genetic background on the reproduction of leptin-deficient obese mice. Endocrinology 140 732–738. [DOI] [PubMed] [Google Scholar]
- Flint, J., W. Valdar, S. Shifman and R. Mott, 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6 271–286. [DOI] [PubMed] [Google Scholar]
- Fraley, C., and A. E. Raftery, 2002. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 97 611–631. [Google Scholar]
- Gelfond, J. A. L., J. G. Ibrahim and F. Zou, 2007. Proximity model for expression quantitative trait loci (eQTL) detection. Biometrics 63 1108–1116. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8 299–309. [Google Scholar]
- Jerez-Timaure, N. C., E. J. Eisen and D. Pomp, 2005. Fine mapping of a QTL region with large effects on growth and fatness on mouse chromosome 2. Physiol. Genomics 21 411–422. [DOI] [PubMed] [Google Scholar]
- Jia, Z., and S. Xu, 2007. Mapping quantitative trait loci for expression abundance. Genetics 176 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., and Z. Zeng, 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnsten, R. J., and S. Keleş, 2008. Mixture models with multiple levels, with application to the analysis of multifactor gene expression data. Biostatistics 9 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, L., and P. Rousseeuw, 1990. Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons, New York.
- Kendziorski, C., and P. Wang, 2006. A review of statistical methods for expression quantitative trait loci mapping. Mamm. Genome 17 509–517. [DOI] [PubMed] [Google Scholar]
- Kendziorski, C. M., M. Chen, M. Yuan, H. Lan and A. D. Attie, 2006. Statistical methods for expression quantitative loci (eQTL) mapping. Biometrics 62 19–27. [DOI] [PubMed] [Google Scholar]
- Lan, H., J. P. Stoehr, S. T. Nadler, K. L. Schueler, B. S. Yandell et al., 2003. Dimension reduction for mapping mRNA abundance as quantitative traits. Genetics 164 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, H., M. Chen, J. B. Flowers, B. S. Yandell, D. S. Stapleton et al., 2006. Combined expression trait correlations and expression quantitative trait locus mapping. PLoS Genet. 2 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian, M., P. Z. Wen, J. Fisler, R. C. Davis and A. J. Lusis, 1998. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J. Clin. Invest. 101 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. A., J. M. Harper, A. Galecki and D. T. Burke, 2002. Big mice die young: early life body weight predicts longevity in genetically heterogeneous mice. Aging Cell 1 22–29. [DOI] [PubMed] [Google Scholar]
- Mitchell, T. J., and J. J. Beauchamp, 1988. Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83 1023–1036. [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, K. G. Devlin, J. L. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggert, J. K., L. D. Fricker, O. Varlamov, P. M. Nishina, Y. Rouille et al., 1995. Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat. Genet. 10 135–142. [DOI] [PubMed] [Google Scholar]
- Ravasz, E., A. Somera, D. Mongru, Z. Oltvai and A. Barabasi, 2002. Hierarchical organization of modularity in metabolic networks. Science 297 1551–1555. [DOI] [PubMed] [Google Scholar]
- Schadt, E. E., S. A. Monks, T. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297–302. [DOI] [PubMed] [Google Scholar]
- Schäfer, J., and K. Strimmer, 2005. Learning large-scale graphical Gaussian models from genomic data, pp. 263–276 in AIP Conference Proceedings 776. Science of Complex Networks: From Biology to the Internet and WWW (CNET 2004), edited by J. F. F. Mendes, S. N. Dorogovtsev, F. V. A. A. Povolotsky and J. G. O. Ira. Aveiro, Portugal. American Institute of Physics, Melville, NY.
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, N., M. G. Tadesse and M. Vannucci, 2006. Bayesian variable selection for the analysis of microarray data with censored outcomes. Bioinformatics 22 2262–2268. [DOI] [PubMed] [Google Scholar]
- Stoehr, J., S. Nadler, K. Schueler, M. Rabaglia, B. Yandell et al., 2000. Genetic obesity unmasks nonlinear interactions between murine type 2 diabetes susceptibility loci. Diabetes 49 1946–1954. [DOI] [PubMed] [Google Scholar]
- Stoehr, J. P., J. E. Byers, S. M. Clee, H. Lan, I. V. Boronenkov et al., 2004. Identification of major quantitative loci controlling body weight variation in ob/ob mice. Diabetes 53 245–249. [DOI] [PubMed] [Google Scholar]
- Stranger, B. E., M. S. Forrest, A. G. Clark, M. J. Minichiello, S. Deutsch et al., 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1 e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, S., M. Moritani, K. Kunika, H. Inoue and M. Itakura, 2006. Diabetic modifier QTLs identified in F2 intercrosses between akita and A/J mice. Mamm. Genome 17 927–940. [DOI] [PubMed] [Google Scholar]
- Wang, S., N. Yehya, E. E. Schadt, H. Wang, T. A. Drake et al., 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., D. L. Delano, N. Mitro, S. V. Su, J. Janes et al., 2008. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 4 e1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M., and C. Kendziorski, 2006. Hidden Markov models for microarray time course data in multiple biological conditions. J. Am. Stat. Assoc. 101 1323–1332. [Google Scholar]
- Yvert, G., R. B. Brem, J. Whittle, J. M. Akey, E. Foss et al., 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35 57–64. [DOI] [PubMed] [Google Scholar]
- Zhang, B., and S. Horvath, 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4 Article 17. [DOI] [PubMed]
- Zhang, Y., R. Proenca, M. Maffei, M. Barone, L. Leopold et al., 1994. Positional cloning of the mouse obese gene and its human homologue. Nature 372 425–431. [DOI] [PubMed] [Google Scholar]
- Zou, H., and T. Hastie, 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 67 301–320. [Google Scholar]