Abstract
S-type cytoplasmic male sterility (CMS-S) in maize is associated with high levels of a 1.6-kb RNA in mitochondria. This RNA contains two chimeric open reading frames (ORFs), orf355 and orf77. The previously described nuclear restorer-of-fertility allele Rf3 causes the processing of all transcripts that contain these chimeric ORFs. The Lancaster Surecrop-derived inbred line A619 carries a restorer that is distinct from Rf3 in that it selectively reduces only the CMS-S-specific 1.6-kb RNA. We have found that 10 additional Lancaster lines carry a single restoring allele traceable to either of two inbred lines, C103 and Oh40B. The C103 and Oh40B restorers are allelic to each other, but not to Rf3. Thus, this restoring allele, designated Rf9, represents a second naturally occurring CMS-S restorer in maize. Rf9 is a less effective restorer of fertility than is Rf3; its expression is influenced by both inbred nuclear background and temperature. Rf9 acts to reduce the amounts of orf355/orf77-containing linear mitochondrial subgenomes, which are generated by recombination of circular subgenomes with CMS-S-specific linear plasmids. The 1.6-kb RNA, which is transcribed only from linear ends, is correspondingly reduced.
MALE-sterile plants usually have normal growth and female fertility but fail to shed functional pollen. There are two types of male sterility: “genic,” caused by alleles of single nuclear genes, and “cytoplasmic,” caused by mutations in mitochondrial DNA (mtDNA). Cytoplasmic male sterility (CMS) is common in higher plants (reviewed by Chase 2007). It is usually caused by the expression of novel chimeric genes that result from mtDNA rearrangements and that are particularly deleterious during pollen development (reviewed by Newton et al. 2004; Chase 2007). Three basic types of CMS have been identified in maize and are designated CMS-C, CMS-T, and CMS-S. Mitochondrial DNA rearrangements have been found to correlate with these classifications. For each maize CMS type, one or more particular nuclear gene(s), termed “restorers of fertility,” that counteract the male-sterile effect of the cytoplasm have been discovered. Restorers of fertility can reduce the expression of CMS-associated chimeric genes by a variety of mechanisms.
Plant mitochondrial genomes have complex organizations due to the presence of actively recombining repeats. The S-type of CMS in maize has an especially complex mitochondrial genome. The S genome can be “linearized” by recombination with a pair of linear double-stranded DNA plasmids (Schardl et al. 1984). These plasmids are designated S1 (6.4 kb) and S2 (5.4 kb) (Pring et al. 1977; reviewed by Gabay-Laughnan et al. 1995) and are related to the R1 and R2 plasmids found in some Latin American races of maize (Weissinger et al. 1982, 1983) and to the M1 and M2 plasmids found in one teosinte accession (Zea luxurians var. Mazoti; Grace et al. 1994). The S1 and S2 plasmids possess 208-bp terminal inverted repeats (TIRs), covalently bound to a terminal protein (Kemble and Thompson 1982; Levings and Sederoff 1983; Paillard et al. 1985).
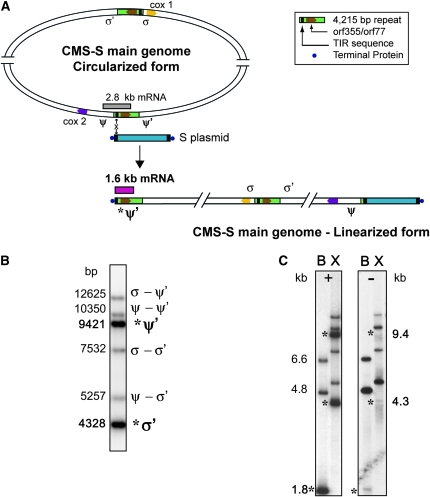
Within the CMS-S main mitochondrial genome, there are two integrated TIR-containing repeats that are very active in intragenomic recombination (see Figure 1A). These TIR sequences also serve as the target sites for homologous recombination between the free linear S plasmids and the main mitochondrial chromosome (Levings et al. 1980; Lonsdale et al. 1984; Schardl et al. 1984). Flanking genomic regions upstream of the integrated TIR sequences have been designated σ and ψ, respectively, and the regions immediately downstream of each repeat have been designated σ′ and ψ′ (Schardl et al. 1984; reviewed by Gabay-Laughnan et al. 1995). The common sequence contained within all four junctions (σ–σ′, σ–ψ′, ψ–ψ′, and ψ–σ′) has been called the “R repeat” (Schardl et al. 1984; Lonsdale et al. 1988) because it contains a portion of the R1 plasmid not present in the S1 plasmid (Houchins et al. 1986). The 4215-bp (4.2-kb) R repeats have been sequenced (Zabala et al. 1997; Xiao et al. 2006; Allen et al. 2007). They contain the adjacent chimeric open reading frames orf355 and orf77, which are unique to CMS-S mitochondrial genomes.
Figure 1.—
Most of the CMS-associated orf355/orf77 sequences are located at linear ends of CMS-S mitochondrial genomes. (A) Schematic showing how recombination between linear S plasmids and integrated TIR sequences next to orf355/orf77 leads to linearized ends of CMS-S mitochondrial genomes (modified from Allen et al. 2007). Two copies of a 4.2-kb R repeat (green) carrying orf355/orf77 sequences (brown inserts) are integrated in the genome and are flanked by different regions designated σ and σ′, and ψ and ψ′. One copy is downstream of cox1 (yellow) and the other downstream of cox2 (purple). Free S1 and S2 (represented by “S plasmid” in blue) recombine through TIR sequences to yield linearized CMS-S genomes. Recombination and the resulting linearized genome is depicted only for the ψ copy of the 4.2-kb repeat, although recombination at the σ copy is equally likely. A 1.6-kb orf355/orf77 transcript (pink) is associated with the presence of the linear ends (Zabala et al. 1997). (B) Most of the CMS-S mtDNA is in linearized form. Hybridization of an orf355-specific probe to a blot of XhoI-digested CMS-S mtDNA; XhoI sites occur outside of the repeats. Band labels to the right indicate the orf355-containing fragments corresponding to those diagrammed above. When recombination occurs with a TIR of an S plasmid (either S1 or S2), a linear end designated *σ′ or *ψ′ results (Schardl et al. 1984). The integrated σ–σ′ and ψ–ψ′ copies recombine with each other to give the σ–ψ′ and ψ–σ′ versions. These various integrated versions occur with approximately equal stoichiometries. Sizes of the XhoI fragments (indicated at left) were determined from the published CMS-S sequence data (Allen et al. 2007). (C) Most of the copies of orf355 are at protein-bound linear ends of CMS-S mitochondrial genomes. mtDNA was isolated either with proteinase K (left) or without proteinase K treatment (right). Samples were digested either with BamHI, which cuts within the 4.2-kb repeat (B), or with XhoI (X), which cuts outside of the repeat. The blot was hybridized with the orf355 probe. Asterisks indicate expected positions of the fragments at linear ends: 9.4 and 4.3 kb for XhoI and 1.8 kb for BamHI. The 1.8-kb BamHI band represents all copies of orf355/orf77 at linear ends.
The TIRs of the S plasmids recombine with homologous sequences in the 4.2-kb repeat, resulting in linearization of the main mitochondrial genome and creating an S-plasmid-like terminal structure just upstream of orf355/orf77 (Levings et al. 1980; Schardl et al. 1984). The S-male-sterile phenotype is correlated with the expression of a 1.6-kb transcript that includes both orf355 and orf77 (Zabala et al. 1997).
With rare exceptions, Rf3, a naturally occurring nuclear restorer of fertility, fully restores CMS-S to fertility. The mode of CMS-S restoration is gametophytic (Buchert 1961); i.e., restoring alleles act in the haploid male gametophyte. Thus, in S cytoplasm a pollen grain of the genotype Rf3 is starch filled and functional while a pollen grain carrying rf3 collapses and is nonfunctional. The Rf3 restorer is present in the public inbred lines Tr, Ky21, and C.I.21E and in the Pioneer Hi-Bred inbred lines CE1, BH2, JG3, and JG5 (Duvick 1956, 1965; Beckett 1971). In genetic crosses and mapping studies, the Rf3 alleles from the different inbred lines act as alleles of a single locus. However, it is possible that these lines carry tightly linked restoring alleles (Duvick 1957; Laughnan and Gabay 1978), as has been shown for the gametophytic restorer Rf-1 of rice Boro II CMS (Wang et al. 2006). The rf3 locus has been mapped to the long arm of chromosome 2 (2L; Laughnan and Gabay 1978; Kamps and Chase 1997; Gabay-Laughnan et al. 2004). Rf3 does not alter the mtDNA organization but specifically decreases the abundance of all of the transcripts from the 4.2-kb repeat region (Zabala et al. 1997).
In addition to the inbred lines of known Rf3/Rf3 genotype listed above, other inbred lines have been reported to restore CMS-S to fertility. Among these is a series of Lancaster Surecrop-derived inbred lines (Beckett 1971; Gracen and Grogan 1974; Gabay-Laughnan and Laughnan 1994). In contrast to the complete restoration of fertility by Rf3, partial fertility has been reported in the CMS-S versions of some of these lines (Beckett 1971; Gracen and Grogan 1974; Gabay-Laughnan and Laughnan 1994).
Here we report that the inbred line A619 carries a single gene capable of gametophytically restoring CMS-S to fertility and that this allele is a weak restorer, sensitive to the environment. We have designated this locus restorer of fertility9 (rf9) and the restoring allele, Rf9. We compare restoration of CMS-S by Rf9 with restoration of CMS-S by Rf3 in F1 progeny. We have also analyzed the DNA organization of, and the transcripts from, the CMS-S-associated region of the mitochondrial genome in the presence of Rf9 and have discovered a novel mechanism by which a nuclear restorer gene affects CMS-S.
MATERIALS AND METHODS
Plant material:
Table 1 lists the inbred lines referred to in this study, the families to which they belong, and the cultivars from which they are derived. The Lancaster Surecrop inbred lines may be classified as pure Lancaster (first cycle), Oh40B-derived, and/or C103-derived inbred lines. All of the second-cycle and later Lancaster Surecrop inbred lines used in this study can be traced back to either one or both of two source inbred lines, Oh40B and C103 (Gerdes and Tracy 1993). Additional inbred lines used in this study trace back to other cultivars (Table 1). The pedigrees of all but four of the inbred lines in Table 1—CE1, SK2, LH38 and M825—can be found in Gerdes et al. (1993). CE1 is a Pioneer inbred line whose pedigree involves a mixture of Golden King, Golden Jewel, and Minnesota 13 (M. Albertsen, personal communication). SK2 is an old Pioneer-developed inbred line. It results from a cross of two inbred lines that were both self-pollinated from the Krug open-pollinated variety (D. Duvick, personal communication). LH38 derives from the cross A619 × L120 (Anonymous 1987). M825 is a starchy version of R825. R825 is a shrunken2 line that was recovered from the Iowa sugary1 line 5125 (J. Laughnan, personal communication).
TABLE 1.
Sources of the inbred lines referred to in this study
| Cultivar | Family | Inbred lines |
|---|---|---|
| Lancaster Surecrop | Pure Lancaster Surecrop | C103; C.I.4-8a; L289a; L317a; Oh40B |
| C103 family of Lancaster Surecrop | C123; H107; H108; H109; Mo17b; Va58 | |
| Oh40B family of Lancaster Surecrop | A619b; H95b; H98; H99b; H107; H108; H109; Hi32b; LH38b; Oh43b; Oh45b; Oh545b; Pa83b; Pa91b; Pa869; Pa870; SD59; Va26b | |
| Reid Yellow Dent | Funk Yellow Dent | 38-11; Ill A; M14 |
| Indiana Station strain | W64A; Wf9 | |
| Krug | SK2; Mo17 | |
| Iowa Stiff Stalk Synthetic | B37; B73 | |
| Troyer strain | Trc | |
| Minnesota #13 | — | CE1c; W23 |
| Pride of Saline | — | K55 |
| Boone County White | — | Ky21c |
| Miscellaneous | — | C.I.21Ec; M825; N6; Oh51A |
An inbred line may derive from more than one source.
This line derives from Richey Lancaster (selected from Lancaster Surecrop) (Troyer 1999).
This line also contains some Reid Yellow Dent germplasm (Gerdes et al. 1993).
Inbred line of genotype Rf3/Rf3.
Maize genetics nomenclature:
In maize genetics nomenclature (http://www.maizegdb.org/maize_nomenclature.php), loci are indicated in lowercase italics (e.g., the rf3 locus). Alleles at a locus are also indicated in italics, with the first letter capitalized for dominant alleles (e.g., the Rf3 allele). Fertility restoration of CMS-S is gametophytic (Buchert 1961); i.e., restorer-of-fertility alleles act in haploid pollen. Due to the gametophytic mode of restoration in CMS-S, dominance of the Rf3 allele cannot be directly assessed. However, Kamps et al. (1996) demonstrated that Rf3 is dominant to rf3 through the use of diploid pollen shed by tetraploid plants. Thus, Rf denotes the restoring allele and rf the nonrestoring allele. On the basis of the work presented below, we adopt the designation Rf9 for the restoring allele characterized in A619 although its dominance has not been determined.
The normal male-fertile cytoplasm of maize is given the designation N, while S male-sterile cytoplasm is designated CMS-S or, briefly, S. Since there have been numerous independent “discoveries” of S-type cytoplasms, subgroups or subtypes of CMS-S have been given unique letter designations (Beckett 1971; Sisco et al. 1985; reviewed by Gabay-Laughnan et al. 1995). The subtypes of CMS-S used in this study are S, I, ML, R, and VG. Male-sterile subtypes were originally given unique designations because they derived from different sources and their relationships were unknown (Beckett 1971). These subtypes were originally classified as belonging to the S group by their restoration to fertility in certain inbred nuclear backgrounds (Beckett 1971). The classifications were later confirmed in studies involving mtDNA analyses as well as restorer-of-fertility alleles (Sisco et al. 1985).
Test for the presence of restoring alleles:
CMS-S male-sterile plants were crossed with pollen from the Lancaster Surecrop inbred lines being tested. The resulting plants are male sterile if the inbred line being tested carries no restorers. The resulting plants are male fertile if the line being tested carries one or more restoring alleles. Due to the gametophytic nature of CMS-S restoration (Buchert 1961), the presence of one restoring allele results in F1 (Rf rf) plants with 50% collapsed (aborted) pollen while the presence of two unlinked restorers results in an F1 with only 25% collapsed pollen. The majority of the Lancaster Surecrop inbred lines were recurrently crossed as pollen parents to a CMS-S nonrestoring inbred line at least five times before they were scored for Table 2. H98 and L317 were each crossed four times while Oh40B was crossed three times. C.I.4-8 and L289 were scored in the F1.
TABLE 2.
Pedigrees of 25 Lancaster Surecrop inbred lines and the restoration of CMS-S to fertility
| Inbred line | Pedigreea | Fertility ratingb | Source of restorer(s), if present |
|---|---|---|---|
| A619 | [(A171 × Oh43) Oh43] | 4–5 | Oh40B |
| C103 | Lancaster Surecrop (from Noah Hershey) | 4 | C103 |
| C123 | C102 × C103 | 4–5 | C103 |
| C.I.4-8 | Lancaster Surecrop | 1 | — |
| H95 | Oh43 × C.I.90A | 4 | Oh40B |
| H98 | Oh45 × Hy | 1 | — |
| H99 | Illinois Synthetic 60C (USDA Blight Resistant Double Double × B8, Ia55:1473, M14, Oh43, Oh45, Oh51A, R160, and R168) | 6c | Oh40B + unknown |
| H107 | [(H99 × H98) H99] | 4–5 | Oh40B |
| H108 | [(Mo17 × H99) Mo17] | 1 | — |
| H109 | [(Mo17 × H99) Mo17] | 1 | — |
| Hi32 | [(Oh545 × MV source) Oh 545 (5)] | 4 | Oh40B |
| L289 | Lancaster Surecrop | 1 | — |
| L317 | Lancaster Surecrop | 1 | — |
| LH38 | A619 × L120 | 4 | Oh40B |
| Mo17 | C.I.187-2 × C10 | 1 | |
| Oh40B | Eight-line composite of Lancaster Surecrop lines | 5 | Oh40B |
| Oh43 | W8 × Oh40B | 4–5 | Oh40B |
| Oh45 | W8 × Oh40B | 4–5 | Oh40B |
| Oh545 | [(M14 × C.I.187-2) Oh45 (2)] × [(Oh45T4 × Cash Synthetic) × ((M14 × C.I.187-2) × Oh45 (2)) × Oh45A] | 1 | — |
| Pa91 | (Wf9 × Oh40B) S4 × [(38-11 × L317) 38-11] S4 | 1 | — |
| Pa869 | 75F-5 × Pa83 | 1 | — |
| Pa870 | 75F-5 × Oh43 | 1 | — |
| SD59 | [(A619 × SD316W) A619] (white conversion of A619) | 1 | — |
| Va26 | Oh43 × K155 | 1 | — |
| Va58 | [C103 × T8 (2)] × [C.I.21E × C103 (2)] | 6c | C103 + C.I.21E |
All pedigrees from Gerdes et al. (1993), except LH38 (Anonymous 1987).
Fertility rating 1 equals sterile; rating 4–6 equals fertile (see materials and methods)
This inbred line carries two restorers.
Test for allelism of two restorers:
If a CMS-S plant is homozygous for a single restoring allele, 100% of the pollen is starch filled and functional. Therefore, once it is determined that two inbred lines each carries a single S-restoring allele, we can determine whether the two restorers are allelic by pollen examination of an F1 plant carrying both restorers. If an F1 carries two allelic restorers from different sources, 100% of the pollen is also starch filled. A CMS-S plant heterozygous for a single restorer exhibits 50% collapsed (aborted) pollen. Note that, in the case of an F1 between two restorers in which one of the two restorers fails to function, plants also exhibit 50% pollen abortion. In contrast, a CMS-S plant heterozygous for two unlinked restorers that are both expressed exhibits only 25% pollen abortion. The percentage of pollen abortion will be <25% if the two restorers being tested are closely linked and will approach 0% in the case of tightly linked restorer loci. Pollen abortion was determined in the field by examining pollen with a ×40 pocket microscope. Fresh pollen from newly exserted, undehisced anthers was dispensed onto a black background (modified from Phillips 1994). Starch-filled pollen grains appeared white, while aborted grains were shriveled and yellow.
Analysis of fertility restoration in F1 progeny:
For the determination of how well Rf9 restores fertility in F1 progeny (Table 3), CMS-S male-sterile inbred plants were crossed once with pollen from the inbred line A619. In the case of the nonrestoring inbred L317, A619 plants with CMS-VG mitochondria were crossed with pollen from the line. For the determination of how well Rf3 restores fertility in F1 hybrids (Table 3), CMS-S male-sterile inbred plants were crossed with pollen from the inbred lines of genotype Rf3/Rf3 (Tr and/or CE1). Alternatively, the CMS-S versions of these restoring inbreds were crossed with pollen from a nonrestoring inbred line. In some cases, Mo17 Rf3-converted strains were used as a source of Rf3 (also derived from CE1 and/or Tr).
TABLE 3.
Comparative restoration of CMS-S to fertility in rf/Rf F1 hybrids
| Subtype of CMS-S | Rf9 fertility ratinga | Rf3 fertility rating | |
|---|---|---|---|
| Lancaster Surecrop inbred lines | |||
| H98 | I | 5 | 6 |
| ML | 5 | 6 | |
| H109 | VG | 1–2 | 6 |
| L317 | S | NDb | 6 |
| VGc | 1 | NAd | |
| Mo17 | S | 4–5 | 6 |
| Oh545 | ML | 4–5 | 6 |
| Pa869 | S | 4–5 | 6 |
| Pa870 | S | 4 | 6 |
| SD59 | ML | 5 | 6 |
| Va26 | S | 3–4 | 6 |
| Non-Lancaster inbred lines | |||
| B37 | S | 2–3 | 6 |
| B73 | R | 3–4 | 6 |
| Ill A | S | 2–4 | 6 |
| K55 | I | 3–4 | 6 |
| M14 | S | 2–3 | 4–6 |
| M825 | ML | 2–3 | 6 |
| N6 | S | 1–2 | 4–5 |
| Oh51A | R | 3–5 | 6 |
| SK2 | I | 5–6 | 6 |
| W23 | VG | 2–3 | 6 |
| W64A | ML | 2–4 | 6 |
| VG | 2–3 | NDb | |
| Wf9 | ML | 3 | 6 |
Source of Rf9 allele is inbred line A619.
ND, not determined.
Inbred line L317 crossed as pollen parent onto CMS-VG A619.
NA, not applicable.
The degree of fertility in the resulting plants was ranked on a scale of 1–6 using a modification of Beckett's scale (Beckett 1971). The ratings are as follows: (1) male sterile, no anthers exserted; (2) sterile anthers exserted, usually less than half the total number, no pollen shed; (3) sterile anthers exserted, usually the entire number, no pollen shed; (4) partially fertile anthers exserted, some pollen shed; (5) slightly subnormal anthers, usually a mixture of fertile and sterile anthers, good pollen; (6) normal anthers, fully fertile, excellent pollen. Thus a rating of 1–3 means that the plant is functionally male sterile while a rating of 4–6 means that the plant sheds at least some pollen and is thus functionally male fertile.
We initially ranked fertility restoration in the Midwest in July and also in Hawaii in January. Due to our preliminary findings (see results), restoration by Rf9 was later scored exclusively in the winter nursery (Molokai, Hawaii).
Genetic tests to locate rf9:
The waxy1 (wx1)-marked reciprocal translocation series was used in an attempt to map rf9 to the chromosome arm. These translocation lines did not carry restorer alleles for S cytoplasm. The use of wx1-marked reciprocal translocations to ascertain the chromosome location of restorer alleles was as previously described (Laughnan and Gabay 1978; Laughnan and Gabay-Laughnan 1994). Briefly, CMS-S plants carrying an unplaced restorer allele are crossed with nonrestoring pollen from plants carrying a wx1-marked translocation. CMS-S plants heterozygous for a restorer exhibit 50% aborted pollen grains due to gametophytic fertility restoration. Because maize plants heterozygous for a reciprocal translocation also exhibit 50% pollen abortion (Patterson 1994), CMS-S plants heterozygous for both a restorer allele and a reciprocal translocation exhibit 75% aborted pollen. Such plants are crossed as males onto wx1/wx1 tester plants. The proportion of waxy kernels on the resulting ears is a function of the recombination between the restorer and wx1. Chi-square values for independence of the restorer and wx1 alleles were calculated using Yates' correction for continuity (Yates 1934).
Isolation and gel blot analysis of mitochondrial DNA and RNA:
Mitochondria were isolated by differential centrifugation as described previously (Newton and Coe 1986; Newton 1994) from either ear shoots or emerging tassels prior to the time of pollen shedding. Mitochondria were treated with proteinase K just prior to lysis with sarkosyl and mtDNA extraction. In some cases, the samples were split into two aliquots and one was not treated with proteinase K. mtDNA restriction digestions, electrophoresis, and blotting onto membrane (Hybond N+, Amersham) followed standard procedures (Sambrook et al. 1989). RNAs were extracted from mitochondria, electrophoresed, and blotted as described previously (Karpova et al. 2002). A 0.24- to 9.5-kb RNA ladder (BRL) was used for molecular size standards. DNA and RNA gel blots were hybridized with 32P-labeled DNA probes or riboprobes. Recombinant plasmids containing mtDNA fragments in the pBluescript II KS− vector (Stratagene) were used for riboprobe preparation (Ausubel et al. 1992). In some experiments, orf355 for DNA probes was PCR amplified from CMS-S mtDNA using the specific primers orf355F1 and orf355R2 (TCAGTATAGAGTCGGGGTACACTC and CAATCCACTCATCGCAGCAGGA). Probes were hybridized to DNA gel blots at 65° and to RNA gel blots in 50% formamide at 45°, followed by stringent washes.
Primer extensions of mitochondrial RNA:
To determine the 5′-ends of three RNAs originating within a TIR sequence, mtRNA samples were reverse transcribed using the following primers: EKPR5-5′-CAGTGTCTCTCGTGTGTAAGAG, annealing to nucleotides (nt) 237–216 downstream from the TIR start in the 4.2-kb R-repeat sequence and also to nt 116–95 of the published M1 plasmid sequence (Grace et al. 1994) and EKPR16-5′-CTGTCAGTGGAGG-ATACTTC (nt 237–218 of the published S2 plasmid sequence; Levings and Sederoff 1983) corresponding to S2-orf1. Aliquots of 5–40 μg mtRNA were typically processed per standard protocols (Ausubel et al. 1992), except that the hybridization was performed for 1 hr at 65° and for 1 hr more at gradually decreasing (65°–35°) temperature in the following solution: 750 mm KCl, 50 mm Tris–HCl, pH 7.8, 10 mm EDTA (Rapp and Stern 1992). 32P-end-labeled extended primers were electrophoresed through 6% sequencing gels along with the sequencing reactions carried out with the same primer for the corresponding cloned mtDNA regions.
RESULTS
Identification of CMS-S restoring alleles in Lancaster Surecrop germplasm:
In addition to the seven inbred lines known to be of genotype Rf3/Rf3, a number of Lancaster Surecrop lines also carry CMS-S restorer alleles. We screened a total of 25 Lancaster Surecrop inbreds to determine those that restore CMS-S. Seven of these lines have already been reported to partially restore S cytoplasm while two have been shown to be maintainers of CMS-S; that is, they carry no S restorers (Beckett 1971; Gracen and Grogan 1974; Gabay-Laughnan and Laughnan 1994). In this study, 10 of the 25 Lancaster Surecrop inbred lines were shown to be maintainers of CMS-S. These are the lines with a fertility rating of 1 in Table 2. Twelve of the Lancaster Surecrop inbred lines tested restored CMS-S to fertility. These are the lines with fertility ratings of 4–6 in Table 2. Ten of these inbred lines carried a single restoring allele, including C103 and C123. C123 traces its pedigree back to C103 (Gerdes and Tracy 1993; see also Table 2), and C103 is assumed to be the source of the restoring allele in these two lines. The remaining eight restoring inbred lines carrying a single restorer are Oh40B and the seven inbred lines A619, H95, H107, Hi32, LH38, Oh43, and Oh45 tracing their pedigrees back to Oh40B (Gerdes and Tracy 1993; see also Table 2). Oh40B is assumed to be the source of the restoring allele carried by each of these lines. Two Lancaster Surecrop-derived lines, H99 and Va58, produce plants with only 25% collapsed pollen in F1 crosses to S-male-sterile plants. Therefore, H99 and Va58 each carry two nonallelic restorers. From the pedigree of Va58 (Table 2) it can be deduced that one of these restorers is Rf3 from C.I.21E and the other restorer is from the Lancaster Surecrop C103 line. The H99 pedigree is more complicated. While one of the restorers comes from either Oh43 or Oh45, the source of the second restorer cannot be determined.
The restorers carried by C103 and Oh40B are allelic or tightly linked:
As described above, either one of the two pure Lancaster first-cycle inbred lines C103 or Oh40B was the source of the restorers identified in the Lancaster Surecrop inbreds that were tested for S restoration (Table 2). Therefore, we made a series of crosses to test allelism of the restorers carried by these two lines. A version of C103 with S-type cytoplasm as the female parent was crossed by pollen from both the Oh40B line and the A619 inbred line. In addition, the CMS-S version of C123 as the female parent was crossed by pollen from the Oh40B line, and the CMS-S version of Oh43 was crossed by pollen from the A619 line. All the resulting F1 plants produced pollen that was 95–100% starch filled. Thus, the restorers carried by the two sources, Oh40B and C103, appear to be allelic, although the possibility of tightly linked loci cannot be ruled out. As mentioned above, assuming allelism, we have named this gene restorer of fertility9 (rf9).
Expression of Rf9 and Rf3 in various inbred nuclear-cytoplasmic combinations:
CMS-S has been discovered in different maize varieties, and the independent sources are referred to as “subtypes” (see materials and methods). Rf3 is an excellent restorer of all subtypes of CMS-S in nearly all F1 progeny tested. Two exceptions are the inbred lines M14 and N6. When subtype S in M14 or in N6 is crossed by a source of Rf3, the F1 progeny are not restored to full fertility (see Table 3), although pollen is usually shed. Because Rf3 does fully restore this subtype in other nuclear backgrounds (Table 3), the nuclear, rather than the cytoplasmic, genotype appears to be responsible for the modifications in Rf3-mediated fertility restoration.
In contrast to restoration by Rf3, restoration by Rf9 is variable. S male-sterile inbred lines were crossed with pollen from the Rf9-carrying line A619. Twenty-three nuclear-cytoplasmic combinations were crossed with inbred A619 as the source of Rf9 (Table 3). Ten of these combinations were with Lancaster Surecrop inbred lines. With the exception of H109 and L317, the F1 progeny were male fertile. Most of the CMS-S lines restored byRf9 are those classified as Lancaster Surecrop inbred lines (Table 3). Interestingly, the highest level of restoration was seen for the Rf9-restored F1 hybrid of CMS-I SK2, a non-Lancaster line.
Nuclear genotype is a factor in restoration by Rf9 (see Table 3). For example, Rf9 is a good restorer of subtype S in Mo17 but does not restore subtype S in N6. Additionally, Rf9 is an excellent restorer of subtype I in SK2, a good restorer of I in H98, but only a partial restorer of I in K55. Restoration by Rf9 of subtype ML in Oh545 is good, but restoration of ML in W64A and in Wf9 is not. Thus, the inbred nuclear background is responsible for the observed differences in the levels of restoration.
Expression of Rf9 under different temperatures and photoperiods:
In our studies, restoration of CMS-S by Rf9 was found to be sensitive to the environment. Over years of observations, Rf9 has been a consistently better restorer in Hawaii in January than it is in Illinois and Missouri in July and August. On Molokai in January, the average high temperature is 25.2°, the average low temperature is 17.8° (http://visitmolokai.com/molokai-climate.html), and the day length is ∼11 hr. Under these conditions, A619 CMS-VG plants are routinely male fertile, with a fertility rating of 5 (see materials and methods). In contrast, such plants are usually male sterile (fertility rating of 1–2) in July and August in Champaign-Urbana, Illinois, and in Columbia, Missouri. During this time, the day length is ∼15 hr and the temperatures commonly reach 30°–35°. Is Rf9 thermosensitive or photoperiod sensitive or both?
The relatively mild summer of 2004 allowed us to distinguish between temperature and photoperiod as the cause of the observed fertility differences between CMS-VG A619 plants grown in Hawaii and those grown in the Midwest. In the summer of 2004, maximum temperatures for June, July, and August were all below average in Champaign-Urbana. Both early (April 28) and later (May 24) sown seed of A619 CMS-VG produced plants that shed pollen, rating 5 on the fertility scale. That summer there were only 5 days total—2 in June, 3 in July, and none in August—with high temperatures of 32°–33° (All Illinois weather data are available at http://www.isws.illinois.edu/atmos/statecli/cuweather/.) July 2004 was the fifth coolest July on record for maximum temperatures. The average daily high temperature was 27.7°, ranging from 22.2° to 32.2°. The average low temperature was 17.4°, ranging from 11.7° to 22.8°.
In contrast to 2004, the summer of 2005 in the Midwest was consistently hot. Temperatures for June, July, and August were all above average. The A619 CMS-VG plants (seed were sown May 24, 2005) in Champaign-Urbana, Illinois, were male sterile, rating 1–2 at maturity (late July–early August). The average high temperature equaled or exceeded 30.0° for all of June, July, and the first 13 days of August. The low temperatures averaged 17.6° in June, 18.6° for July, and 20.0° for the first 13 days of August.
These data indicate that day length had no effect and that temperature was the major factor affecting Rf9 restoration. Our data do not allow us to pinpoint the developmental stage at which Rf9 is sensitive to the heat because temperatures during the growing season in the summers of 2004 and 2005 were either consistently low or consistently high. Additionally, the temperature effect is specific to the interaction of Rf9 with S-type cytoplasm because the fertility of the inbred A619 line itself (N cytoplasm, genotype Rf9/Rf9) is unaffected by high temperatures. No corresponding environmental differences have been observed in the case of Rf3 restoration of CMS-S.
The rf9 locus does not map near rf3:
Since the rf3 locus maps to the long arm of chromosome 2, and because there is a cluster of additional CMS-S restorers, as well as two restorers for CMS-T, in this region (Gabay-Laughnan and Chase 2004), the restoring allele Rf9 as carried by the Lancaster Surecrop inbred line A619 was tested for linkage to 2L. Using the translocation wx1 T2-9d, with breakpoints at 2L.83 and 9L.27, the data for the rf9 locus were compared with the testcross data obtained in the mapping of the rf3 locus (Gabay-Laughnan et al. 2004).
Representative testcross data for the rf9 locus produced recombination values between the restorer and wx1 locus ranging from 41.1% to 48.0% for six different ears. In contrast, using the same wx1 T2-9d translocation, recombination values between rf3 (from four inbred-line sources) and wx1 ranged from 14.5% to 23.6% (Gabay-Laughnan et al. 2004). The Rf9 allele as carried by the Lancaster Surecrop inbred lines C103, C123, H95, Oh40B, Oh43, and Oh45 was also tested for possible linkage to 2L. The results were similar to those obtained with the restorer from A619. Therefore, rf9 and rf3 are genetically distinct loci.
Attempts to place rf9 onto other chromosome arms have been unsuccessful thus far. Many F1 combinations between waxy1-marked translocations and A619 were not fertile and, therefore, could not be tested. We now understand why this is the case. As we have demonstrated, the Rf9 allele is temperature sensitive. Many of the Rf9/translocation F1's were grown in the Midwest during relatively hot summers before we knew of the temperature sensitivity of Rf9. In addition, we found that some inbred lines inhibit the expression of Rf9. Two such lines are the maize inbreds W23 and M14 (Table 3). The majority of the wx1-marked translocation stocks are maintained in W23, M14, and/or a W23/M14 F1. Thus, attempts to map rf9 in Hawaii also failed.
Fertility restoration by Rf9 decreases linearization of the main CMS-S mitochondrial genome:
The 4.2-kb (4215-bp) R repeat within the main CMS-S mitochondrial genome carries the sterility-associated orf355/orf77 region (Figure 1A). Sequences almost identical to the TIR of the S plasmids reside near one end of the repeat (reviewed by Gabay-Laughnan et al. 1995). Recombination between the plasmid TIR and the homologous regions within the main mitochondrial genome leads to linearization as diagrammed in Figure 1A. XhoI cuts outside of the S-specific repeat; therefore, digests with this enzyme and hybridization with an orf355 probe allow us to distinguish each of the copies (Figure 1B). Because the CMS-S mitochondrial genome has been completely sequenced (GenBank accession no. DQ490951; Allen et al. 2007), the sizes of the restriction fragments can be accurately inferred. Linear ends near orf355/orf77 appear to predominate in CMS-S mtDNA.
Linear ends from the CMS-S main mitochondrial genome originating by recombination with the S plasmids should be protected by the same TIR-associated protein(s) that stabilize the ends of the S-plasmids. If terminal proteins protecting the linear ends are not removed by proteinase K treatment during the mtDNA isolations, restriction fragments attached to the proteins are not detected on following gel electrophoresis (Kemble and Thompson 1982). Comparisons of CMS-S mtDNA samples that were treated with proteinase K (Figure 1C, left) with those that were not treated with proteinase K (Figure 1C, right) show us which fragments are located at linear ends.
BamHI cuts within the 4.2-kb repeat after the orf355/orf77 sequences. Following digests of CMS-S mtDNA and hybridization with an orf355-specific probe (Figure 1C, lane B, left and right), 4.8- and 6.6-kb BamHI fragments are detected, corresponding to the integrated σ (cox1-near) and ψ (cox2-near) copies, respectively. The terminal BamHI 1.8-kb (1722-bp) fragment is seen only when proteinase K was used during the mtDNA preparation (Figure 1C, left). It includes both isoforms of the linear ends (*σ′ and *ψ′ in Figure 1B). XhoI cuts outside of the S-specific repeat; therefore, digests with this enzyme allow us to distinguish more versions of the 4.2-kb repeat (Figure 1C, lane X, left and right). The 9.4-kb XhoI fragment (Figure 1C, left) corresponds to the protein-bound ψ′ linear end (*ψ′), whereas the 4.3-kb fragment corresponds to the σ′ linear end (*σ′). These bands are not observed in the absence of proteinase K treatment (Figure 1B). With proteinase K treatment, the 1.8-kb BamHI and the 9.4- and 4.3-kb XhoI fragments are all major bands (Figure 1B, left side). Thus, in the B37 nonrestoring nuclear background, most of the CMS-S main mitochondrial genome is in linear form with the ends protected by a terminal protein.
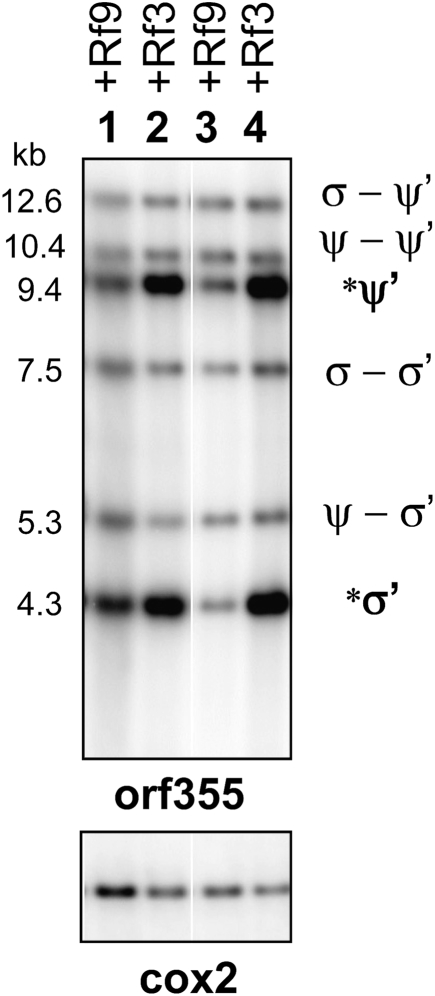
When CMS-S in the inbred B37 line is crossed by the Rf3-containing inbred lines Ky21 and Tr, the ratios of the linearized-to-integrated 4.2-kb S-specific repeats remain high (Figure 2, lanes 2 and 4). In contrast, when the CMS-S plants are crossed by the Rf9-containing A619 inbred line, there is a decrease in linearized repeat copies in the F1 progeny (Figure 2, lane 1). This decrease in linearized copies can be seen also for the CMS-S subtype VG after repeated backcrosses to A619 (Figure 2, lane 3).
Figure 2.—
The presence of Rf9 causes a reduction in the linearized copies of the orf355/orf77 sequences. mtDNAs from ear shoots were digested with XhoI and gel blots were hybridized with an orf355-specific probe. VG and S are different subtypes of CMS-S. Nuclear genotype B37 carries recessive alleles for both rf3 and rf9. A619 carries the dominant Rf9 restorer, whereas Ky21 and Tr carry the dominant Rf3 restorer. (Lane 1) S-B37 × A619 (Rf9/rf9; rf3/rf3). (Lane 2) S-B37 × Ky21 (rf9/rf9; Rf3/rf3). (Lane 3) VG-A619 (Rf9/Rf9; rf3/rf3). (Lane 4) S-B37 × Tr (rf9/rf9; Rf3/rf3).
Rf9 restores fertility by a different mechanism than does Rf3:
We compared the expression patterns of the orf355-orf77 region of CMS-S plants in the absence and presence of the Rf3 and Rf9 restorer genes (Figure 3). Mitochondrial RNA was isolated from emerging tassels (and thus enriched for microspore mitochondria) of sterile plants carrying CMS-S cytoplasm in the B37 inbred nuclear background and from fertility-restored F1 hybrids with either A619 (Rf9) or Ky21 (Rf3). In CMS-S-sterile B37 plants, an orf355-specific probe hybridizes to two major mitochondrial transcripts of 2.8- and 1.6 kb (Figure 3, lane S; see also Zabala et al. 1997). In F1 hybrids produced by crossing CMS-S in inbred B37 with Ky21 (Rf3/rf3), all of the RNAs are severely reduced and new bands, including a 2.1-kb RNA, appear (Figure 3, +Rf3). In contrast, in the F1 hybrids with A619 (Rf9/rf9), a decrease in the level of the 1.6-kb transcript is the only difference observed (Figure 3, +Rf9). Transcripts from cox1 and the S2 plasmid are unaltered by the presence of either restorer.
Figure 3.—
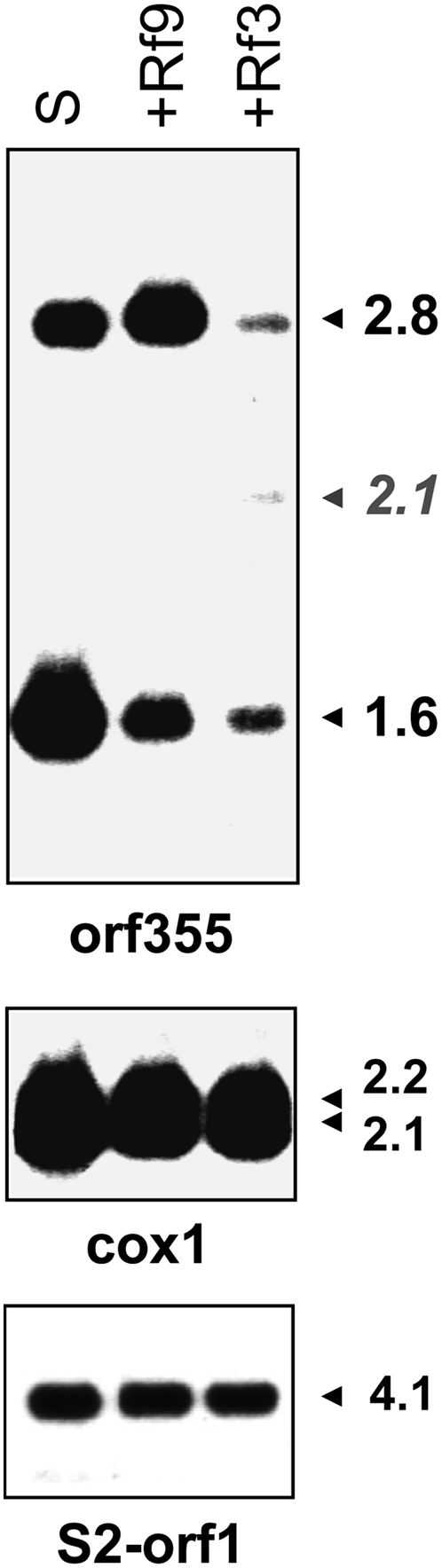
Rf9 mediates a specific reduction of the CMS-associated 1.6-kb RNA. Analysis of mtRNAs isolated from preemergent tassels of (left) S-B37 (rf9/rf9; rf3/rf3) compared with (middle) S-B37 × A619 (Rf9/rf9; rf3/rf3) and (right) S-B37 × Ky21(rf9/rf9; Rf3/rf3). The blot was first hybridized with the orf355-specific DNA probe and then with a cox1 probe. A duplicate blot was probed with the S2 plasmid orf1 control probe.
The presence of Rf3 is correlated with cleavage of all orf355/orf77-containing transcripts (Zabala et al. 1997; Wen and Chase 1999; Xiao et al. 2006). Our evidence corroborates previous findings and further shows that the site of cleavage occurs approximately two-thirds of the way into the reading frame of orf355 (ear shoot data; supporting information, Figure S1). The 5′ products of the processing events could be degraded by a 3′-5′ exonuclease. Faint hybridization signals at 2.1 kb (Figure S1) and at ∼1 kb (see Wen and Chase 1999) could be remnant 5′ cleavage products resulting from the 2.8- and 1.6-kb RNAs, respectively. An abundant 0.6-kb RNA (Figure S1) is the stable 3′ processing product from both the 2.8- and 1.6-kb RNAs.
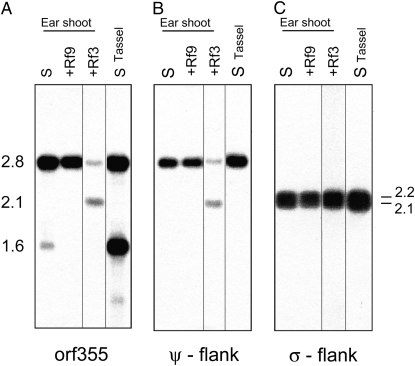
Expression of the 1.6-kb RNA is highest in CMS-S microspores (Wen and Chase 1999; Xiao et al. 2006), and high levels of this RNA can also be detected in mitochondria from total tassels as they emerge from the leaves (Figure 3). Levels of the 1.6-kb RNA are very low in other tissues, including ear shoots (Figure 4A). CMS-S plants show no defects other than male sterility; therefore, low levels of the 1.6-kb RNA are compatible with normal mitochondrial function.
Figure 4.—
The 2.8-kb transcript originates in the ψ region flanking one copy of the 4.2-kb repeat. RNA gel blot analysis of mitochondrial RNA isolated from ear shoots and preemergent tassels. (A–C) Lane S, S-B37 (rf9/rf9; rf3/rf3); lane +Rf9, B37S × A619 (Rf9/rf9; rf3/rf3); and lane +Rf3, S-B37XKy21 (rf9/rf9; Rf3/rf3). The same blot was hybridized sequentially with the orf355-specific probe (A), the ψ-specific probe (B), and the σ-specific probe cox1 (C).
Rf9 mediates a specific reduction of the CMS-associated 1.6-kb mRNA in contrast to Rf3, which causes a reduction of all transcripts containing orf355 sequences (Figure 3 and Figure 4A). Hybridizing mtRNA blots with sequences upstream of the integrated versions of orf355/orf77 shows that the 2.8-kb transcript originates in the ψ region between the cox2 gene and the 4.2-kb repeat (Figure 4B). Rf9 does not affect the levels of the 2.8-kb RNA. The σ region upstream of the other integrated copy of the 4.2-kb repeat contains the cox1 gene; hybridizing probes from this region identify only cox1 transcripts (Figure 4C). Neither Rf9 nor Rf3 affects the cox1 RNAs.
Mapping of the 5′-end of the CMS-specific transcript:
Since the abundance of the CMS-S-associated 1.6-kb RNA is reduced by both the Rf3 and Rf9 nuclear restorers, it is the apparent target for the downregulation imposed by these alleles. Unlike Rf3, which abolishes the overlapping 2.8- and 1.6-kb RNAs, the Rf9 restorer acts specifically to reduce the 1.6-kb RNA. This transcript has a 5′-end located within the TIR of the 4.2-kb R-repeat sequences. The TIR of the R-repeat recombines with an S1 or S2 plasmid TIR to linearize the CMS-S genome (see Figure 1). Thus, the end of a linearized version of the 4.2-kb repeat resembles the ends of linear mitochondrial plasmids: the S, R, or M plasmids.
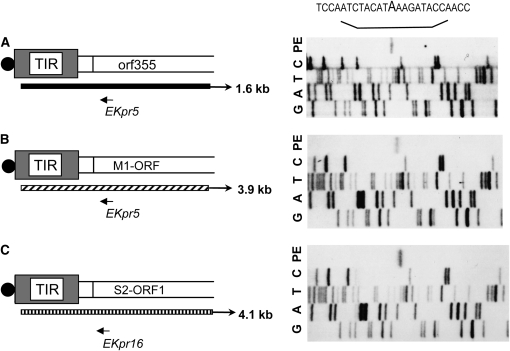
Traynor and Levings (1986) mapped the 5′-ends of S-plasmid transcripts. All transcripts from intact TIRs of both S-1 and S-2 initiate at nucleotide 32, including the 4.1-kb transcript from the S2 plasmid. We conducted primer extension analyses to test the hypothesis that the 5′-end of the 1.6-kb RNA maps to the same nucleotide location. We determined the 5′-end of the 1.6-kb RNA by reverse transcription from a primer within the R1-similar region of orf355 (Figure 5A). This region does not exist on the S1 and S2 plasmids; therefore, in CMS-S, the primer is specific for orf355-containing transcripts. The same primer could be used for the extension of the 3.9-kb transcript (Grace et al. 1994) from the R1-like M1 plasmid from Mazoti's isolate of Z. luxurians (Figure 5B). The 5′ part of this M1 transcript is highly homologous to the CMS-S-specific 1.6-kb RNA. In a control reaction, the 4.1-kb S2 transcript was extended with an S2-orf1-specific primer (Figure 5C). All three RNAs, the S2-orf1 transcript, the M1 transcript, and the CMS-S-specific 1.6-kb RNA, have identical 5′-termini that map to position 32 of the TIR sequence. Thus, transcription appears to initiate at exactly the same nucleotide within the TIR both for the CMS-associated R-repeat region and the linear plasmids. This result suggests that transcription from the TIR promoter may require a specific structural organization of the promoter-containing region, i.e., a linear TIR end. Our results are consistent with the hypothesis that the CMS-S-specific 1.6-kb transcript initiates only at the plasmid-like linear ends.
Figure 5.—
The 1.6-kb CMS-S-specific transcript appears to initiate within the TIR of a linearized CMS-S mitochondrial genome at the same nucleotide as do the linear plasmid-specific transcripts. The 5′-ends of TIR-initiating transcripts were determined by primer extension (in lane PE in A–C). (A) The 1.6-kb transcript end determined with a primer, EKPR5, from the R1-similar region of orf355. (B) The 3.9-kb M1 plasmid-specific transcript end determined using the same primer. (C) The 4.1-kb S2-plasmid-specific transcript end determined with the S2-orf1-specific primer EKPR16. The DNA sequence ladders (in lanes G, A, T, and C in A–C) were generated using the same primers with the corresponding clones of the integrated versions of R1, S2, and orf355. The common start site (boldface A) for all three RNAs within the TIR sequence is indicated.
DISCUSSION
The “standard” CMS-S restorer, Rf3, maps to the long arm of chromosome 2 (Laughnan and Gabay 1978; Kamps and Chase 1997; Gabay-Laughnan et al. 2004). An unlinked S restorer, designated Rf9, is present in some Lancaster Surecrop-derived inbred lines, including A619, the source of Rf9 for our genetic and molecular studies. In this study, 25 Lancaster Surecrop inbred lines were tested or retested for the presence of the Rf9 restoring allele. Twelve of them were determined to carry at least one S restorer. Two of the lines carried restoring alleles at two independent loci, whereas 10 of them were proven to carry an S restoring allele at a single locus. The inbreds C103 and Oh40B are the ultimate progenitors of these 10 lines. Because we demonstrated that the restorers carried by C103 and Oh40B are allelic or very closely linked, these 10 lines probably all carry the same restoring allele, Rf9.
The CMS-S F1 progeny of crosses between nonrestoring (rf/rf) inbred lines and the restorers Rf3 and Rf9 were compared. Rf3 was an excellent restorer in nearly all F1's tested. In contrast to Rf3, Rf9 restoration of CMS-S was more complex. Rf9 was generally a good restorer for CMS-S in Lancaster Surecrop lines; only two of the Lancaster Surecrop F1 progeny (CMS-S Rf9/rf9) shed no pollen. Restoration was generally poorer and more variable when Rf9 was introduced into non-Lancaster lines carrying S-type cytoplasm. We demonstrated that the observed differences are due to inbred nuclear background and not CMS-S subtype.
A dominant or gametophytic inhibitor of Rf9 appears to be present in 3 of 21 tested inbred lines but absent from 8 others. The remaining 10 lines analyzed exhibited variable fertility, suggesting that modifiers may be present. These proposed inhibitors and modifiers require further analysis. It is possible for the CMS-S version of an inbred line to carry the Rf9 (restoring) allele yet fail to exhibit fertility due to the presence of an inhibitor. Thus, a Lancaster Surecrop inbred line, whose CMS-S version has a fertility rating of 1, may in fact be of the genotype Rf9/Rf9.
CMS inhibitors are not without precedent. An inhibitor has recently been identified and mapped in the maize CMS-C-Rf system (Hu et al. 2006). Rf4 and Rf5 are dominant genes, each of which is capable of restoring CMS-C to fertility. Rf4 has the capability of restoring C cytoplasm in all inbred backgrounds tested. Rf5 is able to restore only those inbred versions of CMS-C lacking the dominant inhibitor Rf-I; i.e., Rf5 fails to restore CMS-C in the presence of Rf-I. This inhibitor does not affect restoration of C cytoplasm by Rf4 (Hu et al. 2006).
In contrast to Rf3, Rf9 was determined to be a thermosensitive restorer of CMS-S. When the daily high temperatures during the growing period were consistently <28°, CMS-S plants carrying Rf9 exhibited male fertility. When the daily maximum temperatures were higher, regularly >30°, plants of the same genotype were essentially male sterile. Rf9 is the first reported thermosensitive restorer-of-fertility allele for CMS. However, thermosensitive genic male sterility has been observed in maize, rice, and wheat (Wang et al. 1995; Xing et al. 2003; Tang et al. 2006).
We compared the effects of Rf9 and Rf3 on the organization and expression of the CMS-S-specific mitochondrial region. The actions of these two different nuclear restorer-of-fertility genes reduce the expression of the CMS-S-associated region by different mechanisms. The previously described restorer, Rf3, does indeed condition the cleavage of all orf355/orf77-containing transcripts (as initially suggested by Zabala et al. 1997). Thus, Rf3 is similar to other known restorers of fertility that act to reduce levels of RNA post-transcriptionally (see Chase 2007). In contrast, the Rf9 restorer causes a specific reduction in the amount of the CMS-associated 1.6-kb transcript by decreasing the abundance of its linearized transcription template.
The Rf3 allele induces RNA cleavage within the orf355 coding region (Figure S1) and causes accumulation of a 0.6-kb RNA, which is the 3′ processing product of both the 2.8- and 1.6-kb transcripts. The 0.6-kb RNA contains part of orf355 and all of the orf77 sequences and could be very stable due to a stem-loop structure at its 3′-end. This 3′ stem-loop probably also accounts for the stability of the 1.6-kb sterility-associated RNA (Xiao et al. 2006). Xiao et al. (2006) described a different Rf3-induced cut site within a potential stem-loop structure only 9 nt from the 5′-end of the 1.6-kb RNA (Xiao et al. 2006). This could reflect a secondary action of Rf3 because it can clearly affect multiple sites; Wen and Chase (1999) have shown that Rf3 also affects transcript patterns of cytochrome b and ATPase subunit 6. Our data strongly suggest that the primary cause of Rf3-mediated restoration is the processing event that occurs two-thirds of the way into the orf355 region of the 1.6-kb transcript.
Xiao et al. (2006) proposed that the 2.8-kb transcript is cleaved to generate the 1.6-kb RNA. However, Zabala et al. (1997) have shown that cytoplasmic revertant plants that have lost S plasmids have no 1.6-kb RNA, although they still accumulate the 2.8-kb RNA. Thus, the 2.8-kb transcript is not processed to the 1.6-kb RNA. The promoter of the 2.8-kb RNA lies in the ψ region that flanks one copy of the integrated 4.2-kb R-repeat, and it shares no sequence similarity (supporting information, Figure S2) with the promoter for the 1.6-kb transcript. Our data indicate that the 2.8- and 1.6-kb RNAs are transcribed separately from different promoters.
The 1.6-kb RNA is produced only when either (or both) of the 4.2-kb repeats is linearized by recombination with S plasmids, resulting in the acquisition of a plasmid-like terminal structure just upstream of orf355. The 5-end of the 1.6-kb transcript corresponds exactly to the 5′-ends of TIR-initiated S-plasmid transcripts. These observations strongly suggest that the 1.6-kb transcript is initiated from the plasmid promoter within a linear TIR, apparently by the same mechanism as transcripts of S plasmids. We hypothesize that the S2-plasmid-encoded RNA polymerase (Zabala et al. 1987; Kuzmin et al. 1988) is required for the expression of the CMS-specific 1.6-kb RNA. Terminal protein(s) bound to the TIR 5′-ends might also be required for transcription of these RNAs.
The primary effect of Rf9 is to decrease the copy number of linear mtDNA molecules containing orf355/orf77 at their termini. These linearized subgenomes in CMS-S mitochondria originate from recombination between linear S plasmids and circular mtDNAs. Rf9 might act similarly to the bean CMS restorer Fr (He et al. 1995; Janska et al. 1998), downregulating the stoichiometry of the specific “progenitor” circular subgenomes. Rf9 might affect recombination directly; specific nuclear mutations and backgrounds have been reported to alter mitochondrial recombination and, consequently, mitochondrial genome organization (Kuzmin et al. 2005; Zaegel et al. 2006; Shedge et al. 2007). Also, a nuclear factor could affect the stability of the linearized mitochondrial genomes.
In CMS-S male-sterile anthers, the developing pollen grains abort during the starch-filling stage, and the 1.6-kb transcript is abundant from the prevacuolate microspore stage until pollen collapse (Wen and Chase 1999). Rf3 reduces the amount of the 1.6-kb transcript much more effectively than does Rf9, allowing for full fertility. Rf9 appears to reduce the amount of 1.6-kb RNA in developing microspores to a level that allows for pollen maturation only under optimal conditions. The effect of modifiers could be explained if other nuclear factors affect the stability of the RNA or the availability of the template for its production. A threshold amount of a deleterious product could account for the thermosensitivity of fertility restoration with Rf9. Further studies are needed to evaluate the possibilities.
Acknowledgments
For supplying the inbred lines used in this study, we thank J. B. Beckett, University of Missouri; C. D. Chase, University of Florida; D. N. Duvick, Pioneer Hi-Bred International; M. W. Johnson, Pennsylvania State University; R. J. Lambert, University of Illinois; E. B. Patterson, University of Illinois; E. C. Rossman, Michigan State University; B. Zehr, Purdue University; R. Holden, Holden Foundation Seeds; D. Cochran, Illinois Foundation Seeds; as well as the Ohio Agricultural Research and Development Center and the Purdue Agricultural Alumni Seed Improvement Association. The wx1-marked translocations were supplied by the Maize Genetics Cooperation Stock Center. We thank Natalia Lavrova and Janet Day Jackson for assisting in early aspects of these studies and Louis Meyer for help with later experiments. We thank Ashley Lough and Tiffany Langewisch for comments on the manuscript. We are indebted to James Allen for discussions and assistance. This work was supported by grants from the National Science Foundation (K.J.N.) and the U. S. Department of Agriculture (S.G.-L.). Additional support was provided by Illinois Foundation Seeds (to S.G.-L.). Support for J.M.M was provided by the University of Missouri Life Sciences Undergraduate Research Opportunity Program.
This article is dedicated to Donald N. Duvick, who passed away on May 23, 2006. Don shared our interest in cytoplasmic male sterility, restorer-of-fertility genes, and nonchromosomal stripe mutations.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.099895/DC1.
References
- Allen, J. O., C. M. Fauron, P. Minx, L. Roark, S. Oddiraju et al., 2007. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177 1173–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous, 1987. Genetics Handbook. Mike Brayton Seeds, Ames, IA.
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1992. Short Protocols in Molecular Biology. John Wiley & Sons, New York.
- Beckett, J. B., 1971. Classification of male-sterile cytoplasms in maize (Zea mays L.). Crop Sci. 11 724–727. [Google Scholar]
- Buchert, J. G., 1961. The stage of the genome-plasmon interaction in the restoration of fertility to cytoplasmically pollen-sterile maize. Proc. Natl. Acad. Sci. USA 47 1436–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, C. D., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23 81–90. [DOI] [PubMed] [Google Scholar]
- Duvick, D. N., 1956. Allelism and comparative genetics of fertility restoration of cytoplasmically pollen sterile maize. Genetics 41 544–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick, D. N., 1957. Allelism of FR genes of inbreds which restore pollen fertility to WF9s. Maize Genet. Coop. Newsl. 31 114. [Google Scholar]
- Duvick, D. N., 1965. Cytoplasmic pollen sterility in corn. Adv. Genet. 13 1–56. [Google Scholar]
- Gabay-Laughnan, S., and C. D. Chase, 2004. CMS-S restorers of fertility from multiple sources cluster on chromosome 2L. Maize Genet. Coop. Newsl. 78 67–68. [Google Scholar]
- Gabay-Laughnan, S. J., and J. R. Laughnan, 1994. Male sterility and restorer genes in maize, pp. 418–423 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Gabay-Laughnan, S., G. Zabala and J. R. Laughnan, 1995. S-type cytoplasmic male sterility in maize, pp. 395–432 in The Molecular Biology of Plant Mitochondria, edited by C. S. Levings III and I. K. Vasil. Kluwer Academic, Dordrecht, The Netherlands.
- Gabay-Laughnan, S., C. D. Chase, V. M. Ortega and L. M. Zhao, 2004. Molecular-genetic characterization of CMS-S restorer-of-fertility alleles identified in Mexican maize and teosinte. Genetics 166 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, J. T., and W. F. Tracy, 1993. Pedigree diversity within the Lancaster Surecrop heterotic group of maize. Crop Sci. 33 334–337. [Google Scholar]
- Gerdes, J. T., J. G. Behr, J. G. Coors and W. F. Tracy, 1993. Compilation of North American maize breeding germplasm. Crop Science Society of America, Madison, WI.
- Grace, K. S., J. O. Allen and K. J. Newton, 1994. R-type plasmids in mitochondria from a single source of Zea luxurians teosinte. Curr. Genet. 25 258–264. [DOI] [PubMed] [Google Scholar]
- Gracen, V., and C. Grogan, 1974. Diversity and suitability for hybrid production of different sources of cytoplasmic male sterility in maize. Agron. J. 66 654–657. [Google Scholar]
- He, S., A. Lyznik and S. MacKenzie, 1995. Pollen fertility restoration by nuclear gene Fr in CMS bean: nuclear-directed alteration of a mitochondrial population. Genetics 139 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchins, J. P., H. Ginsburg, M. Rohrbaugh, R. M. Dale, C. L. Schardl et al., 1986. DNA sequence analysis of a 5.27-kb direct repeat occurring adjacent to the regions of S-episome homology in maize mitochondria. EMBO J. 5 2781–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. M., J. H. Tang, H. Yang, H. L. Xie, X. M. Lu et al., 2006. Identification and mapping of Rf-I, an inhibitor of the Rf5 restorer gene for Cms-C in maize (Zea mays L.). Theor. Appl. Genet. 113 357–360. [DOI] [PubMed] [Google Scholar]
- Janska, H., R. Sarria, M. Woloszynska, M. Arrieta-Montiel and S. A. MacKenzie, 1998. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10 1163–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps, T. L., and C. D. Chase, 1997. RFLP mapping of the maize gametophytic restorer-of-fertility locus (rf3) and aberrant pollen transmission of the nonrestoring rf3 allele. Theor. Appl. Genet. 95 525–531. [Google Scholar]
- Kamps, T. L., D. R. McCarty and C. D. Chase, 1996. Gametophyte genetics in Zea mays L.: dominance of a restoration-of-fertility allele (Rf3) in diploid pollen. Genetics 142 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova, O. V., E. V. Kuzmin, T. E. Elthon and K. J. Newton, 2002. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble, R. J., and R. D. Thompson, 1982. S1 and S2, the linear mitochondrial DNAs present in a male sterile line of maize, possess terminally attached proteins. Nucleic Acids Res. 10 8181–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin, E. V., I. V. Levchenko and G. N. Zaitseva, 1988. S2 plasmid from cms-S-maize mitochondria potentially encodes a specific RNA polymerase. Nucleic Acids Res. 16 4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin, E. V., D. N. Duvick and K. J. Newton, 2005. A mitochondrial mutator system in maize. Plant Physiol. 137 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughnan, J. R., and S. J. Gabay, 1978. Nuclear and cytoplasmic mutations to fertility in S male-sterile maize, pp. 427–446 in Maize Breeding and Genetics, edited by D. B. Walden. John Wiley & Sons, New York.
- Laughnan, J. R., and S. Gabay-Laughnan, 1994. The placement of genes using waxy-marked reciprocal translocations, pp. 255–257 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Levings, C. S., III, and R. R. Sederoff, 1983. Nucleotide sequence of the S-2 mitochondrial DNA from the S cytoplasm of maize. Proc. Natl. Acad. Sci. USA 80 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings, C. S., III, B. G. Kim, D. R. Pring, M. F. Conde, R. J. Mans et al., 1980. Cytoplasmic reversion of cms-S in maize: association with a transpositional event. Science 209 1021–1023. [DOI] [PubMed] [Google Scholar]
- Lonsdale, D. M., T. P. Hodge and C. M. Fauron, 1984. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 12 9249–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale, D. M., T. Brears, T. P. Hodge, S. E. Melville and W. H. Rottmann, 1988. The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity. Philos. Trans. R. Soc. Lond. B. 319 149–163. [Google Scholar]
- Newton, K. J., 1994. Procedures for isolating mitochondria and mitochondrial DNA and RNA, pp. 549–556 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Newton, K. J., and E. H. Coe, Jr., 1986. Mitochondrial DNA changes in abnormal growth (nonchromosomal stripe) mutants of maize. Proc. Natl. Acad. Sci. USA 83 7363–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K. J., S. Gabay Laughnan and R. DePaepe, 2004. Mitochondrial mutations in plants, pp. 121–142 in Plant Mitochondria: From Genome to Function, edited by D. A. Day, A. H. Millar and J. Whelan. Kluwer Academic, Dordrecht, The Netherlands.
- Paillard, M., R. R. Sederoff and C. S. Levings, III, 1985. Nucleotide sequence of the S-1 mitochondrial DNA from the S cytoplasm of maize. EMBO J. 4 1125–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, E. B., 1994. Translocations as genetic markers, pp. 361–363 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Phillips, R. L., 1994. Classification of pollen abortion in the field, pp. 297–298 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Pring, D. R., C. S. Levings, III, W. W. Hu and D. H. Timothy, 1977. Unique DNA associated with mitochondria in the “S”-type cytoplasm of male-sterile maize. Proc. Natl. Acad. Sci. USA 74 2904–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp, W. D., and D. B. Stern, 1992. A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J. 11 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schardl, C. L., D. M. Lonsdale, D. R. Pring and K. R. Rose, 1984. Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature 310 292–296. [Google Scholar]
- Shedge, V., M. Arrieta-Montiel, A. C. Christensen and S. A. MacKenzie, 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisco, P. H., V. E. Gracen, H. L. Everett, E. D. Earle, D. R. Pring et al., 1985. Fertility restoration and mitochondrial nucleic acids distinguish at least five subgroups among Cms-S cytoplasms of maize (Zea mays L.). Theor. Appl. Genet. 71 5–15. [DOI] [PubMed] [Google Scholar]
- Tang, J. H., Z. Y. Fu, Y. M. Hu, J. S. Li, L. L. Sun et al., 2006. Genetic analyses and mapping of a new thermo-sensitive genic male sterile gene in maize. Theor. Appl. Genet. 113 11–15. [DOI] [PubMed] [Google Scholar]
- Traynor, P. L., and C. S. Levings, III, 1986. Transcription of the S-2 maize mitochondrial plasmid. Plant Mol. Biol. 7 255–263. [DOI] [PubMed] [Google Scholar]
- Troyer, A. F., 1999. Background of US hybrid corn. Crop Sci. 39 601–626. [Google Scholar]
- Wang, B., W. W. Xu, J. Z. Wang, W. Wu, H. G. Zheng et al., 1995. Tagging and mapping the thermosensitive genic male-sterile gene in rice (Oryza sativa L.) with molecular markers. Theor. Appl. Genet. 91 1111–1114. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Y. Zou, X. Li, Q. Zhang, L. Chen et al., 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissinger, A. K., D. H. Timothy, C. S. Levings, III, W. W. Hu and M. M. Goodman, 1982. Unique plasmid-like mitochondrial DNAs from indigenous maize races of Latin America. Proc. Natl. Acad. Sci. USA 79 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissinger, A. K., D. H. Timothy, C. S. Levings, III and M. M. Goodman, 1983. Patterns of mitochondrial DNA variation in indigenous maize races of Latin America. Genetics 104 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, L., and C. D. Chase, 1999. Pleiotropic effects of a nuclear restorer-of-fertility locus on mitochondrial transcripts in male-fertile and S male-sterile maize. Curr. Genet. 35 521–526. [DOI] [PubMed] [Google Scholar]
- Xiao, H., F. Zhang and Y. Zheng, 2006. The 5′ stem-loop and its role in mRNA stability in maize S cytoplasmic male sterility. Plant J. 47 864–872. [DOI] [PubMed] [Google Scholar]
- Xing, Q. H., Z. G. Ru, C. J. Zhou, X. Xue, C. Y. Liang et al., 2003. Genetic analysis, molecular tagging and mapping of the thermo-sensitive genic male-sterile gene (wtms1) in wheat. Theor. Appl. Genet. 107 1500–1504. [DOI] [PubMed] [Google Scholar]
- Yates, F., 1934. Contingency tables involving small numbers and the χ2 test. J. R. Stat. Soc. 1( Suppl.): 217–235. [Google Scholar]
- Zabala, G., C. O'Brien-Vedder and V. Walbot, 1987. S2 episome of maize mitochondria encodes a 130-kilodalton protein in male sterile and fertile plants. Proc. Natl. Acad. Sci. USA 84 7861–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, G., S. Gabay-Laughnan and J. R. Laughnan, 1997. The nuclear gene Rf3 affects the expression of the mitochondrial chimeric sequence R implicated in S-type male sterility in maize. Genetics 147 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaegel, V., B. Guermann, M. Le Ret, C. Andres, D. Meyer et al., 2006. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 18 3548–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]