Abstract
We developed mice with germline endogenous expression of oncogenic Hras to study effects on development and mechanisms of tumor initiation. They had high perinatal mortality, abnormal cranial dimensions, defective dental ameloblasts, and nasal septal deviation, consistent with some of the features of human Costello syndrome. These mice developed papillomas and angiosarcomas, which were associated with HrasG12V allelic imbalance and augmented Hras signaling. Endogenous expression of HrasG12V was also associated with a higher mutation rate in vivo. Tumor initiation by HrasG12V likely requires augmentation of signal output, which in papillomas and angiosarcomas is achieved via increased Hras-gene copy number, which may be favored by a higher mutation frequency in cells expressing the oncoprotein.
Keywords: allele copy number, angiosarcoma, Costello syndrome, papilloma, senescence
The proto-oncogenes Hras, KrasB, and Nras are ubiquitously expressed plasma membrane GTPases activated by growth factor receptors, nonreceptor tyrosine kinases, and G-protein-coupled receptors, leading to engagement of downstream effector pathways. Although their specificity remains poorly understood, they have discrete functions. Thus, whereas Nras, KrasA, and Hras homozygous knockout mice are viable and grow normally (1, 2), KrasB −/– embryos die between embryonic day 12.5 (E12.5) and gestational term from liver defects and anemia (3).
The molecular basis for oncogenesis by Ras is caused by mutations that favor their constitutively active, GTP-bound, conformation. Additional evidence for functional specificity of Ras isoforms is that many human tumors have a predilection for mutations in one RAS gene family member. HRAS mutations are less common overall, but they have a particularly high prevalence in skin papillomas and cancers of the urinary bladder (4). The molecular mechanism for cell-type specificity for activating mutations of particular RAS genes has recently been proposed to be determined, at least in part, by the expression levels of the individual RAS genes. Thus, urethane induces lung cancers that are associated with Kras mutations in mice, but when an Hras cDNA is knocked into the Kras gene locus, and is thus expressed under the control of the Kras gene promoter, the urethane-induced tumors harbor Hras mutations instead (5). However, there is also evidence that distinct membrane targeting of the different Ras proteins may confer them with specific functions (6).
The consequence of activation of individual oncogenic Ras isoforms on development and tumorigenesis has been explored in mice with targeted knock-in mutations of the respective genes. LSL-KrasG12D mice have a latent allele that, when activated by Cre recombinase in the germline, results in placental insufficiency and embryonic lethality (7, 8). These results differed from those obtained after expression of a bicistronic knock-in KrasG12V allele, in which mice were mostly viable (9). There is no conclusive explanation for this discrepancy. However, mice with germline or lung-specific expression of endogenous mutant Kras developed lung adenomas and eventually adenocarcinomas in both models, cementing the role of oncogenic Kras in initiation of lung carcinogenesis. There is more limited information about the consequences of endogenous expression of oncogenic Nras in mice. In contrast to Kras, expression of a conditional allele of Nras (LSL-NrasG12D) in the distal small intestine and colon of mice expressing a Fabp1-Cre transgene does not result in epithelial hyperplasia. Instead, epithelial cells expressing physiological levels of NrasG12D are protected from dextran-induced apoptosis (10). Of note, in this study the distinct effects of the two oncoproteins could not be accounted for by differences in their steady-state expression levels.
The Hras gene is of particular interest from the standpoint of development, because de novo germline mutations of HRAS cause Costello syndrome (CS) (11), a congenital disorder that belongs to a family of neuro-facio-cardio-cutaneous diseases caused by germline mutations of genes encoding effectors in the Ras-Raf-MEK signaling pathway (12). Human CS is associated with hypotonia, characteristic facies, severe feeding difficulty, and failure to thrive (11). Most patients also have cardiac defects, musculoskeletal anomalies, and neurocognitive impairment (13). Subjects with CS have multiple papillomas and an increased risk of developing rhabdomyosarcomas and bladder carcinomas (14–17). About 80% of individuals affected with CS have HRAS mutations encoding mutant G proteins with comparatively weak transforming activity, primarily HRASG12S. The G12V mutant of HRAS has the lowest GTPase activity among various amino acid substitutions at codon 12 and the highest transformation potential (18, 19), and is by far the most common HRAS mutation of this codon in cancer. Accordingly, there has only been one child reported with a germline HRASG12V mutation, which was associated with a severe form of CS that resulted in death at 18 months of age, suggesting that this mutation may not be well tolerated in humans (11). Unexpectedly, a mouse germline knock-in of a bicistronic HrasG12V allele (HrasG12V-IRES-β-geo) was associated with a mild phenotype (20). Mice were viable, fertile, and survived normally. They had systemic hypertension, and as they aged they developed myocardial and kidney fibrosis. However, they were not tumor-prone and did not develop papillomas, even after exposure of the skin to phorbol esters, leading the investigators to conclude that Hras oncogenes are likely not sufficient for tumor initiation.
Here we show that mice with widespread endogenous expression of HrasG12V, created with a different genetic strategy, have high perinatal lethality, craniofacial defects, and a highly penetrant neoplastic phenotype. Moreover, tumor initiation by oncogenic Hras is invariably associated with a somatic imbalance in Hras-gene copy number that favors the mutant allele, resulting in augmentation of Hras signaling, which may be required for tumor development.
Results and Discussion
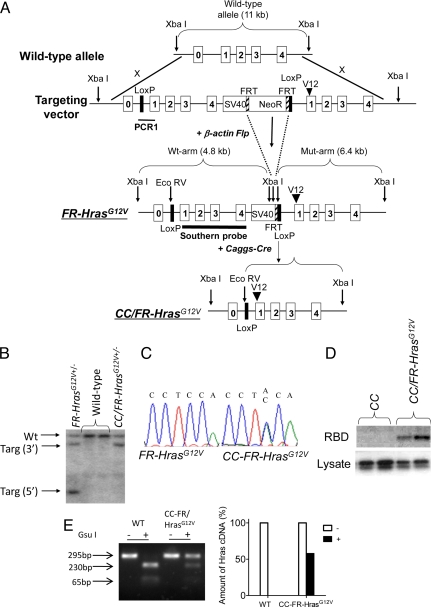
We used “flox-and-replace” gene targeting to obtain a Cre recombinase-dependent switch of expression of WT Hras1 by an HrasG12V allele under the regulatory control of its own gene promoter. The targeted locus consists of 2 tandemly-arrayed Hras genes, with the upstream WT copy flanked by loxP sites, so that the mutant allele is not expressed unless the WT copy is excised. FR-HrasG12V mice were crossed with Caggs-Cre mice to obtain widespread activation of the oncogene (Fig. 1 A and B). Caggs-Cre/FR-HrasG12V (CC/FR-HrasG12V) mouse embryonic fibroblasts (MEFs) expressed WT and HrasG12V mRNA at a ≈1:1 ratio, resulting in expression of a constitutively active Hras protein, as determined with a Raf-binding domain pull-down assay (Fig. 1 C–E).
Fig. 1.
Development of mice with a conditional knock-in activating mutation of Hras. (A) Diagram of Hras targeted allele. (Top) Targeting vector consists of a 5′arm containing the WT Hras gene and a 3′arm containing the mutant Hras gene, which are separated by an Frt-flanked Neomycin minigene. (Middle) Targeted allele after crossing with β actin-Flp mice to remove the Neomycin minigene. (Bottom) Targeted allele after crossing with Caggs-Cre mice to remove the WT Hras copy. (B) Southern blot of tail DNA isolated from WT, FR-HrasG12V, or CC/FR-HrasG12V± mice cut with XbaI and EcoRV and probed with a genomic fragment containing exons 1–4 of Hras. Wt, wild-type allele; Targ, targeted allele. (C) Sequence trace of products generated by RT-PCR of RNA isolated from MEFs from FR-HrasG12V or CC/FR-HrasG12V embryos. (D) Western blot of activated Hras in MEFs from CC/FR-HrasG12V and control mice. Activated Ras proteins were pulled down with an agarose-conjugated Raf-1 Ras-binding domain, followed by SDS/PAGE gel electrophoresis and immunoblotting with a specific anti-Hras antibody. Western blot of total lysate with Hras IgG is shown below. (E) RT-PCR products of RNA isolated from MEFs from WT or CC/FR-HrasG12V embryos were incubated with or without Gsu I, which digests WT but not mutant Hras cDNA. Total Hras cDNA in the absence of Gsu I was normalized to 100%. The right panel shows that the mutant Hras cDNA is ≈50% of total Hras in CC/FR-HrasG12V MEFs.
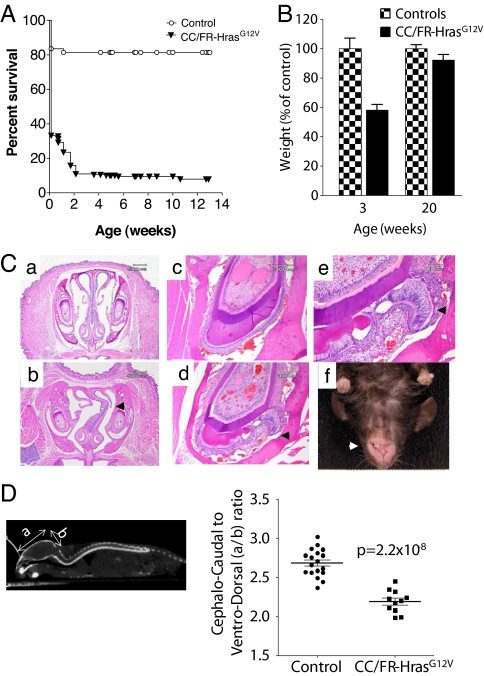
CC/FR-HrasG12V embryos (E19) had no macrosomia and pups were born at the expected Mendelian frequency. However, >80% died within 14 days after birth. Surviving mice were runted and continued to have higher mortality after weaning, although they caught up with the weight of their WT littermates by week 20 (Fig. 2 A and B). The cause of death of neonates could not be determined at necropsy, and there were no abnormalities in the gross or microscopic appearance of the heart muscle, valves, or lungs, or evidence of bleeding. E19 CC/FR-HrasG12V embryos also failed to exhibit any cardiopulmonary anomalies.
Fig. 2.
Increased neonatal mortality and cranio-facial deformities in CC/FR-HrasG12V± mice. (A) Kaplan-Meier survival plot of CC/FR-HrasG12V and control mice. (B) Decreased body weight in CC/FR-HrasG12V mice at weaning. Weight of CC/FR-HrasG12V mice and control littermates at 3 and 20 weeks of age. Bars represent mean ± SE percent-change in body weight versus controls (n = 8; P = 7.7 ×10−7 at 3 weeks, P = 0.25 at 20 weeks). (C) Coronal sections (4×) across the nasal cavity of a representative WT (a) and CC/FR-HrasG12V (b) mouse. The mutant mouse exhibits marked nasal septal deviation. Low power magnification (20×) of an incisor of a WT (C) and a mutant (D) mouse. The tooth of the CC/FR-HrasG12V± mice has an abnormal ameloblast cell lining and defective enamel formation. (e) Higher magnification (40×) demonstrates detachment of the ameloblasts from the adjacent dentin, loss of polarity, and stratification of ameloblasts, as well as areas of enamel sequestration. (f) Mouse with misalignment of incisors and malocclusion. (D) (Left) Whole-body CT scan (sagittal view) illustrating the rectangular region of interest used to determine the overall cephalo-caudal and ventro-dorsal dimensions of the cranial bony structures. (Right) Abnormal cranial dimensions of CC/FR-HrasG12V mice. Mean cephalo-caudal to ventro-dorsal ratio in aged-matched control and CC/FR-HrasG12V mice. All mice studied in this article are HrasG12V heterozygous mice.
CC/FR-HrasG12V mice had facial deformities, including nasal septal deviation, which may represent a severe manifestation of the depressed nasal bridge often seen in children with CS (Fig. 2C). Defects in dentition were observed in all of the CC/FR-HrasG12V mice we surveyed (9 out of 9) between 6 and 12 weeks of age, and characterized by disorganized growth of ameloblasts and irregular deposition of enamel (see Fig. 2C). Interestingly, widespread over-expression of HrasG12V in transgenic mice has been shown to cause odontogenic tumors, which sometimes differentiate into ameloblastomas (21). In addition, CC/FR-HrasG12V mice developed malocclusion (see Fig. 2C), which was not observed in littermates and thus was not a strain effect. Children with CS frequently have relative macrocephaly (22). Indeed, the ratio of the cephalo-caudal to ventro-dorsal cranial axes on microCT discriminated CC/FR-HrasG12V mice from age-matched control littermates (Fig. 2D). Virtually all human neonates with CS have hypotonia and severe feeding difficulties (15). The developmental defects in dentition and facial structure in CC/FR-HrasG12V mice suggest that the high neonatal mortality may also relate to difficulties with suckling.
A comprehensive tissue survey of CC/FR-HrasG12V mice at 5 weeks showed no increase in organ weight or histopathological abnormalities. Moreover, phosphorylation of Ras downstream effectors was not increased in the brain, the tissue with the highest levels of endogenous Hras expression, or in the kidney, colon, or heart [supporting information (SI) Fig. S1 A and B]. These data are consistent with the effects of endogenous expression of activated Kras in MEFs and in hematopoietic cells (7, 23). The lack of detectable effects on Raf/MEK/ERK and PI3 kinase/Akt signaling suggest that normal tissues can restrain oncogenic Ras signaling quite efficiently when these proteins are expressed from the endogenous locus. Liver was a notable exception, as pMEK and pERK, but not pAKT, were markedly increased. Interestingly, Sprouty 2 (a negative regulator of Ras/Raf/MAPK signaling) levels were particularly low in the liver as compared to other tissues, which may have resulted in relatively unopposed activation of MEK-ERK by HrasG12V in this cell type (Fig. S1C).
Cardiac malformations, arrythmias and hypertrophy are common in CS (13). Moreover, HrasG12V-IRES–β-geo mice developed age-dependent systemic hypertension, cardiac fibrosis, and vascular remodeling, particularly in homozygotes (20). There was no increase in pERK or pAKT in cardiac tissue of CC/FR-HrasG12Vmice. However, older animals (40–58 weeks) did develop myocardial fibrosis, but no other significant histological abnormalities in the heart, valves, or arterial wall (Fig. S2).
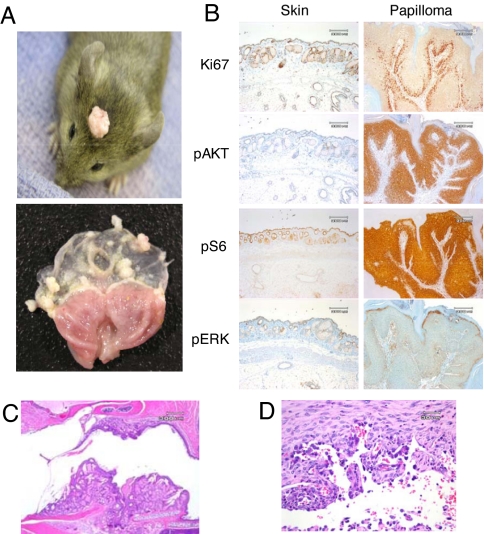
About 88% of CC/FR-HrasG12V mice developed skin papillomas at 40–58 weeks (Table 1). They were frequently found in the skull, face, and external auditory canal, where epidermal and sebaceous hyperplasia were also noted (Fig. 3 A and C). In older animals, multiple papillomas developed with near complete penetrance in the forestomach (see Fig. 3A), and less frequently in the anal epithelium. By contrast, no polyps or hyperplasia developed in the glandular stomach or in intestinal epithelium. All Ras isoforms were expressed at higher levels in squamous than in glandular epithelial cells, which may play a role in conferring susceptibility of these cells to transformation by Hras mutants (Fig. S3). No abnormalities were noted in the transitional epithelium of the bladder through 52 weeks of age. Older animals (> 30 weeks) did develop s.c. angiosarcomas (Fig. 3D), with a prevalence of ≈40% (see Table 1). These tumors have been associated with oncogenic RAS activation in humans (24). Moreover, 7,12-dimethylbenz[a]anthracene, besides inducing papillomas when applied topically, results in development of hemangiosarcomas when given systemically, and these tumors harbor HrasQ61L substitutions (25). Two mice developed hepatocellular adenomas, an insufficient number to conclude whether or not this was associated with mutant Hras expression.
Table 1.
Tumor incidence in CC/FR-HrasG12V mice
| Line | Age (w) | Skin Papilloma | Gastric Papilloma | Angiosarcoma | Hepatocellular Adenoma |
|---|---|---|---|---|---|
| HrasWT | 4–7 | 0/13 | 0/13 | 0/13 | 0/13 |
| 9–12 | 0/14 | 0/14 | 0/14 | 0/14 | |
| 40–52 | 0/7 | 0/7 | 0/7 | 0/7 | |
| HrasG12V | 4–7 | 0/2 | 0/2 | 0/2 | 0/2 |
| 9–12 | 1/7 | 0/7 | 0/7 | 0/7 | |
| 40–52 | 15/17 | 15/17 | 6/17 | 2/15 |
Fig. 3.
Squamous papilloma and angiosarcoma development in CC/FR-HrasG12V mice is associated with augmented Hras signaling. (A) (Upper) Representative papilloma in a CC/FR-HrasG12V mouse. Papillomas frequently formed in areas exposed to friction. (Lower) Stomach from a 28-week-old CC/FR-HrasG12V mouse showing multiple papillomas in the stomach fundus, but not in the cardia or antrum. (B) Representative IHC staining for Ki67, pAKT, pS6, and pERK1/2 in sections of papilloma and adjacent non-neoplastic skin from CC/FR-HrasG12V mice. (C) H&E-stained sections (10×) of the external auditory canal of a CC/FR-HrasG12V mice with epidermal and sebaceous hyperplasia. (D) Representative H&E section (100×) of angiosarcoma developing in CC/FR-HrasG12V mice.
Although nominally CC/FR-HrasG12V and HrasG12V-IRES–β-geo mice express the same mutant allele at endogenous levels, the 2 mouse models have profound phenotypic differences. Their genetic strains differed only slightly, as both were maintained in a 129/Sv/J-C57BL/6J background, although CC/FR-HrasG12V had ≈25% contribution from NIH Black Swiss. Recently, a polymorphic Patched allele present in the FVB/N, but not in C57BL/6 strain, was shown to promote skin carcinogenesis by oncogenic Hras (26). Although strain differences may account for some of the observed differences, the high frequency of perinatal lethality and tumor development in our model argues against this. It is also possible that the IRES–β-geo cassette may interfere in a subtle way with the expression level of HrasG12V, and that a critical threshold required for tumor initiation was not achieved. Indeed, the magnitude of oncogenic Hras expression was recently shown to be critical for bladder tumorigenesis (27). The fact that KrasG12D- IRES–β-geo mice also had a markedly attenuated phenotype compared to mice with a conventional latent oncogenic Kras allele is consistent with this interpretation (7–9).
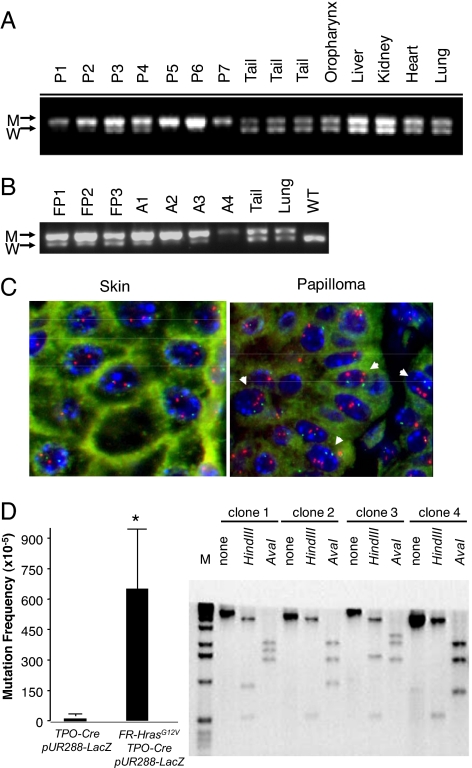
Papillomas developing in CC/FR-HrasG12V mice had higher mitotic activity in the basal layer compared to adjacent epidermis. Moreover, there was a marked increase in pAKT, phospho-ribosomal S6, and to a lesser degree, pERK staining, consistent with augmentation of Hras signaling (Fig. 3B). We next determined whether this was associated with Hras-gene copy number changes and found that the mutant Hras allele was over-represented in 7 out of 7 skin and 3 out of 3 forestomach papillomas, providing further evidence for a critical role for HrasG12V expression levels in papilloma development (Fig. 4 A and B). This was also seen in 4 out of 4 angiosarcomas we analyzed. We confirmed that this was because of increased HrasG12V-gene copy number in papillomas by interphase FISH (Fig. 4C). A subset of papillomas had loss of the WT Hras allele (Fig. S4). WT Ras has been proposed to function as a tumor suppressor in cells with a corresponding mutant Ras allele (28, 29). However, the fact that this was not found in all of the tumors suggests that this is not required for tumor development. A requirement for intensification of mutant Hras signaling through copy-number imbalances is also supported by studies in Rat 1 fibroblasts with an oncogenic Hras knock-in mutation, in which endogenous HrasG12V expression did not by itself induce transformation; yet when transformed clones developed after serial passaging, they commonly had excess copies of the mutant Hras gene (30).
Fig. 4.
Hras allelic imbalance in papillomas and angiosarcoma from CC/FR-HrasG12V mice. (A) PCR of genomic DNA of papillomas with primers that distinguish mutant from WT Hras alleles (see Fig. 1A). W: 622 bp WT allele; M: 666 bp targeted allele because of insertion of loxP site. P1-P7: DNA from papillomas, or indicated nontumoral tissues. (B) PCR of DNA from forestomach papillomas (FP1–FP3), angiosarcomas (A1–A4), or indicated nontumoral tissues. (C) FISH of a representative section from papilloma tissue (P2) using a mouse BAC containing the Hras gene. Papilloma nuclei have 3 to 5 fluorescent signals corresponding to Hras (red), whereas adjacent skin is diploid. Mouse chromosome 7 centromeres are labeled in green. (D) (Left) Increased mutation rate in thyroid cells from FR-HrasG12V/TPO-Cre mice. Plasmids rescued from DNA extracts of thyroid glands of TPO-Cre/pUR288-LacZ or FR-HrasG12V/TPO-Cre/pUR288-LacZ were screened for alterations in LacZ, as described in Methods. Bars represent the mean mutation frequency ± SE of pooled samples from TPO-Cre/pUR288-LacZ (n = 3; 10 thyroids per pool) and FR-HrasG12V/TPO-Cre/pUR288-LacZ (n = 4; 10 thyroids per pool). P < 0.05. (Right) Representative Southern blot of LacZ-negative clones. The type of mutation was determined by PCR amplification and restriction digestion of LacZ-negative clones. Clone 1 contained an inactivating point mutation of LacZ, whereas the other 3 show distinct restriction profiles consistent with recombination events, insertions, or deletions.
The Hras-gene copy number changes in papillomas may be a result of selection of random events occurring by chance or, alternatively, expression of endogenous HrasG12V may increase the frequency of somatic mutations. To explore this, we determined the mutation rate in vivo by intercrossing FR-HrasG12V/TPO-Cre mice, in which recombination and expression of mutant Hras occurs selectively in thyroid follicular cells, with pUR288-lacZ mutation reporter mice, a plasmid-based transgenic model used to study spontaneous mutation frequency in vivo (31). Thyroid cells were selected because they are susceptible to transformation by oncogenic Hras, yet had no increase in mitotic rate, pERK, pAKT, or histological abnormalities through 28 weeks of age. The mutation frequency was determined with a plasmid-rescue procedure applied to genomic DNA derived from thyroid tissue of 7-week-old TPO-Cre/FR-HrasG12V/pUR288-lacZ animals, with TPO-Cre/pUR288-lacZ age-matched littermates as controls, with a subsequent selection for LacZ-negative clones (32). Thyroid cells expressing HrasG12V had a higher mutation frequency. Restriction mapping of the LacZ-negative clones showed that 97% of the mutants had undergone rearrangements, insertions or deletions (Fig. 4D). The mechanisms accounting for this are unknown. Activated Hras has been shown to lead to premature entry of cells into S phase, increase permissivity for gene amplification, and generation of aberrant chromosomes within a single cell cycle (33–37). Conditional activation of HRasG12V in thyroid PCCL3 cells, which are WT for p53, allows cells to bypass the G2 DNA damage and mitotic spindle checkpoints (38), which results in genomic destabilization (39). It is unlikely that endogenous expression of HrasG12V in vivo induces such profound cell-cycle checkpoint defects in a ubiquitous way, yet it may increase the frequency of these occurrences in cooperation with other cell-intrinsic or environmental triggers. Mutant Ras has been shown to increase the generation of reactive oxygen species, which could also contribute to this effect (40).
Activation of oncogenic Ras or its downstream effectors has been shown to induce a DNA damage response (DDR), which in turn may trigger cellular senescence (41). Papillomas of CC/FR-HrasG12V mice showed abundant focal nuclear positivity for γH2AX and p53 (Fig. S5A), which was not apparent in the adjacent epidermis, suggesting that endogenous expression of the oncoprotein is not sufficient to induce a DDR that is, however, present in the tumor cells. Papillomas did not stain with SA-β gal or Dcr2 (data not shown), yet expression of other key markers of senescence, such as p16, p19, and p21, was increased compared to uninvolved skin (Fig. S5 A and B). Hence, a DDR and induction of senescence is only apparent when Hras-gene dosage and signaling is augmented, and may serve to restrain further growth. This is consistent with observations in mice expressing endogenous levels of oncogenic Kras (7, 9), and with evidence that oncogene-induced senescence may only be triggered in vivo by higher levels of mutant Ras expression, as recently shown following doxycycline-inducible expression of HrasG12V in mouse mammary cells (42).
MEFs expressing HrasG12V (Hrasm-MEFs) did not initially have a growth advantage compared to WT controls (Hraswt-MEFs). However, at later passages the aggregate cell number and the growth rate were higher in all Hrasm-MEF lines compared to controls (Fig. S6 A–C). This is consistent with a premature escape from senescence, although no clear differences in SA-β-gal, p16 or p19 levels were seen between lines at various passage numbers (not shown). Senescence also manifests as a response-refractoriness to growth factor activation of signaling effectors, which was evident in late passage Hraswt-MEFs compared to Hrasm-MEFs (Fig. S6D). Escape from senescence was also reported in MEFs expressing endogeneous KrasG12V or KrasG12D (7, 9), through unclear mechanisms. This was not associated with loss-of-function of p53, as treatment of late-passage Hrasm-MEFs with ionizing radiation (data not shown) or doxorubicin resulted in p53 phosphorylation and transactivation of p21 (Fig. S6E).
Our data point to a requirement for a threshold of HrasG12V expression to promote transformation in vivo. This was also apparent in vitro, as transduction with c-Myc or T antigen cooperated with HrasG12V to transform MEFs when the latter was over-expressed, but not when expressed at endogenous levels (Fig. S6F). Thus, although HRAS mutations are rare in comparison to KRAS and NRAS in cancer, this animal model shows that the highly transforming G12V mutant is capable of initiating tumor development in susceptible cell types. In these, endogenous expression is not sufficient to activate a signature of DDR or oncogene-induced senescence, or to promote clonal initiation. This may require augmentation of signaling by the oncoprotein, which in all tumor types developing in this model was strongly associated with Hras allelic imbalance, which may in turn be favored by an increased mutation frequency.
Methods
Construction of Targeting Vector.
We used recombineering to introduce a DNA fragment containing the entire Hras gene and 2.3 kb upstream of Hras exon 0 (5′ arm) or a fragment consisting of the entire Hras gene and 2.5 kb downstream of Hras exon 4 (3′ arm) into pBSIISK+ or pBSIIKS+(Stratagene), respectively. To complete the 5′ arm a loxP site was introduced upstream of exon 1, and a SV40 poly(A)/translation stop signal 3′ of Hras exon 4. To complete the 3′ targeting arm we changed the nucleotide sequence at codon 12 to encode a valine, and replaced exon 0 with a Frt-flanked Neo cassette with a 3′ loxP site added upstream of exon 1. The final targeting construct was generated by ligating the 5′ and 3′ arms, and was ≈11.8 kb in size (see Fig. 1A).
Development of FR-HrasG12V Mice.
The targeting construct was transfected into 2 different 129SvEv (129S6) ES cell lines. One clone (D14) with appropriate targeting of Hras was injected into D2B6F1 X B6 (Jackson Labs) blastocysts to produce chimeric mice. The founders were crossed with WT Swiss-Black mice. The Neo cassette in FR-HrasG12V-Neo mice was removed by crossing with β-actin-Flp mice, and offspring containing the FR-HrasG12V allele without the Neo cassette and that were negative for β-actin-Flp were mated with WT littermates and the line expanded.
Clinical and Anatomical Pathology.
Mice were killed by CO2 asphyxiation. Complete postmortem evaluations were performed, and tissues were weighed and prepared for histology and IHC using standard techniques. All studies were approved by the Institutional Animal Care and Utilization Committee.
IHC staining.
Tissues were promptly excised after killing, and either embedded in paraffin, or immediately frozen with dry ice for frozen sectioning. Next, 5-μm paraffin sections were immunostained with different antibodies (for antibody details see the SI Text) Staining for SA-β-gal was performed using Biovision's senescence detection kit (Biovision).
MicroCT Imaging.
Animals were anesthetized by inhalation of a mixture of 1.5% isoflurane in air at a flow rate of 1 to 2 L/min. Ungated whole-body CT images were acquired without contrast using a small-animal CT scanner (MicroCAT-IITM, ImTek).
Isolation of MEFs.
MEFs were isolated from E13.5-day embryos as described (43). The fetal heads were used for genotyping. Cells were grown to confluence and split once before freezing. To determine cumulative cell counts after serial passaging, MEFs (106) of the indicated genotypes from passage 3 onwards were plated on 10-cm dishes, grown in 10% FCS for 3 days, trypsinized, counted, and replated at 106 cells per well. This was repeated for each passage. The cumulative cell number was calculated based on the formula T n = T n-1 × P n/106, where T = cumulative cell number; n = passage number; P = cell number of current passage after 3 days in culture. For short-term growth curves, MEFs at passages 3 and 15 were plated at 5 × 104, grown in 2% FCS, and counted every other day for 7 days.
Assay for Activated Hras.
The Ras activation assay was performed using an agarose-bound GST-fused Ras-binding domain (RBD) of Raf-1 (Upstate). Briefly, MEFs grown in 10% FCS were washed with cold PBS and lysed. The activated Ras was pulled down with agarose-conjugated Raf-1 RBD, followed by SDS/PAGE gel electrophoresis and immunoblotting with a specific anti-Hras antibody (see SI Text).
Western Blotting.
Cells were washed 3 times with ice-cold PBS and lysed in RIPA buffer containing a mixture of protease and phosphatase inhibitors (Sigma). Tissue samples were homogenized and lysed with the same buffers, and following centrifugation, soluble protein was quantified by the Bradford method. Western blots were performed according to standard procedures (for antibody details see the SI Text).
Determination of Mutation Frequency in Vivo.
Mutation frequency was determined using a plasmid rescue procedure applied to genomic DNA derived from thyroid tissues of ≈7-week-old TPO-Cre/FR-HrasG12V/pUR288-LacZ mice. Plasmids were recovered from mouse thyroid genomic DNA by restriction digestion and magnetic bead separation. After ligation, plasmids were electroporated into Escherichia coli C (ΔLacZ, gal E-). Transformants were plated in titration plates (containing X-gal), and LacZ-mutant plates (containing P-gal). Only Lac-Z mutants can form colonies in the presence of P-galactose, as described (31). The ratio of colonies in selective vs. titration plates was used to calculate mutation frequency. The type of mutation was determined by PCR amplification and restriction digestion of LacZ-negative clones, to discriminate between point mutations or large-scale events, such as recombinations.
Transformation Assays.
Early passage MEFs were transduced with retrovirus vectors expressing Myc, T antigen, or HrasG12V, as indicated, for 48 h. Cells were plated on 10-cm plates and maintained for 14 days, with media changed every 2 days. Foci were scored after staining with crystal violet.
Supplementary Material
Acknowledgments.
We thank Dr. Phil Sanford and the Mouse Gene Targeting facility of the University of Cincinnati, and the Molecular Cytology, Molecular Cytogenetics and Comparative Pathology Core facilities of Memorial Sloan Kettering Cancer Center. This work was supported in part by National Institutes of Health Grants CA72597, CA50706, and T32 DK07313. Technical services provided by the Memorial Sloan Kettering Cancer Center Small-Animal Imaging Core Facility were supported in part by National Institutes of Health Small-Animal Imaging Research Program Grant R24 CA83084 and National Institutes of Health Center Grant P30 CA08748.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900343106/DCSupplemental.
References
- 1.Esteban LM, et al. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21:1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plowman SJ, et al. While K-ras is essential for mouse development, expression of the K-ras 4A splice variant is dispensable. Mol Cell Biol. 2003;23:9245–9250. doi: 10.1128/MCB.23.24.9245-9250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 5.To MD, et al. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40:1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuveson DA, et al. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AT, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 10.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki Y, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 12.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 13.Lin AE, et al. Further delineation of cardiac abnormalities in Costello syndrome. Am J Med Genet. 2002;111:115–129. doi: 10.1002/ajmg.10558. [DOI] [PubMed] [Google Scholar]
- 14.Kerr B, et al. Genotype-phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J Med Genet. 2006;43:401–405. doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gripp KW, et al. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 16.Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 17.Gripp KW, et al. Five additional Costello syndrome patients with rhabdomyosarcoma: proposal for a tumor screening protocol. Am J Med Genet. 2002;108:80–87. doi: 10.1002/ajmg.10241. [DOI] [PubMed] [Google Scholar]
- 18.Colby WW, Hayflick JS, Clark SG, Levinson AD. Biochemical characterization of polypeptides encoded by mutated human Ha-ras1 genes. Mol Cell Biol. 1986;6:730–734. doi: 10.1128/mcb.6.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 20.Schuhmacher AJ, et al. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118:2169–2179. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardiff RD, Leder A, Kuo A, Pattengale PK, Leder P. Multiple tumor types appear in a transgenic mouse with the ras oncogene. Am J Pathol. 1993;142:1199–1207. [PMC free article] [PubMed] [Google Scholar]
- 22.Hennekam RC. Costello syndrome: an overview. Am J Med Genet C Semin Med Genet. 2003;117:42–48. doi: 10.1002/ajmg.c.10019. [DOI] [PubMed] [Google Scholar]
- 23.Braun BS, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rennel E, et al. Regulation of endothelial cell differentiation and transformation by H-Ras. Exp Cell Res. 2003;291:189–200. doi: 10.1016/s0014-4827(03)00347-1. [DOI] [PubMed] [Google Scholar]
- 25.Doi ST, Kimura M, Katsuki M. Site-specific mutation of the human c-Ha-ras transgene induced by dimethylbenzanthracene causes tissue-specific tumors in mice. Jpn J Cancer Res. 1994;85:801–807. doi: 10.1111/j.1349-7006.1994.tb02951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakabayashi Y, Mao JH, Brown K, Girardi M, Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- 27.Mo L, et al. Hyperactivation of Ha-ras oncogene, but not Ink4a/Arf deficiency, triggers bladder tumorigenesis. J Clin Invest. 2007;117:314–325. doi: 10.1172/JCI30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 29.Diaz R, et al. The N-ras proto-oncogene can suppress the malignant phenotype in the presence or absence of its oncogene. Cancer Res. 2002;62:4514–4518. [PubMed] [Google Scholar]
- 30.Finney RE, Bishop JM. Predisposition to neoplastic transformation caused by gene replacement of H-ras1. Science. 1993;260:1524–1527. doi: 10.1126/science.8502998. [DOI] [PubMed] [Google Scholar]
- 31.Boerrigter ME, Dolle ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 32.Geiger H, et al. Mutagenic potential of temozolomide in bone marrow cells in vivo. Blood. 2006;107:3010–3011. doi: 10.1182/blood-2005-09-3649. [DOI] [PubMed] [Google Scholar]
- 33.Denko N, Stringer J, Wani M, Stambrook P. Mitotic and post mitotic consequences of genomic instability induced by oncogenic Ha-ras. Somat Cell Mol Genet. 1995;21:241–253. doi: 10.1007/BF02255779. [DOI] [PubMed] [Google Scholar]
- 34.Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci USA. 1994;91:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J Biol Chem. 1999;274:38083–38090. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 36.Wani MA, Xu X, Stambrook PJ. Increased methotrexate resistance and dhfr gene amplification as a consequence of induced Ha-ras expression in NIH 3T3 cells. Cancer Res. 1994;54:2504–2508. [PubMed] [Google Scholar]
- 37.Agapova LS, et al. P53-dependent effects of RAS oncogene on chromosome stability and cell cycle checkpoints. Oncogene. 1999;18:3135–3142. doi: 10.1038/sj.onc.1202386. [DOI] [PubMed] [Google Scholar]
- 38.Knauf JA, et al. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem. 2006;281:3800–3809. doi: 10.1074/jbc.M511690200. [DOI] [PubMed] [Google Scholar]
- 39.Saavedra HI, et al. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19:3948–3954. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- 40.Rassool FV, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007;67:8762–8771. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- 41.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 42.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 43.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.