Abstract
The microRNA miR-7 is perfectly conserved from annelids to humans, and yet some of the genes that it regulates in Drosophila are not regulated in mammals. We have explored the role of lineage restricted targets, using Drosophila, in order to better understand the evolutionary significance of microRNA-target relationships. From studies of two well characterized developmental regulatory networks, we find that miR-7 functions in several interlocking feedback and feedforward loops, and propose that its role in these networks is to buffer them against perturbation. To directly demonstrate this function for miR-7, we subjected the networks to temperature fluctuation and found that miR-7 is essential for the maintenance of regulatory stability under conditions of environmental flux. We suggest that some conserved microRNAs like miR-7 may enter into novel genetic relationships to buffer developmental programs against variation and impart robustness to diverse regulatory networks.
Keywords: microRNA, Drosophila, photoreceptor, robustness, networks, development
Introduction
Biological systems are imbued with the property of robustness. Perturbation of such systems is buffered such that the output or response is invariant or uniform. Numerous examples abound in which robust systems can compensate for remarkably large genetic or environmental perturbations (Kitano, 2004). How this occurs is not well understood and is currently the focus of intense study. Buffering is thought to be an epigenetic process, and it has been speculated to play a role in evolution by canalizing or masking genetic variation at the level of phenotypic expression (Meiklejohn and Hartl, 2002; Siegal and Bergman, 2002). A variety of mechanisms provide stability and robustness (Hartman et al., 2001). Functional redundancy buffers processes against genetic and environmental noise (Kitano, 2004). Complex networks of interacting regulatory molecules also generate robustness for diverse biological processes (Lee et al., 2002; Milo et al., 2002; Spirin and Mirny, 2003).
In this study, we examine the role of microRNAs (miRNAs) in biological robustness. These small non-coding RNAs are transcribed from plant, algal, and animal genomes where their gene numbers range in the hundreds (Griffiths-Jones et al., 2006). Animal miRNAs typically repress translation of mRNAs that are complementary in sequence, and repression increases additively with miRNA occupancy on messages (Bushati and Cohen, 2007). Most targeted genes are only modestly repressed by miRNAs, which indicates that miRNAs primarily tune gene expression (Baek et al., 2008; Nakahara et al., 2005; Selbach et al., 2008).
It has been speculated that miRNAs provide robustness to programs of gene expression (Hornstein and Shomron, 2006). Stark and colleagues (Stark et al., 2005) observed anti-correlative expression of miRNAs and their target mRNAs. This suggests that transcription primarily controls gene expression while miRNAs lend further reinforcement to gene regulation by attenuating unwanted transcripts. MicroRNAs could provide robustness a second way. Feedback and feedforward motifs impart robustness to complex networks (Milo et al., 2002). Bioinformatic analysis has indicated that miRNAs frequently collaborate with transcription factors in feedback and feedforward loops to regulate their targets (Martinez et al., 2008; Tsang et al., 2007), and there are several experimentally defined examples of these kinds of regulatory relationships (Hobert, 2006). Despite these provocative speculations about miRNAs and robustness, to date there has been no direct evidence that a miRNA buffers gene expression against fluctuation or noise.
To explore the issue, we have focused on one of the most highly conserved animal miRNAs, miR-7. The miR-7 gene is found in most sequenced Urbilateria species, and the sequence of its mature miRNA product is perfectly conserved from annelids to humans (Prochnik et al., 2007). We find that in Drosophila, miR-7 acts within two complex gene networks that regulate the determination of photoreceptor cells, proprioceptor organs, and olfactory organs. MiR-7 acts within several interlocking feedback and feedforward loops theoretically implicated as network stabilizers. Thus, we provide a mechanistic picture of miR-7 working in networks to buffer gene expression against perturbation. To directly demonstrate this function for miR-7, we subjected the networks to temperature fluctuation and show that miR-7 is essential for stable gene expression and cell fate determination in the face of this perturbation. Thus, we have demonstrated that this miRNA imparts robustness to diverse regulatory networks.
Results
Novel functional and target acquisition by miR-7 during evolution
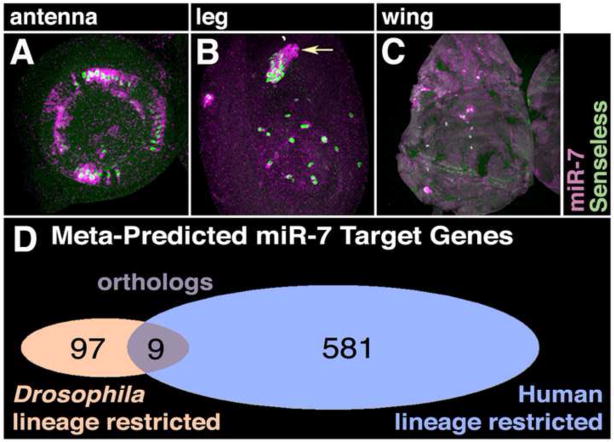
The mature miR-7 RNA sequence is perfectly conserved from annelid to human, indicating a strong functional conservation (Prochnik et al., 2007). In support of this notion, miR-7 is specifically expressed in neurosecretory cells of the vertebrate brain and in homologous cells of the annelid nervous system (Tessmar-Raible et al., 2007; Wienholds et al., 2005). In Drosophila, miR-7 is additionally expressed in retinal photoreceptor cells during post-embryonic development (Li and Carthew, 2005). Other Drosophila sensory organs also express miR-7, including proprioceptor and olfactory organs located on the antenna, leg, and wing (Fig. 1A–C). Strikingly, miR-7 is not expressed in the homologous sensory organs of vertebrates, implying that miR-7 function has differentially evolved (Landgraf et al., 2007; Wienholds et al., 2005). To examine the issue more closely, we focused on genes whose expression is regulated by miR-7 in developing sensory organs of Drosophila.
Figure 1. Non-conserved expression and targeting of miR-7.
(A–C) Localization of miR-7 RNA (purple) and Sens protein (green) in developing antenna (A), leg (B) and wing (C) discs. The miR-7 RNA is detected in the cytoplasm of proprioceptor and olfactory SOP cells, which are marked with Sens-positive nuclei. Comparable sensory organs in vertebrates do not express miR-7.
(D) Overlap of predicted miR-7 targets in Drosophila and human is limited to nine orthologous genes.
Expression of yan is inhibited by miR-7 in photoreceptor cells due to four miR-7-binding sites in its transcript 3′UTR (Li and Carthew, 2005). The E(spl) gene family are direct targets of miR-7 mediated repression in other sensory organs (Stark et al., 2003; Lai et al., 2005). Yan and E(spl) are direct targets of miR-7, and these factors are essential for development of insect sensory organs. Are their vertebrate orthologs also targets of miR-7? We compared the predicted miR-7 targets from Drosophila and humans using six different prediction algorithms. Based on this meta-analysis, 97 genes were predicted with high or moderate stringency to be miR-7 targets in Drosophila (Fig. 1D and STable 1). A total of 581 miR-7 targets were predicted with high or moderate stringency in humans (STable 2). We then compared the overlap between the two datasets, and observed that only 9 targets from both datasets were defined orthologs (STable 3). Strikingly, the mammalian orthologs of yan and E(spl) were not predicted to be targets of miR-7. Therefore, these miR-7 targets were either differentially acquired or lost in different evolutionary lineages.
Questions of robustness
We asked what function miR-7 played in regulating these non-conserved gene targets in Drosophila. We were not able to assay E(spl) protein expression. However, we had previously found that miR-7 mutants had only minor defects in Yan protein expression (Li and Carthew, 2005). Moreover, though miR-7 is expressed in developing sensory organs, loss of miR-7 had little or no detectable impact on their development under uniform laboratory conditions (Li and Carthew, 2005; data not shown). One possible explanation is that miR-7 is functionally redundant with other miRNAs. However, loss of all mature miRNAs within Dicer-1 clones had negligible effects on determination of these structures (T. Hayashi and R.W.C., unpublished data).
These results are consistent with miR-7 providing robustness to gene expression programs in development. It was especially intriguing to consider that this function could evolve in some animal lineages and not others. If robustness is a miR-7 function, we had two predictions. First, miR-7 would act in gene networks as a stabilizing factor. Second, miR-7 would prevent development from being perturbed when the environment of the animal was perturbed. We embarked on a systematic test of these two predictions.
The network controlling photoreceptor determination
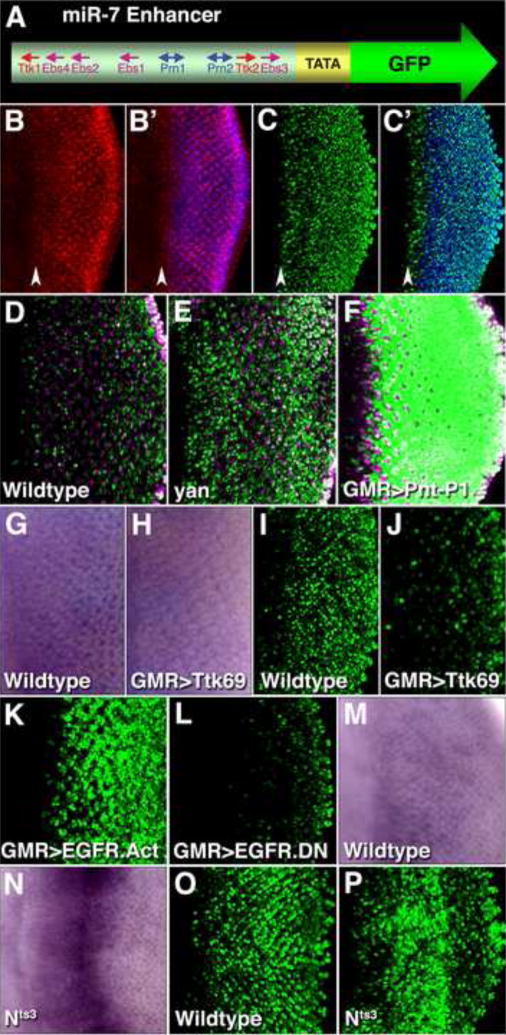
We initially focused on the interaction between miR-7 and yan, which encodes a transcription repressor (Voas and Rebay, 2004). We had identified a cluster of Yan-binding sites in DNA located 2 kb upstream of the miR-7 sequence (Li and Carthew, 2005 and SFig. 1). To show that the cluster acts as a miR-7 transcription enhancer, we placed it into a transgenic expression reporter (Fig. 2A), and observed strong reporter expression in photoreceptor cells and weak expression in their precursors (Fig. 2C,C′). This pattern resembled the endogenous miR-7 RNA expression pattern (Fig. 2B,B′). Therefore, the cluster behaves as a miR-7 transcription enhancer.
Figure 2. Regulation of miR-7 expression in photoreceptors.
(A) Schematic representation of the transgenic reporter for miR-7 enhancer activity in vivo. The enhancer contains binding sites for Ttk69 (Ttk1–2), Yan and Pnt-P1 (Ebs1–4), and Ato/Da (Prn1–2). The enhancer was placed upstream of a minimal promoter and nuclear GFP coding sequence.
(B,B′) Localization of miR-7 RNA (red) and Elav protein (blue) in a developing eye disc. The vertical red stripe in (B′) corresponds to cells in the morphogenetic furrow (arrowhead), which is the zone of R8 photoreceptor determination. To the right of this zone other photoreceptors are then determined, as marked by their expression of Elav.
(C,C′) miR-7 enhancer activity in a developing eye disc, as detected by the (miR-7)E>GFP reporter (green). Elav (blue) marks photoreceptor cells. The vertical green stripe in (C′) corresponds to cells in the morphogenetic furrow (arrowhead). Expression is weakly variegated, suggesting additional regulatory elements might be missing.
(D–F) miR-7 enhancer activity as detected by the reporter (green) in wildtype (D), yan1 (E), GMR≫Pnt-P1 (F) eye discs. Enhancer activity is stronger in yan1 and GMR≫Pnt-P1 precursor cells, which are not marked with Elav (purple). The GMR driver expresses genes (in this case Pnt-P1) in precursor and photoreceptor cells.
(G,H) miR-7 RNA detected by colorimetric in situ hybridization in wildtype (G), and GMR≫Ttk69 (H) eye discs.
(I,J) miR-7 enhancer activity as reported (green) in wildtype (I) and GMR≫Ttk69 (J) eye discs.
(K,L) miR-7 enhancer activity as reported (green) in GMR≫EGFR.Act (EGFR.λtop) (K) and GMR≫EGFR.DN (L) eye discs. These mutants drive constitutively active EGFR and dominant negative EGFR, respectively, in photoreceptors and their precursors.
(M–P) Wildtype (M and O), and Nts3 (N and P) larvae were shifted to the restrictive temperature (31°C) for 19 hrs before analysis. (M,N) miR-7 RNA detected by in situ hybridization. (O,P) miR-7 enhancer activity as detected by the reporter.
We next examined enhancer activity in a yan mutant. Enhancer activity was greatly increased in precursor cells, indicating that the enhancer is repressed by Yan in these cells (Fig. 2D,E). Yan competes with a transcription activator called Pnt-P1 for the same DNA-binding sites in enhancers (Flores et al., 2000; Xu et al., 2000). To determine if Pnt-P1 activates the miR-7 enhancer, we misexpressed Pnt-P1 in precursor cells and observed a tremendous increase in enhancer activity (Fig. 2F). Altogether, these data indicate that Yan and Pnt-P1 regulate the miR-7 enhancer in opposing directions.
Yan indirectly regulates two other transcription repressors, Ttk88 and Ttk69. Yan represses the transcription of phyllopod (phyl), which encodes an E3 ubiquitin ligase subunit that targets Ttk69 and Ttk88 proteins for degradation (Li et al., 1997; Tang et al., 1997; Treier et al., 1995). Thus, the presence of Yan stabilizes these repressors. We wondered if Yan might also act through these repressors to inhibit the miR-7 enhancer. Examination of the enhancer DNA sequence revealed two Ttk69 binding sites (Fig. 2A and SFig. 1). Misexpression of Ttk69 in photoreceptor cells led to decreased miR-7 RNA (Fig. 2G,H) and enhancer activity (Fig. 2I,J). Misexpression of Ttk88 in photoreceptor cells had no effect on miR-7 RNA expression, consistent with the absence of Ttk88 binding sites in the enhancer (data not shown). These data suggest that Ttk69 and not Ttk88 can bind to the miR-7 enhancer and repress its activity.
Retinal precursor cells receive extracellular signals that influence their fates. Yan plays a central role in transducing these signals (Voas and Rebay, 2004). To ascertain how these extracellular signals regulate the miR-7 enhancer, we used signaling mutants. When enhancer activity was monitored in precursor cells containing constitutively active EGFR, activity was strongly upregulated (Fig. 2I,K). Conversely, activity was greatly reduced in photoreceptor cells carrying a dominant-negative EGFR mutant (Fig. 2I,L). EGFR signaling activates Pnt-P1 synthesis and inhibits Yan by stimulating degradation of Yan protein (Voas and Rebay, 2004). Thus, EGFR signaling activates the miR-7 enhancer, most probably through its effects on Pnt-P1 and Yan.
We also determined how Notch signaling regulates the enhancer. We observed an increase in miR-7 expression in precursor cells carrying a temperature sensitive Notch mutation (Fig. 2M,N). Enhancer activity was also up-regulated (Fig. 2O,P), indicating that Notch signaling represses the miR-7 enhancer. Notch signals are transduced through the transcription effector Su(H) (Mumm and Kopan, 2000). It was previously found that Su(H) activates yan transcription (Rohrbaugh et al., 2002). Thus Yan is the most likely mediator of the repressive effect of Notch on the miR-7 enhancer. Consistent with this idea, a constitutively active Su(H) mutant repressed enhancer activity, and Notch mutant cells with greater enhancer activity had reduced Yan protein levels (SFig. 2).
Our genetic analysis revealed a network-like architecture acting in photoreceptor determination. Yan represses miR-7 transcription directly, and also represses transcription indirectly through Ttk69. This mode of direct and indirect repression is an example of a coherent feed-forward loop (Fig. 3A). miR-7 is involved in a second coherent feed-forward loop. Pnt-P1 directly activates miR-7 transcription, which in turn represses Yan. Pnt-P1 also directly represses yan transcription (Rorbaugh et al., 2002). This coherent feed-forward loop between Pnt-P1 and Yan interlocks with the other coherent feed-forward loop between Yan and miR-7 (Fig. 3A). Coherent feed-forward loops of this type, in which X regulates Y, and both negatively regulate Z, create stability against fluctuations in X. It generates a delay or persistence that rejects fluctuating dips in X and only accepts persistent decreases in X (Mangan and Alon, 2003; Mangan et al., 2003). Thus, we can hypothesize that levels of miR-7 and Yan are buffered against fluctuating drops in Yan and Pnt-P1. This buffering would ensure that a cell only switches from one state (Yan ON) to the other state (Yan OFF) when there is a persistent decrease in Yan. The Yan OFF state would also be buffered against switching back to Yan ON due to Pnt-P1 fluctuations. This mechanism likely functions in collaboration with degradation of Yan protein to promote zero-order ultrasensitivity (Melen et al., 2005), which ensures that a cell’s fate change is not spontaneously induced or reverted.
Figure 3. miR-7 stabilization of gene regulatory networks.
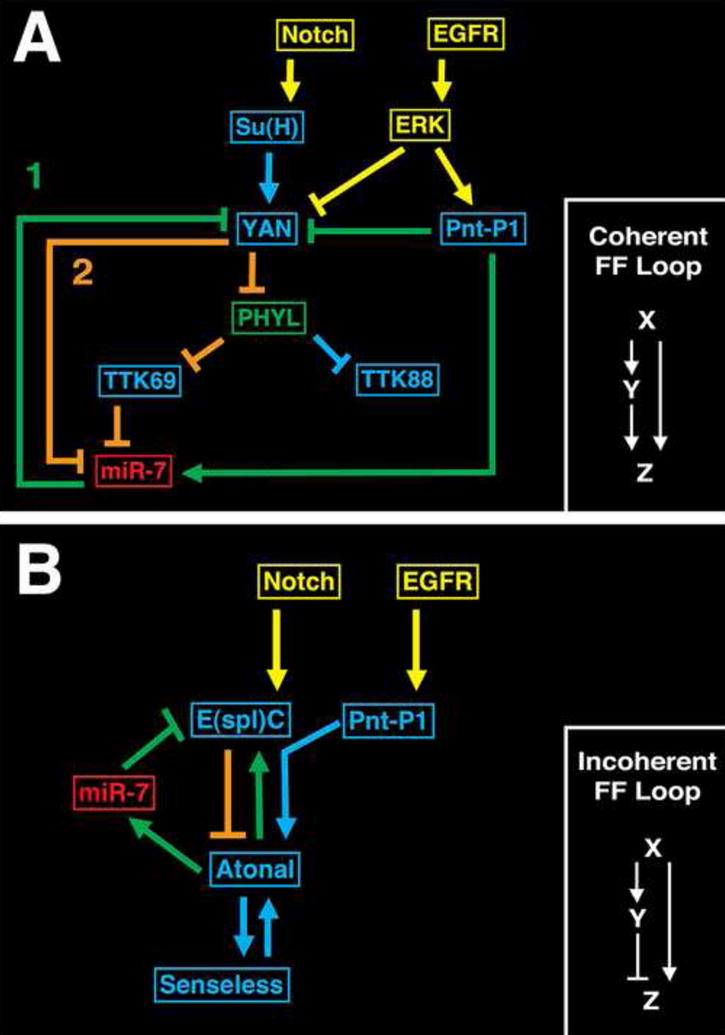
(A) The network controlling photoreceptor determination. Shown are signal transduction components (yellow), transcription factors (blue) and miR-7 (red) in the network. The miRNA participates in two interlocking coherent feedforward loops, labeled 1 and 2. Loop 1 is highlighted in green and loop 2 is in orange. A typical coherent feedforward loop of this type is shown to the right. The interlocked loops together construct a double-negative feedback loop between miR-7 and Yan.
(B) The network controlling SOP determination. Components are color-coded as in (A). miR-7 participates in an incoherent feedforward loop highlighted in green. A typical incoherent feedforward loop of this type is shown to the right. The feedforward loop is also interconnected with a double-negative feedback loop between Atonal and E(spl), with miR-7 as an effector of Atonal, and E(spl) directly inhibiting Atonal (orange).
The network controlling SOP determination
MiR-7 is expressed in developing proprioceptor and olfactory organs within the antenna, leg and wing (Fig. 4A–C). The miR-7 enhancer is also specifically active in these organs (Fig. 4D–F). Precursor cells of proprioceptor and olfactory organs transiently express the atonal (ato) gene in a zone called the proneural cluster (PNC) (Artavanis-Tsakonas et al., 1999). Ato protein activates transcription of genes that enable a subset of PNC cells to adopt a sensory organ precursor (SOP) fate (Jafar-Nejad et al., 2003). SOPs then proceed to form the sensory organs. Since Ato is present in cells with an activated miR-7 enhancer (Fig. 4G– I″), we wondered if this transcription factor might directly regulate the enhancer. Ato protein binds to DNA as a heterodimer with the ubiquitously expressed bHLH protein Daughterless (Da). An Ato/Da binding consensus sequence has been deduced (Powell et al., 2004). We identified two conserved sequences that matched the Ato/Da consensus in the miR-7 enhancer (Fig. 2A and SFig. 1A). To determine if Ato/Da activates the enhancer by binding these sequences, we misexpressed Ato or another proneural protein in the leg, antenna, and wing, and observed ectopic enhancer activation in those cells (Fig. 5A–F). We then constructed a mutant form of the enhancer in which the Ato/Da sequences were mutated. The resulting enhancer was completely inactive in the leg, antenna, and wing (Fig. 5J–L). Taken together, our results argue that Ato directly activates the miR-7 enhancer.
Figure 4. The enhancer drives miR-7 expression in SOP cells.
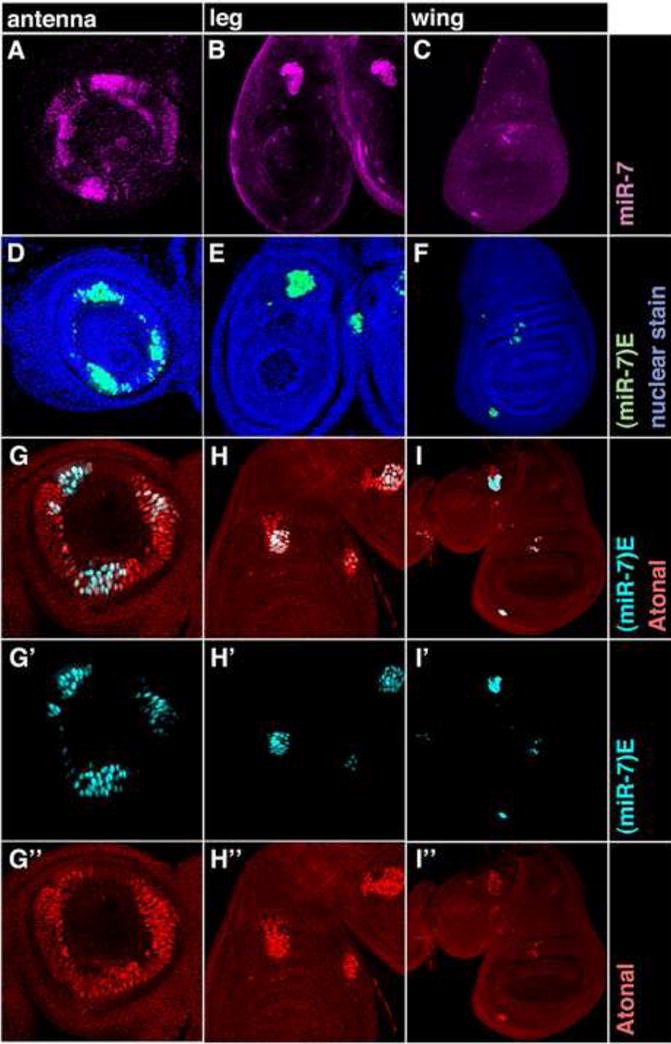
(A–C) miR-7 RNA (purple) in antenna (A), leg (B) and wing (C) discs.
(D–F) miR-7 enhancer activity (green) in antenna (D), leg (E) and wing (F) discs that are counterstained for nuclei in blue.
(G–I″) miR-7 enhancer activity as reported (cyan) in antenna (G,G′), leg (H,H′) and wing (I,I′) discs. (G,G″,H,H″,I,I″) Discs were counterstained for Ato protein (red). Merged fluorescence due to reporter GFP and Ato colocalization appears white.
Figure 5. Regulation of miR-7 expression in photoreceptors.
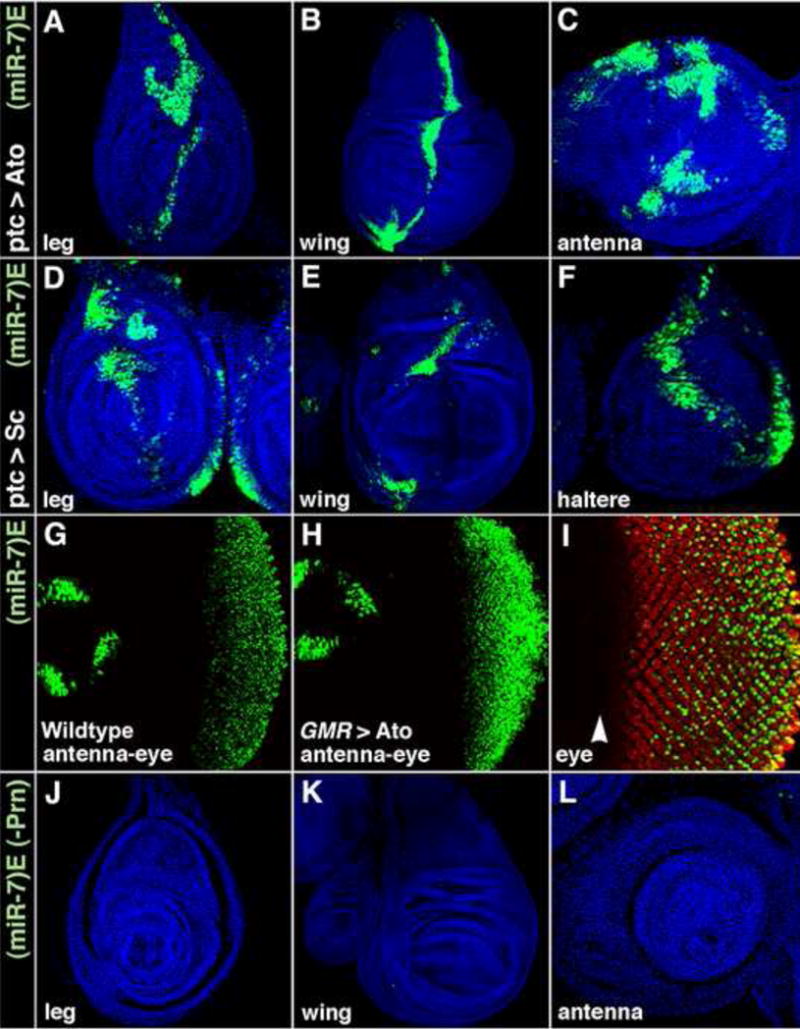
(A–F) miR-7 enhancer activity (green) in ptc>Ato (A–C), and ptc>Sc (D–F) imaginal discs that are counterstained for nuclei in blue. The ptc driver expresses Ato and Sc in a stripe of cells along the anteroposterior (vertical) midline of the discs.
(G and H) miR-7 enhancer activity (green) in wildtype (G), and GMR>Ato (H) eye discs.
(I–L) Activity of the mutated miR-7 enhancer with altered Ato-binding sites. (miR-7)E>GFP(-Prn) reporter expression (green) in eye (I), leg (J), wing (K), and antenna (L) discs. The leg, wing, and antenna were counterstained for nuclei in blue. The eye disc was counterstained for Elav protein in red. Note the enhancer is inactive in the morphogenetic furrow (arrowhead) of the eye where R8 photoreceptors are determined.
E(spl) genes can be directly repressed by miR-7. E(spl) genes encode proteins that directly repress transcription of the ato gene. Taken together, these data suggest that miR-7 can stimulate ato transcription and it would do so by repressing E(spl)-mediated repression. In support of this idea, we observed ectopic ato expression in cells that misexpressed miR-7 RNA (SFig. 3B, D, F-F″,H-H″). To determine if this effect was mediated through E(spl), we misexpressed miR-7 RNA along with mutant E(spl) mRNAs that lacked miR-7 binding sites in their 3′UTRs. Under these circumstances, we saw little or no ectopic Ato in cells misexpressing both miR-7 RNA and E(spl) proteins (SFig. 3J-J″,L-L″,N-N″).
This regulatory pathway should also affect SOP fate determination. As predicted, misexpression of either miR-7 RNA or Ato protein induced SOP determination (Lai et al., 2005 and Fig. 6A–F), and misexpression of miR-7-resistant E(spl) genes inhibited SOP determination (Fig. 6G,I,K). When we misexpressed both miR-7 RNA with different miR-7-resistant E(spl) proteins, we saw inhibition of SOP determination (Fig. 6H,J,L). Similar effects were observed when external sensory organ formation was assayed in adults (Fig. 6M and STable 4). Altogether, these data indicate that E(spl) genes act downstream of miR-7 to mediate its effects on ato expression and SOP fate determination.
Figure 6. miR-7 regulates Ato expression and SOP determination.
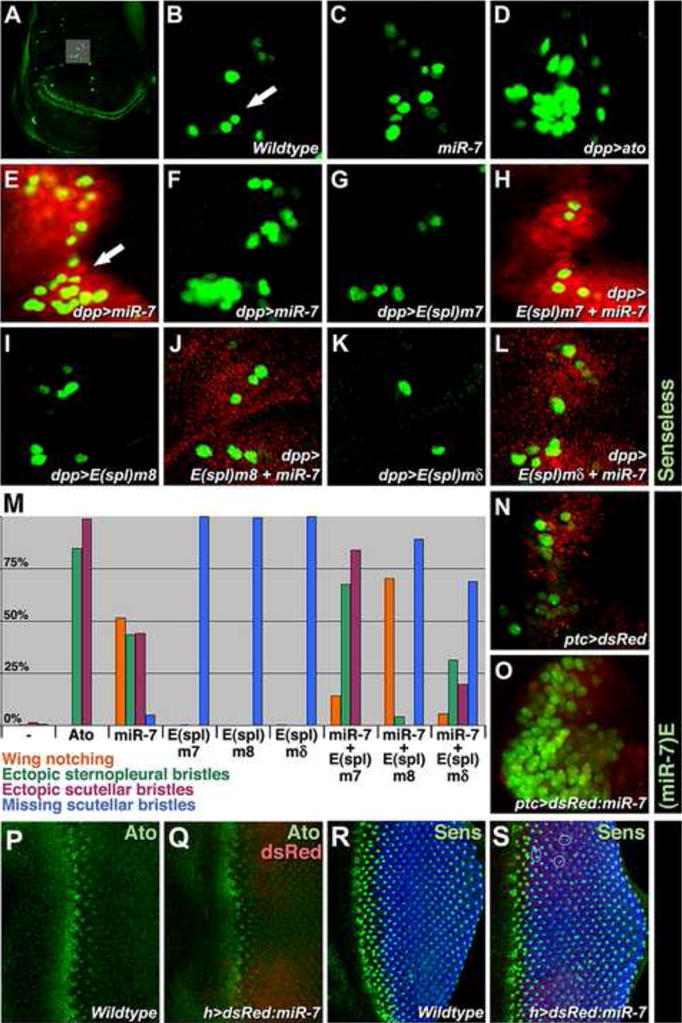
(A–L) SOP cells are marked with Sens protein in green. (A) A wing disc at low magnification shows the pattern of all wing SOPs, with a grey box highlighting the dorsal radius SOP group. (B–L) High magnification view of the dorsal radius group from (B) wildtype, (C) miR-7Δ1/Df(2R)exu1, (D) dpp≫Ato, (E) dpp≫dsRed-miR-7, (F) dpp≫miR-7-1401, (G) dpp≫E(spl)m7, (H) dpp≫dsRed-miR-7≫E(spl)m7, (I) dpp≫E(spl)m8, (J) dpp≫dsRed-miR-7≫E(spl)m8, (K) dpp≫E(spl)mδ, (L) dpp≫dsRed-miR-7≫E(spl)mδ wing discs. The dpp driver expresses miR-7 and E(spl) genes in a stripe of cells along the anteroposterior midline of the wing, which is visualized by the dsRed fluorescence from the dsRed-miR-7 chimera gene, observed in panels (E,H,J,L). The arrow in (E) points to an expanded cluster of dorsal radius SOP cells where miR-7 is misexpressed, relative to a cluster of SOP cells in wildtype, as highlighted with the arrow in (B). The miR-7-1401 transgene, when misexpressed, gives a comparable phenotype but is not marked by dsRed.
(M) Percentage of adults with ectopic or missing external sensory bristles (scutellar and sternopleural) observed in various mutants. N indicates total animals scored for wildtype (n=198), dpp≫Ato (n=89), dpp≫dsRed-miR-7 (n=118), dpp≫E(spl)m7 (n=193), dpp≫E(spl)m8 (n=166), dpp≫E(spl)mδ, dpp≫dsRed-miR-7≫E(spl)m7 (n=111), dpp≫dsRed-miR-7≫E(spl)m8 (n=168), and dpp≫dsRed-miR-7≫E(spl)mδ(n=313)
(N, O) miR-7 enhancer activity as reported (green) in the wing dorsal radius group counterstained with dsRed (red) from control ptc≫dsRed (N) and ptc≫dsRed-miR-7 (O) animals.
(P,Q) Ato protein (green) in wildtype (P) and hairy≫dsRed-miR-7 (Q) eye discs. dsRed (red) indicates where miR-7 is misexpressed.
(R,S) R8 photoreceptors marked with Sens (green) and other photoreceptors marked with Elav (blue) in wildtype (R) and hairy≫dsRed-miR-7 (S) eye discs. dsRed (red) indicates area where miR-7 is misexpressed. Photoreceptor clusters normally have a single R8 cell. Circles in (S) highlight some mutant clusters with more than one R8 cell.
Since we found that Ato activates miR-7 transcription, it would suggest the existence of a feedback loop in which ato activates miR-7, which then represses E(spl), which otherwise represses ato. The feedback loop would imply that miR-7 RNA positively activates its own transcription. As confirmation of this prediction, we observed activation of the miR-7 enhancer in cells misexpressing miR-7 RNA (Fig. 6N,O).
This mechanism is not restricted to proprioceptors and olfactory organs alone. It also operates during R8 photoreceptor fate determination at the earliest stages of eye patterning. We observed miR-7 RNA expression and miR-7 enhancer activity in cells where R8 determination occurs (Fig. 2B′,C′). Enhancer activity was not detected in this region when Ato/Da binding sites were mutated (Fig. 5I). This suggests that Ato activates the enhancer in the eye, and is consistent with our observation that misexpressed Ato activates the miR-7 enhancer (Fig. 5G,H). We also found that miR-7 feeds back onto Ato in the eye. Ato expression was modestly activated in cells misexpressing miR-7 RNA (Fig. 6P,Q), and these cells adopted an R8 cell fate (Fig. 6R,S), consistent with previous observations that Ato triggers determination of R8 photoreceptors (Jarman et al, 1994).
Our analysis of SOP determination has uncovered network-like features. Ato activates miR-7, which in turn represses E(spl). Ato also directly activates transcription of E(spl) (Cave et al., 2005; Cooper et al., 2000; Nellesen et al., 1999). Therefore, Ato both directly activates and indirectly represses E(spl) (Fig. 3B). This is an example of an incoherent feedforward loop. Incoherent feedforward loops of this type impart an accelerated and transient pulse of downstream gene expression (Mangan and Alon, 2003). In addition, E(spl) feeds back to ato to create a double-negative feedback loop that is interconnected with the feedforward loop (Fig. 3B). The overall effect is a network in which fluctuating peaks of Ato would result in transient pulses of Ato repression by E(spl), but sustained increase of Ato would result in sustained repression of E(spl) by miR-7 and stabilization of Ato (Fig. 3B).
miR-7 stabilizes developmental processes against temperature perturbation
If miR-7 provides biological robustness, then miR-7 should prevent development from being perturbed when the environment of the animal is perturbed. Environmental fluctuation is one type of perturbation against which gene expression can be remarkably stable (Freeman, 2000). We speculated that miR-7 may stabilize gene expression under fluctuating conditions, and that this would not be apparent under uniform conditions. Indeed, ato expression is normal in miR-7 loss-of-function mutants under uniform laboratory conditions (Fig. 7A,B and data not shown). We then perturbed the environment around developing Drosophila larvae by fluctuating the environmental temperature between 31°C and 18°C every ~ 1.5 hours. When wild-type larvae were challenged with such a temperature fluctuation, they exhibited no defects in expression of ato and yan (Fig. 7C,C′). In contrast, miR-7 mutant eyes exhibited a strong decrease in ato expression under fluctuating temperature conditions (Fig. 7D). Yan expression was abnormally strong and irregular in miR-7 mutant eyes (Fig. 7D′). The directions of expression change were consistent with the mutant failing to activate ato and repress yan.
Figure 7. miR-7 stabilizes gene expression and SOP determination under temperature fluctuation.
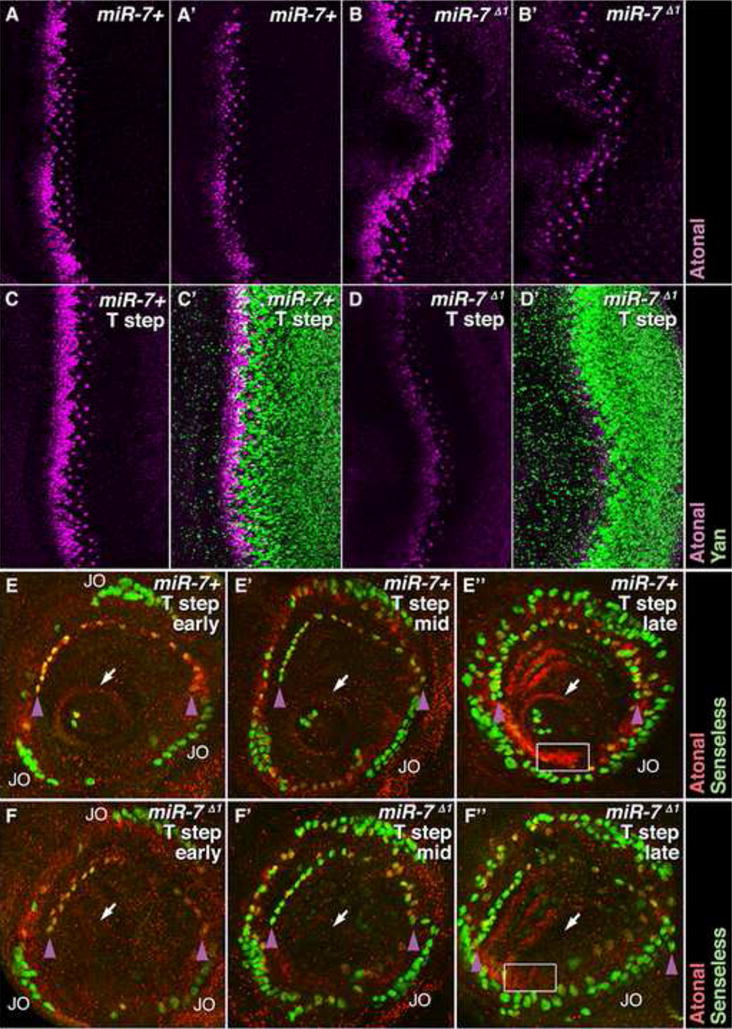
(A–B′) Ato protein (purple) in wildtype (A,A′), and miR-7Δ1/Df(2R)exu1 mutant (B,B′) eye discs from animals grown under uniform temperature conditions. (A,B) show maximal projections of confocal z stacks. (A′,B′) show single focal planes.
(C–D′) Ato (purple) and Yan (green) proteins in wildtype (C,C′) and miR-7Δ1/Df(2R)exu1 mutant (D,D′) eye discs from animals grown under fluctuating temperature conditions. Images are maximal projections of confocal z stacks.
(E-E″) Ato (red) and Sens (green) proteins in wildtype antennal discs from animals grown under fluctuating temperature steps. Sens marks the SOPs while Ato marks the PNCs. Sensory organs are progressively more developed in each panel. (E) An arc of coeloconic sensilla SOPs co-expressing Sens and Ato is first evident (purple arrowheads), along with the nascent Johnston’s organ, marked JO. A ring of cells expressing Ato surrounds the initial arista SOPs (arrow). (E′) SOP numbers increase within each organ system, and expression of Ato in these cells is reduced. (E″) There appears new rows of SOPs that are enveloped by cells with upregulated Ato (box).
(F-F″) miR-7Δ1/Df(2R)exu1 mutant antennal discs from animals grown under fluctuating temperature conditions. (F) The nascent Johnston’s organ (JO) appears normal, but the arc of coeloconic sensilla SOPs (purple arrowheads) is depleted at the top of the arc. Cells in the arista domain do not express a ring of Ato and do not form arista SOPs (arrow). (F′) Deficits in SOP cell number and spacing in the arista and coeloconic SOPs are further seen. (F″) In addition to reduced SOP numbers, there is little or no up-regulation of Ato in cells enveloping new SOPs (box).
We also examined the capacity of miR-7 to stabilize proprioceptor and olfactory SOP determination when perturbed for temperature. We subjected wildtype and miR-7 mutant animals to temperature fluctuations, and then followed the formation of antennal SOPs. Groups of SOPs that constituted the Johnston’s Organ appeared near-normal. However, the arista SOP group failed to form in the miR-7 mutant (Fig. 7E-E″, F-F″). The number of SOPs that form the coeloconic sensillae were reduced, and those that did develop were abnormally patterned. These defects were correlated with a reduction in ato expression within antennal cells (Fig. 7E–F″). Altogether, our experiments indicate that miR-7 buffers specific gene expression and cell fates against environmental perturbation. This function appears dispensable under uniform environmental conditions.
Discussion
Two features of miRNAs have suggested that they could potentially play a role in generating biological robustness. First, they regulate gene expression additively and thus tune gene expression rather than switch expression. Graduated output modulation in response to variable input is a mechanism for simple stabilization. Second, bioinformatic analysis suggests that many miRNAs act in feedback and feedforward network motifs (Martinez et al., 2008; Tsang et al., 2007). Some of these motifs have been theoretically and experimentally implicated to stabilize networks (Milo et al., 2002). However, direct experimental evidence that a miRNA promotes robustness (stability against noise or perturbation) has been missing. Here, we provide such evidence for miR-7 in Drosophila. This miRNA is required to maintain normal gene expression and sensory organ fate determination under fluctuating temperature conditions. We interpret this to mean that miR-7 buffers gene expression against environmental fluctuation. The fact that this function of miR-7 is exposed under fluctuating conditions underscores its primary role as a stabilizer for sensory organ development.
The robustness that miR-7 provided was most apparent for its proximate gene targets, yan and ato. Determination of R8 and SOP sensory cells was less dependent upon miR-7 under the fluctuation paradigm, although it led to defects in patterning of these in the eye (data not shown) and the antenna. Not surprisingly, it hints that there are mechanisms in place downstream or in parallel to ensure further robustness when there is fluctuation. These likely compensate and normalize the outcome. However, since certain SOP cell types were considerably more sensitive to fluctuation when miR-7 was absent, perhaps it underscores the mechanistic diversity that different cell types utilize for generating robustness.
The conceptual significance of the robustness-miRNA connection is several-fold. Their dynamic kinetic properties help answer the question of “why miRNA gene regulation” instead of just using more transcription factors. Their rate of biogenesis is more rapid than proteins, and they affect expression with less delay than factors that regulate nuclear events. These features enable miRNAs to produce rapid responses, something that is expected to counteract rapid and variable fluctuations. It also explains why miRNAs frequently appear dispensable under uniform laboratory conditions (Bushati and Cohen, 2007; Leaman et al., 2005; Miska et al., 2007).
Our analysis of two gene networks explains how miR-7 can buffer gene expression against perturbation. The miRNA acts in feedforward and feedback loops that are theoretically implicated as network stabilizers. Stability is experimentally apparent under conditions of temperature fluctuation though there is no reason a priori why stability cannot be expressed under other variable conditions. Another key point is that tight regulation of miRNAs is crucial. Misexpression of miRNAs frequently mimic loss-of-function phenotypes for their targets (Bushati and Cohen, 2007). Our results with miR-7 hint at how this is normally prevented. Namely, miR-7 has a restricted expression pattern that is strictly controlled by its targets. The restricted expression pattern can also explain how off-targeting effects are carefully limited.
Canalization and miRNAs
Waddington coined the word canalization to describe how development is buffered against perturbation (Siegal and Bergman, 2002; Waddington, 1942). Despite considerable genetic or environmental variation, organisms develop traits that are remarkably uniform in phenotype. Indeed, the insect compound eye and sensory organs appear to be deeply canalized systems (Jander and Jander, 2002; Meir et al., 2002; Rendel, 1959). It has been speculated that miRNAs might be important for canalization (Hornstein and Shomron, 2006). Certainly miR-7 has many attributes that suggest it helps canalize development in Drosophila.
There is an evolutionary implication to canalization. If canalization masks the phenotypic expression of genetic variation, then individuals within a species appear highly uniform (Waddington, 1953). This lack of diversity limits the number of traits upon which selection can act, resulting in stabilization of a species and reduced evolution. Conversely, lack of canalization results in enhanced phenotypic variation and the possibility of selection to evolve new forms. Theoretical and experimental studies indicate that canalization itself can evolve, that is, increase or decrease over evolutionary time (Gibson and Hogness, 1996; Proulx and Phillips, 2005; Rendel and Sheldon, 1960; Siegal and Bergman, 2002). In this light, it is interesting to consider miR-7. Several lines of evidence indicate that miR-7 has acquired a novel role in sensory organ development specifically within insects and not other animals. The miRNA is expressed in these Drosophila organs but not the orthologous organs of vertebrates. The enhancer that drives its expression in Drosophila sensory organs is not conserved in vertebrates. We found strong conservation of the miR-7 enhancer in Drosophila species divergent over 30 Myrs (SFig. 1A). A cluster of binding sites is also present upstream of the mosquito miR-7 sequence, (SFig. 1B,C), which implies conserved miR-7 transcription in the eyes of other insects. In contrast, the human miR-7-1 gene lacks a cluster of binding sites for the Yan ortholog TEL1, indicating divergent regulation of the human miR-7 ortholog (SFig. 4). Moreover, the vertebrate orthologs of E(spl) and Yan are not predicted targets of miR-7. Indeed, only a few vertebrate/drosophilid orthologs have been conserved as miR-7 targets, and most of these conserved targets have no known role in sensory organ development.
We propose that miR-7 was recruited into insect sensory organ development specifically for the purposes of canalization of those systems. As such, it has helped stabilize the remarkable uniformity of sensory organ form within different insects, particularly observed in the compound eye (Strausfeld and Nassel, 1981). If miR-7 is typical of highly conserved animal miRNAs, then it would imply that the acquisition of novel targets by these miRNAs is not necessarily to generate new traits but to stabilize pre-existing traits.
Experimental Procedures
Assaying miR-7 enhancer activity
A 349 bp DNA fragment located 1711 bp upstream of the 5′ end of the Drosophila pre-miR-7 sequence was PCR amplified and inserted into the transgenic expression vector pH-Stinger (Barolo et al., 2004). This contains a minimal promoter driving nuclear GFP. The resulting (miR-7)E>GFP construct was transformed into Drosophila. To make the reporter with mutated Ato/Da binding sites, the two predicted binding sites were mutated from CAGCTG to CCGCTA, and from CATCTG to CCTCTA. The mutated 349 bp enhancer was cloned into pH-Stinger to make (miR-7)E>GFP(-Prn) and transformed into Drosophila. Enhancer activity was assayed in vivo by visualizing GFP fluorescence or GFP protein localization by immunofluorescence.
Genetics
We used Drosophila stocks carrying miR-7Δ1; yan1; Nts3; GMR-Gal4; dpp-Gal4; ptc-Gal4; UAS-PntP1; UAS-Ttk88; UAS-Ttk69; UAS-EGFR.λtop (UAS-λDER); UAS-EGFR.DN; UAS-Ato; UAS-Sc; UAS-dsRed-miR-7; UAS-miR-7-1401; UAS-E(spl)m7; UAS-E(spl)m8; UAS-E(spl)mδ. Nts3 flies were grown at 18°C and shifted to 31°C for 19 hr before dissection. Flies carrying Gal4 or UAS constructs were grown at 25° or 29°C. Wing notching and ectopic posterior sternopleural bristles were scored twice per animal (once for each left and right side) whereas any lack of or extra scutellar bristles were scored once.
Temperature perturbation
w or CantonS (wildtype) and miR-7Δ1/Dfexu1 stocks were grown in bottles at a uniform temperature of 18 to 25°C for several days. They were shifted to 31°C for 16–24 hr. They were then subjected to two to five rounds of temperature cycles. Each round consisted of a shift to 18°C for 1.5 – 2 hours, and then back to 31°C for 1.5 – 2 hours. Bottles were incubated in air-circulating incubators for each temperature step. At the completion of the final round, either wandering third-instar larvae or white pre-pupae were harvested for analysis.
In situ hybridization and immunofluorescence
In situ hybridization against miR-7 mature RNA was performed as described (Li and Carthew, 2005) using an antisense miR-7 LNA probe 5′AAATCACTAGTCTTCCA-3′ned from Exiqon (Vedbaek, Denmark). To detect RNA by fluorescence, TSA Plus Fluorescence Systems from NEN was used following manufacturer’s instructions. Immunofluorescence of third-instar larval and pupal discs was performed as described (Li and Carthew, 2005). Antibodies used were guinea pig anti-Ato, guinea pig anti-Sens, rabbit anti-Ato, rat anti-Elav, mouse anti-GFP, mouse anti-Yan, and mouse anti-Ttk88.
Acknowledgments
We thank H. Bellen, S. Bray, C. Delidakis, S. Cohen, T. Hayashi, G. Mardon and the Bloomington stock center for fly strains; N. Baker, H. Bellen, T. Hayashi, Z.C. Lai and the Developmental Studies Hybridoma Bank for antibodies; T. Hayashi for sharing unpublished data; R. Gejman for advice and help with all stages of the informatic analysis; Carthew lab members and C. Labonne for discussions; the Biology Imaging Facility at Northwestern University for imaging. This work was supported by the by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust, the NIH (GM077581), the Northwestern Vision Training Grant (C.A.R.), the CMDB Training Grant (J.J.C.), and the Malkin Scholars program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques . 2004;36:436–440. 442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Tyler DM, Furriols M, Chalkiadaki A, Delidakis C, Bray S. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev Biol. 2000;221:390–403. doi: 10.1006/dbio.2000.9691. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- Gibson G, Hogness DS. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science. 1996;271:200–203. doi: 10.1126/science.271.5246.200. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JLt, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Hobert O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol. 2006;71:181–188. doi: 10.1101/sqb.2006.71.006. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet . 2006;38(Suppl):S20–24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander U, Jander R. Allometry and resolution of bee eyes (Apoidea) Arthropod Struct Dev. 2002;30:179–193. doi: 10.1016/s1467-8039(01)00035-4. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol Evol. 2002;17:468–473. [Google Scholar]
- Meir E, von Dassow G, Munro E, Odell GM. Robustness, flexibility, and the role of lateral inhibition in the neurogenic network. Curr Biol. 2002;12:778–786. doi: 10.1016/s0960-9822(02)00839-4. [DOI] [PubMed] [Google Scholar]
- Melen GJ, Levy S, Barkai N, Shilo B. Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol Syst Biol. 2005:0028. doi: 10.1038/msb4100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Nakahara K, Kim K, Sciulli C, Dowd SR, Minden JS, Carthew RW. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci U S A. 2005;102:12023–12028. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. The opportunity for canalization and the evolution of genetic networks. Am Nat. 2005;165:147–162. doi: 10.1086/426873. [DOI] [PubMed] [Google Scholar]
- Rendel JM. Canalization of the scute phenotype of Drosophila. Evolution. 1959;13:425–439. [Google Scholar]
- Rendel JM, Sheldon BL. Selection for canalization of the scute phenotype in Drosophila melanogaster. Aust J Biol Sci. 1960;13:36–47. [Google Scholar]
- Rohrbaugh M, Ramos E, Nguyen D, Price M, Wen Y, Lai ZC. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr Biol. 2002;12:576–581. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Siegal ML, Bergman A. Waddington’s canalization revisited: developmental stability and evolution. Proc Natl Acad Sci U S A. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLOS Biology. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ, Nassel DR. Neuroarchitectures Serving Compound Eyes of Crustacea and Insects. In: Autrum H, editor. Handbook of Sensory Physiology. Berlin: Springer-Verlag; 1981. pp. 1–132. [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Treier M, Bohmann D, Mlodzik M. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell. 1995;83:753–760. doi: 10.1016/0092-8674(95)90188-4. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Canalisation of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [Google Scholar]
- Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]