Abstract
Wound healing is a complex process involving multiple cellular events, including cell proliferation, migration, and tissue remodeling. ADAM12 (a disintegrin and metalloprotease 12) is a membrane-anchored metalloprotease, which has been implicated in activation/inactivation of growth factors that play an important role in wound healing, including heparin-binding EGF-like growth factor (HB-EGF) and insulin growth factor (IGF) binding proteins. Here we report that expression of ADAM12 is fivefold up-regulated in the non-healing edge of chronic ulcers compared to healthy skin, based on microarrays of biopsies taken from five patients and from healthy controls (p=0.013). The increase in ADAM12 expression in chronic ulcers was confirmed by quantitative real time-PCR. Moreover, immunohistochemical analysis demonstrated a pronounced increase in the membranous and intracellular signal for ADAM12 in the epidermis of chronic wounds compared to healthy skin. These findings, coupled with our previous observations that lack of keratinocyte migration contributes to the pathogenesis of chronic ulcers, prompted us to evaluate how the absence of ADAM12 affects the migration of mouse keratinocytes. Skin explants from newborn ADAM12−/− or WT mice were used to quantify keratinocyte migration out of the explants over a period of seven days. We found a statistically significant increase in the migration of ADAM12−/− keratinocytes compared to WT control (P=.0014) samples. Taken together, the upregulation of ADAM12 in chronic wounds, and the increased migration of keratinocytes in the absence of ADAM12 suggest that ADAM12 is an important mediator of wound healing. We hypothesize that increased expression of ADAM12 in chronic wounds impairs wound healing through the inhibition of keratinocyte migration, and that topical ADAM12 inhibitors may therefore prove useful for the treatment of chronic wounds.
Keywords: ADAM12, chronic ulcers, wound healing, keratinocyte migration
INTRODUCTION
Chronic and acute wound healing disorders represent a serious health problem that affects more than 8 million people in the United States alone. Faced with epidemic numbers of chronic wounds, we encounter limitations in early diagnosis and treatment deriving from the lack of knowledge of wound healing mechanisms at the cellular and molecular level (1). The majority of chronic wounds fall into three categories: pressure, venous, and diabetic ulcers. They mostly affect elderly individuals, and the average age of patients with chronic wounds is over 60 (2–5).
Disruption of the epidermal integrity after wounding activates a homeostatic response that proceeds in a highly regulated manner. Wound healing is tailored to rapidly restore a functional epithelial lining over the injured site (6). The entire process is a dynamic continuum with an overlap of each phase and sustained remodeling. Such complexity, which involves keratinocyte and fibroblasts proliferation and migration, matrix deposition, changes in vascular permeability, angiogenesis and immune responses, depends on multiple synchronized signaling mechanisms (7, 8). These include growth factors, such as transforming growth factor β (TGFβ), epidermal growth factor (EGF), transforming growth factor α (TGFα), vascular endothelial growth factor (VEGF), platelet derived growth factor (PDGF), keratinocyte growth factor (KGF); cytokines, such as interleukin-1 (IL-1), tumor necrosis factor alpha (TNFα) and interferon gamma (IFNγ); enzymatic activation of matrix metalloproteinases MMPs (MMP1, 3, 9, 10, 11, and 13) and tissue inhibitors of metalloproteinase (TIMPs), hormones and vitamins (9, 10).
The epidermal morphology of chronic wounds differs from that of the normal epidermis. The epidermis of a non-healing edge of chronic wounds is hyperproliferative and hyper- and para-keratotic, suggesting that keratinocytes at the non-healing edges do not successfully complete either of the two possible pathways that can result in activation or differentiation (11, 12). Keratinocytes at the non-healing edge of chronic wounds are hyperproliferative but non-migratory, suggesting that lack of migration leads to inability to epithelialize and plays an important role in pathogenesis of chronic ulcers. The intricate role of cellular growth factors and cytokines in wound healing may be altered by several processes. Of importance may be the processing by a group of transmembrane metalloproteases termed ADAMs, which can process membrane anchored growth factors and cytokines, and thereby act as post-translational regulators of the function of several of their cleaved substrate proteins. This process, which is referred to as ‘protein ectodomain shedding’, has been implicated in cell-cell and cell-matrix interactions during neurogenesis, heart development, angiogenesis and cancer, to name a few examples (13). Substrates cleaved by ADAMs include the pro-inflammatory cytokine TNF-α and all ligands of the EGF-receptor, such as TGF-α, amphiregulin, and betacellulin. ADAM12, in particular has been reported to cleave HB-EGF and has been shown to increase longitudinal bone growth by modulating chondrocyte proliferation and maturation (14). Moreover, ADAM12 is a marker of skeletal muscle regeneration (15), and it is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression (16). However, very little is currently known about the function of ADAM12 in skin. In the current study, we used microarray analysis as well as quantitative RT-PCR to evaluate the expression of ADAM12 in chronic and acute wounds, and tested how deletion of ADAM12 in mice affects the migration of mouse keratinocytes.
MATERIALS AND METHODS
Human Specimens
Human skin specimens were obtained from reduction mammoplasty in accordance to an approved Institutional Review Board (IRB) protocol and used to generate acute wounds as previously described (17). A 3 mm biopsy punch was used to create an acute wound and skin specimens were maintained at the air-liquid interface with Dulbecco’s Modified Eagle Medium (DMEM) (Cambrex, Walkersville, MD), antibiotic/antimycotic and fetal bovine serum (FBS) (Gemini Bio – Products, West Sacramento, Ca) for 0, 4, 24, 48, and 96 hours. In addition, specimens were obtained from non-healing edges of venous ulcers from consenting patients having surgical debridement in accordance to the approved IRB protocol (11, 12).
Venous ulcers biopsies were obtained from normally discarded tissue from patients. None of the patients had diabetes. Evaluation for ischemia was performed either by non-invasive flow exams (ankle-brachial index of <0.9) or arteriogram and it was ruled out in all patients. Proper diagnosis was made following previously published guidelines and protocols (18, 19). All patients were debrided in the operating room. After the wound was prepped in betadine, a #20 sterile surgical scalpel was used to debide the wound edges. The non-healing edges used in this study were clinically identified by a surgeon (Dr Brem) as the edge of the most proximal skin edge to the ulcer bed. Routine histolpathological analysis with Hemotoxylin & Eosin were performed to verify typical morphology of the non- healing edge as described by Stojadinovic et al. (11). In addition, all patients had a venous duplex exam and confirmed the presence of venous valvular incompetence and venous reflux.
A small portion of the specimens was fixed in formalin and processed for paraffin embedding whereas the majority of the samples were stored in RNAlater ® (Ambion, Austin, TX) for RNA isolation.
Preparation and Hybridization of Probes
Three skin specimens deriving from healthy skin and five skin specimens deriving from chronic wounds were homogenized and total RNA was isolated using an RNeasy Mini Kit ® (Qiagen, Valencia, Ca) according to the manufacturer’s protocol. Approximately 5μg of total RNA was reverse transcribed, amplified, and labeled as described (18). Labeled cRNA was hybridized to HG-U133A microarrays ® (Affymetrix, Santa Clara, CA), which contain more than 22,000 probe sets. Arrays were washed, stained with avidin- biotin streptavidin-phycoerythin labeled antibody using an Affymetrix fluidics station and then scanned using the Agilent GeneArray Scanner ® system (Hewlett-Packard, Palo Alto, CA) following a protocol provided by Affymetrix.
Gene Array Data Analysis
Microarray Suite 5.0 ® (Affymetrix ) was used for data extraction and for further analysis, data mining tool 3.0 (Affymetrix) and GeneSpring™ software 7.3.1 ® (Silicon Genetics, Redwood City, CA) was used for normalization to median, filtering on Volcano plot for fold change and p-value calculations. Samples were normalized to the 50th percentile per chip, and to a median for each gene. Statistical comparisons of expression level, between each condition were performed using ANOVA test. A change in expression with a p-value of less than 0.05 was considered to be statistically significant. P-vales were generated as an average value from five venous ulcers and three normal skin samples. Differential expressions of transcripts were determined by calculating the fold change. Genes were considered regulated if the expression levels differed more than 2-fold relative to healthy skin. As part of this analysis, we developed a gene annotation table describing the molecular function and biological category of many genes present on the chip, primarily based on data from J. M. Ruillard and the Gene Ontology Consortium Data (available on the World Wide Web at cgap.nci.nih.gov/Genes/GOBrowserdot.ped.med.umich.edu:2000/ourimage/pub/shared/JMR_pub_affyannot.html)
Immunohistochemistry
Seven μm thick tissue sections were serially cut on a microtome (Carl Zeiss, Thornwood, NY) and mounted on slides. Sections were de-waxed in xylene, re-hydrated and washed with 1 × PBS. For antigen retrieval, paraffin sections were heated in a 95° C water bath in Target Retrieval Solution (DAKO Corporation, Carpinteria, Ca). Histological slides were treated with 0.1% H2O2 in methanol for 30 minutes, rinsed with H2O, and blocked with normal serum for 30 minutes (Vectastain Elite Universal Kit ABC, Vector Labs, Burlingame, Ca). Sections were then incubated with an anti-human ADAM12 antibody (rb122, rabbit polyclonal, gift from Dr Ulla Wewer, University of Copenhagen, Denmark) in a commercially available antibody diluent (DAKO Antibody Diluent with Background Reducing Components, DAKO Corporation) over night at +4° C. A biotinylated rabbit secondary antibody was added and the avidin-biotin complex was visualized using DAB (Vectorlabs, DAB Peroxidase Substrate Kit,). Slides were counterstained with hematoxylin. Negative controls were prepared by substitution of the primary antibody with PBS. Staining was analyzed using a Nikon Eclipse E800 microscope (Nikon, Inc. Melville, NY) and digital images were collected using the SPOT- Camera Advanced program.
Quantitative Real Time -PCR
RNA isolation and purification was performed using Trizol (Invitrogen, Carlsbad, Ca) extraction and subsequent Qiagen RNeasy Kit column purification (Qiagen). For quantitative RT- PCR, 0.5 μg of total RNA from healthy skin, chronic and acute wounds was reverse transcribed using an Omniscript Reverse Transcription kit (Qiagen,). Real time PCR was performed in triplicate using an iQ SYBR Green Supermix detection system (Bio-Rad, Hercules, Ca). The samples were amplified and quantified using an Opticon 2 thermal cycler ® (Bio-Rad) under the following conditions: 10 minutes at 95° C; and 35 cycles with 30 seconds at 95°C; 30 seconds at 58° C followed by 30 seconds at 60° C. Relative expression was normalized for levels of hypoxanthine-guanine phosphoribosyltransferase (HPRT1). The primer sequences used were: HPRT1, forward (5′-AAAGGACCCCACGAAGTGTT -3′) and reverse primer (5′-TCAAGGGCATATCCTACAACAA-3′) and for ADAM12, forward (5′-CGAGGGGTGAGCTTATGGAAC -3′) and reverse primer (5′-CACTCCGAACAGAGGCACTG -3′). The data were analyzed and samples quantified using the software Opticon Monitor3 ® (Bio-Rad).
Isolation of skin biopsies from newborn ADAM12-deficient mice and wild type littermates
ADAM12+/− mice (19) were mated to generate ADAM12−/− or ADAM12+/+ littermate offspring. A total of four litters with between 5 and 7 pups were analyzed in this study. For each litter, all newborn mice were sacrificed and their tails used for genotyping by Southern blot. A total of 25 newborn pups were sacrificed, and found to have the following genotypes: 6 ADAM12+/+, 7 ADAM12−/−, 12 ADAM12+/−. Since the results of the Southern blot analysis only became available after one week, skin biopsies were cultured from all embryos, but only those from wild type and ADAM12 knockout littermates were selected for further experimental analysis. All animals were maintained in an accredited animal facility at the Hospital for Special Surgery according to the guidelines of the American Veterinary association, and all procedures were approved by the HSS Institutional Animal Care and Use Committee.
Keratinocyte migration out of skin explants
Skin explants were prepared from 1 day-old mice as described (20, 21). Briefly, one day-old mice were sacrificed and submerged in a 10% povidine/iodine solution, then rinsed in phosphate buffered saline. The epidermis, dermis, and underlying adipose tissue and fascia were completely removed from the body of the animal by incision on the ventral aspect of the specimen and careful dissection over the dorsal aspect. The adipose tissue/fascia was removed in order improve the adherence of the dermis to the tissue culture plate. The tissue specimen was once again washed in 10% povidine/iodine solution followed by PBS. Six full-thickness biopsies with a diameter of 3-mm were taken from the dorsal midline of each animal and placed individually in 24 well plates. Each sample was submerged in 200 μL of nutrient-rich media containing adenine (24.4 μg/ml) (Sigma, St. Louis, MO), insulin (5 μg/mL) (Sigma), transferrin (5 μg/mL) (Sigma,), hydrocortisone (0.4 mM) ( Sigma), 10% fetal calf serum(Sigma), cholera toxin (0.5 μg/ml) (Sigma), penicillin G (100 IU/mL) (Sigma), T3 (1mM) (Sigma), gentamicin (25 μg/mL) ( Gibco, Gran Island, NY), 75% Dulbecco’s Modified Eagle’s Medium (Cambrex), and 25% F12 medium ( Invitrogen). Samples were incubated overnight at 37° C and the following day fed with 1.5 ml of media. Keratinocyte migration was observed over the course of 7 days, with changes of media on days 1, 3, and 5. To document keratinocyte migration, images were acquired on days 3, 5, and 7 using a Nikon Coolpix 35mm camera (Nikon Inc.) with manual zoom mounted on a Nikon Eclipse TS100 inverted microscope (Nikon Inc.). The objective was a Plan-UW 2x/.06 (Nikon Inc.), and Picasa 2.0 ® (Microsoft Corporation, Redmond, WA) and Powerpoint ® 2007 (Microsoft Corporation) was used to process images.
Quantification of keratinocyte migration
Pictures taken on days 3, 5, and 7 were used for data analysis. Each experiment was performed in triplicates, and keratinocyte migration was quantified independently by two laboratory members who did not know the genotype of the samples. The distance of keratinocyte migration was calculated using the image program, ImageJ ® (NIH, Bethesda, MD). Briefly, the center of the punch biopsy was determined by digital image measurement and all other measurements were made from that point. Using the line tool, the radius from the center point to the edge of the punch biopsy was determined, using an average of 10 line measurements per sample. The values obtained from the center to the edge of the punch biopsy were then averaged to give a digital measurement (in pixels) of the average radius of the biopsy, which corresponded to 1.5mm in all samples. Then an average of 11 line measurements was taken from the center point to the edge of the greatest confluent cell migration at different days (i.e. center to punch biopsy edge and center to keratinocyte migration front). The values from the center point to the edge of confluent cell migration were averaged to give a digital measurement (in pixels) of the average radial distance of cell migration (Fig. 1). The values graphed in Fig. 4b represent the distance from the center of the biopsy to the edge of the migration front of keratinocytes divided by the radius of the biopsy at day 0. A total of 9 untreated wild-type and 18 untreated ADAM12−/− biopsies, taken from 7 newborn ADAM12−/− mice and 6 newborn wild type littermate control mice were included in the statistical analysis using Student’s t-test available at http://www.physics.csbsju.edu/. P values <.05 were considered statistically significant.
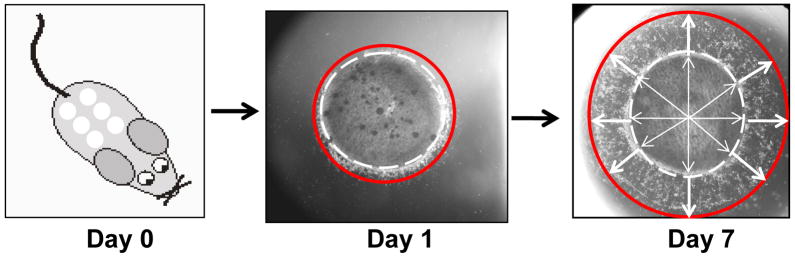
Figure 1. Acute wound model method and migration quantification.
One day-old littermates from the mating of ADAM12+/− and ADAM12+/− adult mice were sacrificed and six 3mm punch biopsies were removed from the back skin. Each biopsy was individually placed in a well of a 25 well-plate and submerged in supplemented growth media as described in Materials and Methods. Pictures were taken on days 1, 3, 5, and 7 to document keratinocyte outgrowth from the biopsy edge. The white lines represent the radius from a point determined to be the center of the punch biopsy through the use of the imaging program Image J and the red lines represent keratinocyte outgrowth from the edge of the biopsy. Using Image J an average of 10 white line measurements and 11 red line measurements were documented per sample. Values represent a ratio of the [average of white line distance + red line distance)/(average of white line distances)]; essentially giving a ratio of migration distance from the center of the punch biopsy to the edge of the migrating keratinocytes divided by the distance from the center to the edge of the punch biopsy (or radius of 1.5 mm, this latter value remaining static among all samples).
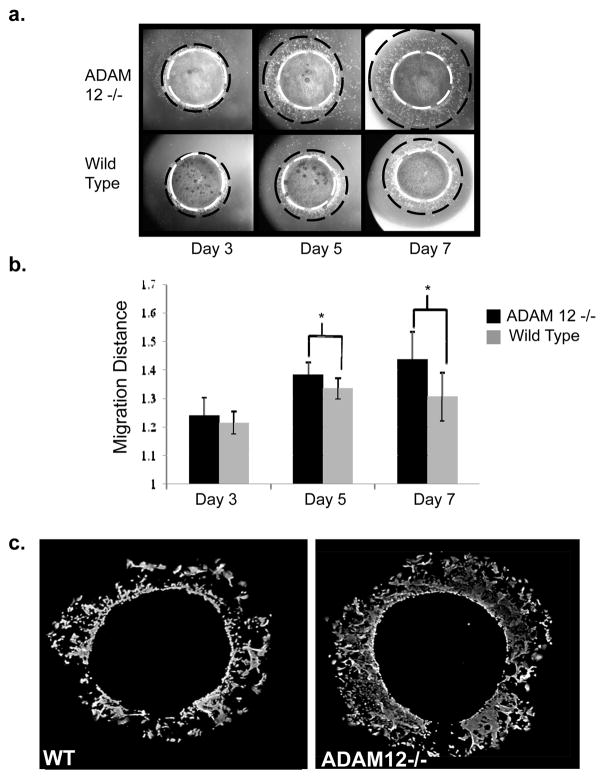
Figure 4. Acute wound model: A pronounced increase in keratinocyte migration in ADAM12−/− mice skin biopsies seven days after wound initiation.
a. Documentation of keratinocyte outgrowth by light microscopy shows the cell migration front (red solid circle) beginning from the biopsy edge (white dashed circle) on day 3 and increasing to a maximum documented distance on day 7. An increase in keratinocyte migration from ADAM12−/− biopsies was generally noted among samples on day 5 and this difference was accentuated on day 7 at termination of experiment. b. Keratinocyte migration was quantified from a total of 9 biopsies from 6 wild type mice and from 18 biopsies from 7 ADAM12−/− mice. A statistically significant increase in migration of 12.1% on day 5 (p=.01) and 35.4% on day 7 (p=.0014) was documented in keratinocytes derived from ADAM12−/− skin biopsies compared to wild type controls. c. Immunofluorescent staining of samples with keratin17, a marker for activated keratinocytes, on day 7 shows that the migrating cells are keratinocytes, and corroborates that keratinocyte migration extended further from the biopsy edge in samples from ADAM12−/− mice compared to wild type controls.
Immunoflourescent staining of keratinocytes
After 7 days of tissue culture the punch biopsy was removed from each well and the remaining adherent cells were washed in phosphate buffered saline (PBS) and fixed for 10 minutes in methanol at +4°C. Following fixation, cells were washed with PBS and permealized with 0.1% Triton X-100 in PBS for 10 minutes at room temperature. Cells were then washed in PBS and further blocked with 5% BSA in PBS for 30 min. Cells were incubated overnight in 200μl/well of a 1:1000 dilution of a rabbit polyclonal antibody against human keratin 17 (kindly provided by Dr. Coulombe) (22) in 5%BSA in 1 × PBS. Wells were then rinsed in PBS and incubated with a 1:150 dilution of a secondary fluorescent - conjugated anti-rabbit IgG (Sigma) for three hours at room temperature. After a final wash in PBS, the stained cells were examined under a Carl Zeiss microscope (Carl Zeiss) and digital images were collected using Adobe Photoshop 4.0 TWAIN 32 ® program (Adobe Systems Incorporated, San Jose, Ca) and processed using Powerpoint® 2007 (Microsoft, Corporation)
RESULTS
Gene chip expression analysis shows up-regulation of ADAM12 in chronic wounds
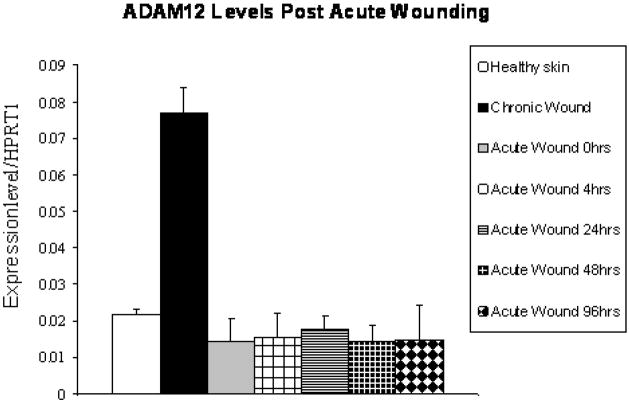
In order to identify molecules whose expression is dysregulated in wound healing, and that might therefore play a role in pathogenesis of wound healing, we performed a microarray gene expression analysis on mRNA isolated from biopsies from five patients with chronic wounds (venous ulcers) and from three normal controls. Using Volcano plot software, we identified a subset of 1557 genes with more than 2-fold changes in gene expression that were statistically significant (p value< 0.05) in comparisons between healthy skin and non–healing edges from chronic wounds (see materials and methods for details). Interestingly, we found that expression of ADAM12 was significantly induced in all patients’ samples (Table 1), raising the possibility that this molecule may contribute to pathogenesis of chronic wounds. The increase in ADAM12 expression in chronic wounds compared to controls was confirmed using quantitative RT-PCR (Fig. 2). When we evaluated the expression of ADAM12 in acute wounds, we found no significant changes in ADAM12 expression levels at 0, 4, 24, 48 and 96 hours post wounding by quantitative RT-PCR compared to healthy skin (Fig. 2).
Table 1. Microarray data shows upregulation of ADAM12 mRNA in human chronic wounds.
Illustrated are the results of the microarray analysis of chronic wounds showing the regulation of various ADAMs in chronic wounds versus healthy skin. ADAM12 is up-regulated 5.88 fold in chronic wounds (p=.013).
| Unigene Symbol | Fold Change | p-value |
|---|---|---|
| ADAM12 | 5.88 | .013 |
| ADAM23 | 5.31 | .088 |
| ADAMTS1 | −4.85 | .048 |
| ADAMTS5 | −3.32 | .012 |
| ADAMTS20 | −2.10 | .005 |
Figure 2. Quantitative RT -PCR shows induction of ADAM12 in chronic wounds.
Quantitative RT-PCR shows expression of ADAM12 in healthy skin, chronic wounds and acute wounds at 0, 4, 24, 48 and 96 hrs post wounding. Expression levels were normalized to HPRT1.
Evaluation of ADAM12 expression in chronic wounds by immunohistochemistry
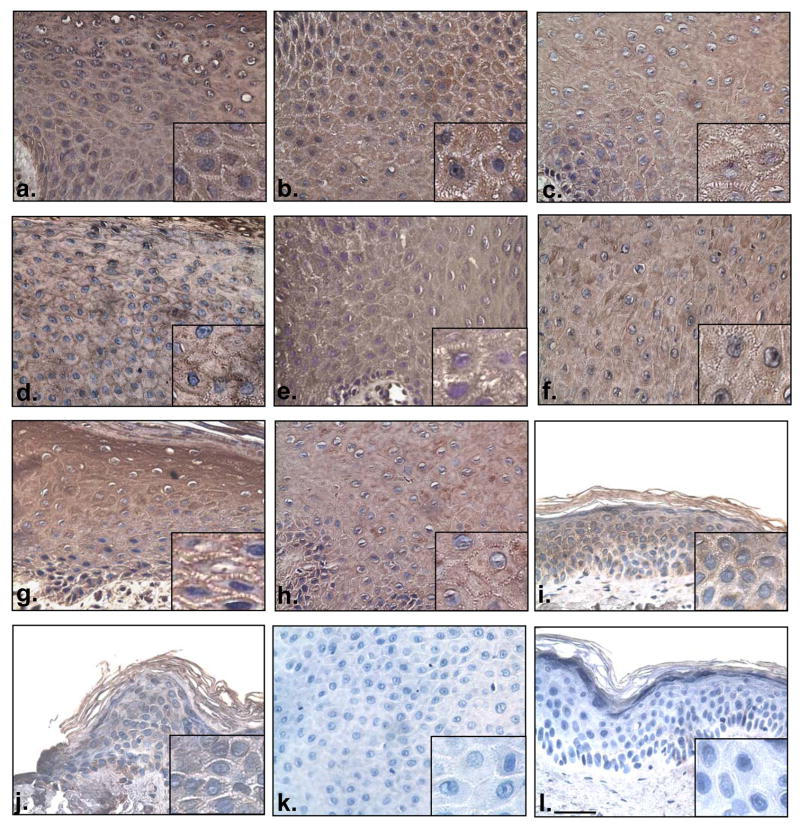
To further corroborate that ADAM12 is over expressed in chronic wounds, we used an ADAM12-specific antibody to perform immunohistochemistry on chronic wound biopsies (Fig. 3a–h). Strong staining with anti-ADAM12 was detected in the cytoplasm and cell membrane of the normal epidermis and acute wound. We found a robust increase in the anti-ADAM12 signal in biopsies originating from the non-healing edges of chronic ulcers, thus confirming the data obtained by microarrays. Membranous staining was patchy and present throughout the epidermis in chronic wound samples while in healthy skin and acute wound, membranous staining was c present only on the membrane surrounding a small number of keratinocytes. We conclude that, consistent with microarray analyses, mRNA and protein expression data, the expression of ADAM12 is induced in chronic wounds, suggesting that this molecule could be involved in impairing wound healing.
Figure 3. ADAM12 is increased in chronic wounds in comparison to healthy skin.
ADAM12 is present in the epidermis of chronic wound skin (a–h). The expression is seen in both cytoplasmic and membranous fractions. Consistent with the microarray and real–time PCR data, ADAM12 expression is increased in chronic wounds when compared to either healthy skin (i) or acute wound (j). Negative controls of chronic wound (k) and normal skin (l) show staining in the absence of the primary antibody. Scale bar 250 microns (magnification 40 x).
Analysis of the migration of keratinocytes from ADAM12-deficient mice
To further evaluate the potential role of ADAM12 in wound healing and to test how lack of ADAM12 affects keratinocyte migration, we used an established mouse skin explant wound healing model (21). Specifically, we measured the distance of keratinocyte migration out of ex vivo cultures of skin explants from newborn wild type pups and their ADAM12−/− littermates over a period of seven days (Fig. 1 and 4a). When plated in tissue culture, keratinocytes derived from both ADAM12−/− and WT mouse explants migrated out from the edge of the punch biopsy, and moved steadily during seven-day period (Fig. 4a). However, the migration of keratinocytes from ADAM12−/− samples was increased compared to samples isolated from wild type control littermates on day 5 (p= 0.01) and day 7 (p=0.0014) (Fig. 4b). On average, keratinocytes derived from ADAM12−/− mice displayed a 35.4% increase in migration compared to wild type control keratinocytes after 7 days. Finally, when we stained the cells migrating out of skin explants with an antibody specific for activated keratinocytes (anti-K17), we found that almost the entire cell population was K17 positive (Fig. 4c). In addition, the anti-K17 staining revealed a more confluent pattern of keratinocytes derived from ADAM12−/− cells when compared to wild type controls.
DISCUSSION
Non-healing wounds such as venous leg ulcers, diabetic foot ulcers, and pressure ulcers represent a major worldwide health burden. Yet much remains to be learned about the mechanism underlying the pathogenesis of chronic wounds, which is thought to depend, at least in part, on dysregulation of cell-cell and cell-matrix interactions. The data presented in this study suggest that the membrane-anchored metalloprotease ADAM12, which has been implicated in cell-cell and cell matrix interactions, could play an integral role in wound healing. Gene array data and immunohistochemistry from human venous reflux ulcers demonstrated that ADAM12 is dysregulated in the non-healing edge of chronic skin ulcers, with at least five-fold greater expression than in healthy skin. In addition, studies with skin explants from ADAM12-deficient mice revealed a significant increase in keratinocyte migration and/or proliferation compared to skin explants from wild-type littermates. Based on these findings, it is tempting to speculate that increased expression of ADAM12 in chronic wounds impairs wound healing through the inhibition of keratinocyte migration and/or proliferation.
ADAM12 could be an attractive target for treatment of chronic wounds because mice lacking this protein have no major defects in development or adult homeostasis, with the exception of a reduction in brown adipose tissue and an impaired formation of interscapular and neck muscles in some, but not all ADAM12-deficient mice (19). This suggests that inhibitors of ADAM12 expression or function should be well tolerated, especially if applied to wounds instead of taken systemically. With respect to the potential mechanism underlying the role of ADAM12 in chronic wound healing, several possibilities must be taken into consideration. Since ADAM12 is a modular protein, consisting of a catalytically active metalloprotease domain as well as a disintegrin and cystein-rich domain and an EGF-like repeat, the role of ADAM 12 in keratinocyte migration and/or proliferation could depend on its catalytic activity, or on the function of its ancillary domains and their role in cell-cell interaction, or a combination of both. With respect to the role of ADAM12 as a metalloprotease, it is possible that ADAM12 either activates an inhibitor of keratinocyte migration and/or wound healing, or alternatively, that ADAM12 inactivates an inhibitory molecule. ADAM12 is already known to be able to participate in ectodomain shedding of several potential substrates on the cell surface, including HB-EGF (19) and EGF (23), and so it is likely to also be able to cleave other proteins on the cell surface, which could be critical for regulating keratinocyte migration. In addition, ADAM12 has been implicated in cell-cell interactions through initial binding to syndecans (24). Moreover, ADAM12 transfected Chinese hamster ovary cells showed a 35%–50% decrease in migration on full-length fibronectin mediated by the α4β1 integrin compared to controls in transwell and scratch wound assays (25). ADAM12 was also recently shown to associate with the TGFβ-type II receptor (TGFβRII), thereby facilitating TGFβ signaling in a manner that does not require the catalytic activity of ADAM12, so the lack of ADAM12 could also affect TGFβ signaling (26). Future studies, including the generation of mice expressing a catalytically inactive ADAM12 will be necessary to dissect the relative importance of these different pathways with respect to the role of ADAM12 in keratinocyte migration and healing of chronic wounds. However, it is noteworthy that the level of ADAM12 expression does not change during acute wound healing, suggesting that its up-regulation is a specific pathogenic marker of non-healing ulcers.
In conclusion, our data demonstrate that human chronic wounds exhibit a marked increase in the expression of ADAM12 when compared to healthy skin. We have shown that the absence of ADAM12 enhances keratinocyte migration and/or proliferation in mice, suggesting that increased ADAM12 expression in chronic wounds directly or indirectly results in the delayed migration/proliferation of human keratinocytes. Based on these findings, we hypothesize that reducing the expression of ADAM12 in chronic wounds may release this “break” on keratinocyte function, thereby prompting wounds to epithelialize faster. Depending on whether the function of ADAM12 relies on its catalytic activity, or its role in cell-cell or cell matrix interactions, or a combination of these mechanism, selective inhibitors of these functions, such as hydroxamate-type metalloprotease inhibitors, or inhibitors that prevent ADAM12 from binding to syndecans, or approaches that reduce ADAM12 expression, could be tested for their ability to promote healing of chronic wounds. With more information on ADAM12’s mechanism of inhibiting keratinocyte migration and the molecular characteristics of certain non-healing wounds, a multi-faceted-patient tailored therapy may reduce the morbidity, mortality, and health care burden associated with chronic wounds.
Acknowledgments
Our research is supported by the National Institutes of Health grants GM64750 to CB, and NR008029 to MT-C. We thank Irena Pastar for help with quantitative RT-PCR and all members of Tomic’s and Blobel’s lab for their support.
References
- 1.Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crovetti G, Martinelli G, Issi M, et al. Platelet gel for healing cutaneous chronic wounds. Transfus Apher Sci. 2004;30:145–151. doi: 10.1016/j.transci.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Moreo K. Understanding and overcoming the challenges of effective case management for patients with chronic wounds. Case Manager. 2005;16:62–63. 67. doi: 10.1016/j.casemgr.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Supp DM, Boyce ST. Engineered skin substitutes: practices and potentials. Clin Dermatol. 2005;23:403–412. doi: 10.1016/j.clindermatol.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Mustoe T. Understanding chronic wounds: a unifying hypothesis on their pathogenesis and implications for therapy. Am J Surg. 2004;187:65S–70S. doi: 10.1016/S0002-9610(03)00306-4. [DOI] [PubMed] [Google Scholar]
- 6.Broughton G, 2nd, Janis JE, Attinger CE. The basic science of wound healing. Plast Reconstr Surg. 2006;117:12S–34S. doi: 10.1097/01.prs.0000225430.42531.c2. [DOI] [PubMed] [Google Scholar]
- 7.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 8.Stadelmann WK, Digenis AG, Tobin GR. Physiology and healing dynamics of chronic cutaneous wounds. Am J Surg. 1998;176:26S–38S. doi: 10.1016/s0002-9610(98)00183-4. [DOI] [PubMed] [Google Scholar]
- 9.Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 10.Ravanti L, Kahari VM. Matrix metalloproteinases in wound repair (review) Int J Mol Med. 2000;6:391–407. [PubMed] [Google Scholar]
- 11.Stojadinovic O, Brem H, Vouthounis C, et al. Molecular pathogenesis of chronic wounds: The role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am J Pathol. 2005;167:59–69. doi: 10.1016/s0002-9440(10)62953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brem H, Stojadinovic O, Diegelmann RF, et al. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol Med. 2007;13:30–39. doi: 10.2119/2006-00054.Brem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 14.Kveiborg M, Albrechtsen R, Rudkjaer L, Wen G, Damgaard-Pedersen K, Wewer UM. ADAM12-S stimulates bone growth in transgenic mice by modulating chondrocyte proliferation and maturation. J Bone Miner Res. 2006;21:1288–1296. doi: 10.1359/jbmr.060502. [DOI] [PubMed] [Google Scholar]
- 15.Galliano MF, Huet C, Frygelius J, Polgren A, Wewer UM, Engvall E. Binding of ADAM12, a marker of skeletal muscle regeneration, to the muscle-specific actin-binding protein, alpha -actinin-2, is required for myoblast fusion. J Biol Chem. 2000;275:13933–13939. doi: 10.1074/jbc.275.18.13933. [DOI] [PubMed] [Google Scholar]
- 16.Peduto L, Reuter VE, Sehara-Fujisawa A, Shaffer DR, Scher HI, Blobel CP. ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene. 2006;25:5462–5466. doi: 10.1038/sj.onc.1209536. [DOI] [PubMed] [Google Scholar]
- 17.Tomic-Canic M, Mamber SW, Stojadinovic O, Lee B, Radoja N, McMichael J. Streptolysin O enhances keratinocyte migration and proliferation and promotes skin organ culture wound healing in vitro. Wound Repair Regen. 2007;15:71–79. doi: 10.1111/j.1524-475X.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 18.Brem H, Kirsner RS, Falanga V. Protocol for the successful treatment of venous ulcers. Am J Surg. 2004;188:1–8. doi: 10.1016/S0002-9610(03)00284-8. [DOI] [PubMed] [Google Scholar]
- 19.Robson MC, Cooper DM, Aslam R, et al. Guidelines for the treatment of venous ulcers. Wound Repair Regen. 2006;14:649–662. doi: 10.1111/j.1524-475X.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 20.Stojadinovic O, Lee B, Vouthounis C, et al. Novel genomic effects of glucocorticoids in epidermal keratinocytes: Inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J Biol Chem. 2007;282:4021–4034. doi: 10.1074/jbc.M606262200. [DOI] [PubMed] [Google Scholar]
- 21.Kurisaki T, Masuda A, Sudo K, et al. Phenotypic analysis of Meltrin alpha (ADAM12)-deficient mice: involvement of Meltrin alpha in adipogenesis and myogenesis. Mol Cell Biol. 2003;23:55–61. doi: 10.1128/MCB.23.1.55-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzalupo S, Wong P, Martin P, Coulombe PA. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev Dyn. 2003;226:356–365. doi: 10.1002/dvdy.10245. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. The nuclear hormone receptor co-activator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGowan KM, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horiuchi K, Le Gall S, Schulte M, et al. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iba K, Albrechtsen R, Gilpin B, et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J Cell Biol. 2000;149:1143–1156. doi: 10.1083/jcb.149.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thodeti CK, Frohlich C, Nielsen CK, et al. Hierarchy of ADAM12 binding to integrins in tumor cells. Exp Cell Res. 2005;309:438–450. doi: 10.1016/j.yexcr.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Atfi A, Dumont E, Colland F, et al. The disintegrin and metalloproteinase ADAM12 contributes to TGF-beta signaling through interaction with the type II receptor. J Cell Biol. 2007;178:201–208. doi: 10.1083/jcb.200612046. [DOI] [PMC free article] [PubMed] [Google Scholar]