The heart must adapt its mechanical activity to the prevailing hemodynamic demands. When an increased demand is brought about by sustained stimuli such as growth and development, pressure overload, or mutations in sarcomeric proteins, the heart will typically undergo an increase in size caused by myocyte hypertrophy. Underlying this hypertrophic response is the coordinated interaction of distinct signaling modules capable of transmitting and executing modifications in gene expression that lead to alterations in myocyte physiology and long-term cardiac adaptation.1
One of the more intriguing characteristics of the hypertrophic response is that despite the ability of a wide variety of both pathologic and physiologic stimuli to induce cardiac hypertrophy, distinct cytoplasmic signaling cascades that initiate changes in gene expression converge on a common set of nuclear factors. These factors will then transactivate or repress cardiac genes through cis-regulatory elements. One of these transcription factors, myocardin, that appears to be capable of relaying hypertrophic signals to the genome is the subject of the studies performed by Xing et al2 detailed below.
An important hypertrophic transcriptional regulator is the zinc finger– containing transcription factor, GATA4. GATA4 is widely expressed and has been identified as a pivotal regulator of developmental and stress-induced changes in cardiac gene expression.3,4 GATA4 has been shown to activate numerous genes in the heart by divergent signaling molecules including protein kinase C, calcineurin, and members of the mitogen-activated protein kinase signaling axis.1 Although GATA4-null embryos arrest before birth,5 a point mutation in GATA4 results in septation and coronary vasculature defects,6 whereas GATA4 mutations are linked to congenital heart defects in humans.7 GATA4 has also been shown to activate a cardiac enhancer/promoter construct in nonmuscle cells.8
Another central transcriptional regulator of cardiac gene expression is serum response factor (SRF), which is also widely expressed in many cell types and binds to a DNA consensus sequence known as the CArG (CC[A/T]6GG) box.9,10 SRF belongs to the MADS (MCM1, Agamous, Deficiens, SRF) family of transcription factors that includes myocyte enhancer factor 2 (MEF2).11 Highlighting the importance of SRF is the fact that many cardiac muscle genes require CArG boxes for proper expression,12 and disruption of SRF DNA binding by cardiac overexpression of SRF that contains 2 mutations located within the domain important for SRF dimerization leads to an early postnatal death (12 days) from a severe dilated cardiomyopathy.13 In addition, cardiac-specific ablation of SRF is embryonic lethal because of cardiac insufficiency,14 whereas cardiac overexpression of wild-type SRF results in hypertrophic cardiomyopathy.15 Apart from its role in cardiac development and hypertrophy, SRF also mediates the expression of numerous growth-inducible developmental genes and is critical for smooth muscle development.16 Moreover, activation of SRF is induced by numerous hypertrophic signals, many of which overlap with GATA4, including calcineurin-dependent signaling17 and the alpha-adrenergic agonist, phenylephrine (PE).18,19 In fact, GATA4 can interact with SRF, and it has been suggested that they may act synergistically through recruitment of each other to bind DNA20,21
More recently, a novel and potent SRF transcriptional coactivator, myocardin, was discovered and subsequently shown to transactivate CArG box-dependent cardiac promoters. 22 Myocardin belongs to the SAP (scaffold-attachment factor A/B, Acinus, PIAS) domain family of nuclear proteins, which are implicated in chromatin remodeling.23 It has been shown to be necessary and sufficient for both cardiac and smooth muscle differentiation.22,24 Interestingly, GATA4 modulates myocardin transcriptional activity either positively on genes such as Nkx2.5 or negatively such as ANF.21 Moreover, forced expression of myocardin can induce both cardiac and smooth muscle genes in nonmuscle cells.25,26 As mentioned above, GATA4 can activate a transfected cardiac promoter but its ability to activate endogenous genes is unknown. Although GATA4 and SRF null mice have early developmental myocardial defects, myocardin null mice show no myocardial defects at midgestation. However, the role of myocardin in the adult heart has not yet been evaluated because of embryonic lethality caused by vascular abnormalities. 27 It will be necessary to evaluate adult cardiac-specific ablation of myocardin before it is possible to determine its role in the adult heart. Inconsistent with the observation in mice is the finding that morpholino knockdowns of myocardin in Xenopus embryos inhibit cardiac development, so future experiments need to be aimed at these differences in the two species.26
In this issue of Circulation Research, Xing et al2 address and confirm a role for myocardin in the hypertrophic response of cardiac myocytes and in the intact heart. In this study, both myocardin mRNA and protein were upregulated by two hypertrophic agonists, fetal bovine serum or PE, in cultured cardiac myocytes. In two mouse models of cardiac hypertrophy, thoracic aortic banding and calcineurin overexpression, myocardin expression levels in the heart were significantly upregulated with concomitant elevations in fetal genes. Finally, the translation of these data to human diseased hearts was validated by demonstrating increased myocardin expression in protein extracts from patients with idiopathic dilated cardiomyopathy. The necessity of myocardin in the hypertrophic response was supported by studies in which myocardin was introduced into cardiac myocytes by adenoviral delivery and subsequently exhibited a hypertrophic phenotype. In contrast, cardiac myocytes expressing a dominant negative myocardin that maintains the ability to interact with SRF but lacks the transcriptional activity domain showed a small decrease in size and were unable to respond to hypertrophic stimuli. The authors demonstrated an additional level of myocardin regulation by the attenuation of myocardin-induced hypertrophy by histone deacetylase, HDAC5.
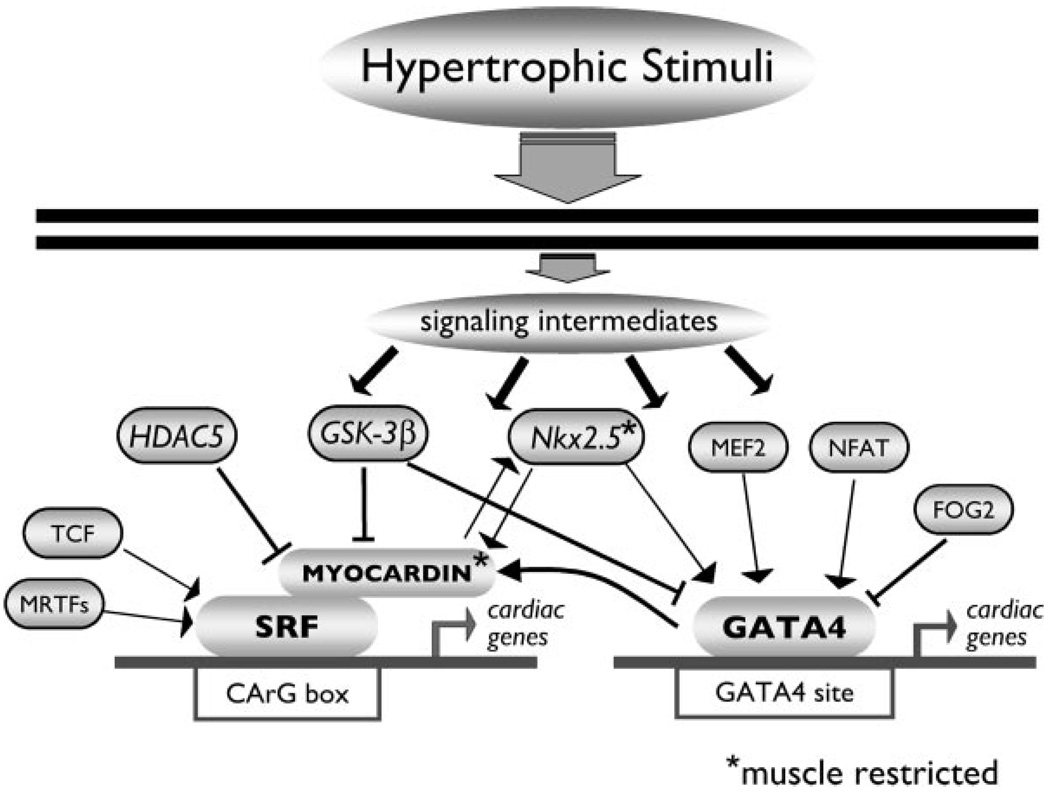
Given that hypertrophic stimuli initiate a reexpression of fetal gene isoforms in the heart28 and that myocardin is necessary for cardiac development in Xenopus, it is tempting to conclude that myocardin causes the reappearance of these fetal genes in the pathological heart. However, the developing heart does not progress to a decompensated state as occurs with persistent pathological stimuli such as hypertension. This fact alone highlights the importance of temporal activation of these signaling intermediates and the necessity for additional “players” that may impart selectivity to gene target activation by posttranslational modification of specific transcription factors and cofactors by intracellular signals. For example, it was recently shown that the transcriptional activity of myocardin is regulated by site-specific phosphorylation by glycogen synthase kinase-3β.29 A schematic of the transcriptional regulatory network is illustrated in the Figure. Another looming question is whether myocardin mediates hypertrophy attributable to alternative physiological hypertrophic stimuli such as exercise. Consequently, the emerging picture is that, although distinct hypertrophic and developmental stimuli appear to converge on common transcription factors such as GATA4 and SRF, activation of these factors does not result in indiscriminate expression of GATA4 or SRF-dependent genes. Simultaneous activation of cofactors is required but, in addition, there are many indications that one or more additional levels of regulation exist that allow a cell to further distinguish between different stimuli.
Figure 1.
Schematic representation of GATA4- and SRF-dependent gene regulation. Hypertrophic stimuli initiate a signaling cascade that converges on the transcriptional regulators GATA4 and SRF in the heart. The activation and/or repression of gene expression by GATA4 and SRF depend on the interaction with specific cofactors that are spatially and temporally activated. MRTF indicates myocardin-related transcription factor; TCF, ternary complex factor; HDAC5, histone deacetylase 5; GSK-3β, glycogen synthase kinase 3β; MEF2, myocyte enhancer factor 2; NFAT, nuclear factor of activated T-cells; FOG2, friend of GATA2.
Acknowledgments
This work was supported by NIH grant to L.A. Leinwand (HL 56510) and by a National Research Service Awards from the NIH awarded to J.P. Konhilas (F32 HL 70509).
Footnotes
Reprints: Information about reprints can be found online at http://www.lww.com/static/html/reprints.html
References
- 1.Molkentin JD, Dorn GW., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 2.Xing W, Zhang TC, Cao D, Wang Z, Antos CL, Li S, Wang Y, Olson EN, Wang DZ. Myocardin induces cardiomyocyte hypertrophy. Circ Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- 3.Liang Q, Molkentin JD. Divergent signaling pathways converge on GATA4 to regulate cardiac hypertrophic gene expression. J Mol Cell Cardiol. 2002;34:611–616. doi: 10.1006/jmcc.2002.2011. [DOI] [PubMed] [Google Scholar]
- 4.Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 6.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001;15:839–844. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 8.Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 9.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 10.Sprenkle AB, Murray SF, Glembotski CC. Involvement of multiple cis elements in basal- and alpha-adrenergic agonist-inducible atrial natriuretic factor transcription. Roles for serum response elements and an SP-1-like element. Circ Res. 1995;77:1060–1069. doi: 10.1161/01.res.77.6.1060. [DOI] [PubMed] [Google Scholar]
- 11.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 12.Reecy J, Belaguli NS, Schwartz RJ. SRF/Homeobox protein interactions. In: Rosenthal RHaN., editor. Heart Development. San Diego: Academic Press; 1998. pp. 273–290. [Google Scholar]
- 13.Zhang X, Chai J, Azhar G, Sheridan P, Borras AM, Furr MC, Khrapko K, Lawitts J, Misra RP, Wei JY. Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J Biol Chem. 2001;276:40033–40040. doi: 10.1074/jbc.M104934200. [DOI] [PubMed] [Google Scholar]
- 14.Niu Z, Yu W, Zhang SX, Barron M, Belaguli NS, Schneider MD, Parmacek M, Nordheim A, Schwartz RJ. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J Biol Chem. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Azhar G, Chai J, Sheridan P, Nagano K, Brown T, Yang J, Khrapko K, Borras AM, Lawitts J, Misra RP, Wei JY. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280:H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, Nadal-Ginard B. Molecular cloning and sequence analysis of smooth muscle calponin. J Biol Chem. 1991;266:13284–13288. [PubMed] [Google Scholar]
- 17.Lin K, Wang D, Sadee W. Serum response factor activation by muscarinic receptors via RhoA. Novel pathway specific to M1 subtype involving calmodulin, calcineurin, and Pyk2. J Biol Chem. 2002;277:40789–40798. doi: 10.1074/jbc.M202745200. [DOI] [PubMed] [Google Scholar]
- 18.Cheng G, Hagen TP, Dawson ML, Barnes KV, Menick DR. The role of GATA, CArG, E-box, and a novel element in the regulation of cardiac expression of the Na+-Ca2+ exchanger gene. J Biol Chem. 1999;274:12819–12826. doi: 10.1074/jbc.274.18.12819. [DOI] [PubMed] [Google Scholar]
- 19.Morimoto T, Hasegawa K, Kaburagi S, Kakita T, Wada H, Yanazume T, Sasayama S. Phosphorylation of GATA-4 is involved in alpha 1-adrenergic agonist-responsive transcription of the endothelin-1 gene in cardiac myocytes. J Biol Chem. 2000;275:13721–13726. doi: 10.1074/jbc.275.18.13721. [DOI] [PubMed] [Google Scholar]
- 20.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J Biol Chem. 2002;277:25775–25782. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 21.Oh J, Wang Z, Wang DZ, Lien CL, Xing W, Olson EN. Target genespecific modulation of myocardin activity by GATA transcription factors. Mol Cell Biol. 2004;24:8519–8528. doi: 10.1128/MCB.24.19.8519-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 25.van Tuyn J, Knaan-Shanzer S, van de Watering MJ, de Graaf M, van der Laarse A, Schalij MJ, van der Wall EE, de Vries AA, Atsma DE. Activation of cardiac and smooth muscle-specific genes in primary human cells after forced expression of human myocardin. Cardiovasc Res. 2005;67:245–255. doi: 10.1016/j.cardiores.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadal-Ginard B, Mahdavi V. Molecular basis of cardiac performance. Plasticity of the myocardium generated through protein isoform switches. J Clin Invest. 1989;84:1693–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badorff C, Seeger FH, Zeiher AM, Dimmeler S. Glycogen synthase kinase 3beta inhibits myocardin-dependent transcription and hypertrophy induction through site-specific phosphorylation. Circ Res. 2005;97:645–654. doi: 10.1161/01.RES.0000184684.88750.FE. [DOI] [PubMed] [Google Scholar]