Summary
Heterogeneous nuclear ribonucleoproteins (hnRNPs) have been traditionally seen as proteins packaging RNA nonspecifically into ribonucleoprotein particles (RNPs), but evidence suggests specific cellular functions on discrete target pre-mRNAs. Here we report genome-wide analysis of alternative splicing patterns regulated by four Drosophila homologues of the mammalian hnRNP A/B family (hrp36, hrp38, hrp40 and hrp48). Analysis of the global RNA binding distributions of each protein revealed both small and also extensively bound regions on target transcripts. A significant subset of RNAs were bound and regulated by more than one hnRNP protein, revealing a combinatorial network of interactions. In vitro RNA binding site selection experiments (SELEX) identified distinct binding motif specificities for each protein that were over-represented in their respective regulated and bound transcripts. These results indicate that individual heterogeneous ribonucleoproteins have specific affinities for overlapping, but distinct, populations of target pre-mRNAs controlling their patterns of RNA processing.
Keywords: alternative splicing, hnRNP proteins, RNA binding proteins, microarray, Drosophila melanogaster
Introduction
The majority of the genes in higher eukaryotic genomes are interrupted by non-coding sequences, introns, that are removed to allow the coding sequences or exons, to be spliced together producing functional mRNAs. The splicing reaction is performed by a large multi-component machinery, the spliceosome, consisting of ~200 proteins and 5 different small nuclear RNAs (snRNAs). One surprising observation that has arisen from the recent sequencing of higher eukaryotic genomes is that there is little correlation between the number of genes in a given genome and organismal complexity. It has been proposed that the paucity of genes in higher eukaryotes could be compensated by the production of different mRNA isoforms, where portions of the initial pre-mRNA are differentially included in the final mRNA, through a mechanism called alternative splicing of pre-mRNA. Alternative splicing has been found to be almost ubiquitous among multi-exon genes in mammals (Wang et al., 2008). These mRNA isoforms frequently encode different proteins, and can differ in their 5′ or 3′ untranslated regions (UTR) to include cis-acting elements required for the correct expression of the corresponding protein (for review see Hughes, 2006).
Most of our understanding of the mechanisms controlling pre-mRNA splicing comes from molecular approaches focused on single-gene studies. These studies have led to the molecular characterization of only a relatively small number of alternatively spliced genes but provide a general understanding of the basic mechanisms of splicing and its regulation.
The sequences recognized by the splicing machinery are highly degenerate and frequently embed in introns that are significantly larger than the flanking exons, and necessitate auxiliary proteins to promote their use. Conversely, several protein factors have been found to modify the interaction of the splicing machinery with the pre-mRNA. Several of these regulatory protein factors have been characterized and found to recognize cis-acting elements within pre-mRNAs that generally fall into two major categories, splicing enhancers and splicing silencers, found in either exons or introns (reviewed in Black, 2003; Cartegni et al., 2002; House and Lynch, 2008; Pagani and Baralle, 2004; Smith and Valcarcel, 2000).
Two group of proteins with apparently antagonistic effects on splicing regulation have been well characterized to date: the SR family and the hnRNP proteins. Several SR proteins recognize splicing enhancers and stimulate the use of nearby splice sites through interactions with components of the basal splicing machinery, like the U2AF factor and the U1 snRNP 70K protein (for review see Matlin et al., 2005). HnRNP proteins, which have more than 20 different members in most mammals and ten to fifteen members in Drosophila (Dreyfuss et al., 2002) tend to interact with splicing silencers (for review see Matlin et al., 2005). Some hnRNP proteins prevent binding of other splicing factors to the pre-mRNA by forming complexes with RNA and/or other proteins and prevent the association with RNA splicing control elements (Zhu et al., 2001). HnRNP proteins also mediate long range interactions between distant RNA regions flanking alternative exons, thus looping out the intervening region of the pre-mRNA and preventing splicing of the excluded RNA region (Blanchette and Chabot, 1999; Martinez-Contreras et al., 2006). In several cases hnRNP proteins have been found to antagonize, both in vitro and in vivo, the activity of SR proteins (Mayeda et al., 1993; Zahler et al., 2004; Zhu et al., 2001)
High-density oligonucleotide microarray technologies provide the opportunity to profile RNA splicing patterns and the interaction of RNA-binding proteins with RNA transcripts at the whole-transcriptome level, which offers an insight into the post-transcriptional control of gene expression at the level of mRNA maturation and alternative splicing (for review see Blencowe, 2006). We have developed a splicing-sensitive microarray platform to monitor changes in alternative splicing in Drosophila (Blanchette et al., 2005). Our first analysis identified the alternatively spliced genes controlled by four different splicing regulators, two SR proteins SC35 and B52/SRp55 and two hnRNP members from different protein families, PSI and hrp48. This initial analysis suggested that very few genes are co-regulated by these SR proteins and hnRNP proteins while identifying a significant number of genes co-regulated by members of the same protein family. These experiments raised several important questions, namely the extent of co-regulated genes within a family of splicing regulators, the specificity, organization and distribution of the cis-acting elements recognized by those proteins, and the molecular mechanisms used to regulate these alternative splicing events.
The hnRNP A/B family of proteins is well conserved from C. elegans to humans and is believed to carry out similar functions across animal phyla. In this work, we have studied four closely related members of the five known Drosophila hnRNP A/B family, hrp36, hrp38, hrp40, and hrp48. Using splicing-sensitive microarrays, between 200 and 300 genes were detected as specifically regulated by each individual hrp protein. The specific RNA sequence motifs recognized by individual hrp proteins were identified using purified proteins and in vitro selection (SELEX) and found to be different for all four proteins tested. The identified binding sites were significantly over-represented in the genes specifically, as well as co-regulated, by the four hnRNP proteins indicative of a complex regulatory network. The genome-wide RNA binding distributions of the four hrp proteins were characterized using a newly developed nuclear RNA-protein (RNP) complex immunopurification approach in conjunction with Drosophila whole-genome tiling arrays (RIP-Chip). Discrete binding locations on expressed RNA transcripts were characterized for all four proteins and found to be predominantly intronic for hrp36, hrp38 and hrp40, while largely exonic binding regions were found for hrp48. Finally, similarly to what was found in our previous analysis, there was no significant overlap between genes regulated by the four hrp proteins and two of the eight different Drosophila SR proteins. In summary, our data suggest that hnRNP proteins bind and control alternative splicing of specific subsets of pre-mRNAs through sequence-specific binding to their target RNAs. Together these studies provide mechanistic insights into how these subsets of specific pre-mRNAs are regulated in vivo.
Results
HnRNP proteins regulate both unique and overlapping subsets of splice junctions
Several hnRNP proteins have been found to specifically regulate the alternative splicing patterns of transcripts in all metazoans. HnRNP A1 is one of the best characterized hnRNP proteins and is part of a larger family of related proteins, the hnRNP A/B RNA binding proteins (Dreyfuss et al., 2002). Previous reports suggest overlapping activities among the mammalian family members (Bilodeau et al., 2001; Caputi et al., 1999; Hutchison et al., 2002; Mayeda et al., 1994) as well as the Drosophila hnRNP A/B family members (Zu et al., 1996). In order to examine the extent of this overlap, we used a microarray-based approach (Blanchette et al., 2005) to globally identify the specific splicing events on target transcripts regulated by four highly related members of the fly hnRNP A/B family, namely hrp36, hrp38, hrp40 and hrp48 (see Supplemental Figure 1 for a description of the proteins and their degree of similarity to each other).
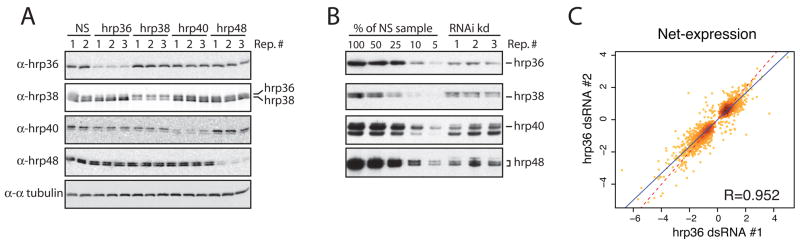
To accommodate the release of Flybase version 4.2.1, a redesigned microarray was used to interrogate the 2,797 annotated Drosophila genes having EST evidence of alternative splicing (see Exp. Procedures and Supplemental Materials). These alternatively spliced genes are predicted to produce on average 2.82 isoforms per genes to generate 7,892 different mRNAs corresponding to 9,434 and 10,676 alternative and constitutive exon-exon junctions simultaneously monitored. First, we investigated the effect of changing the levels of specific hnRNPs on global splicing patterns in Drosophila cell culture. Expression of specific splicing regulators was reduced by RNAi knockdowns in three independent replicates and the splice junction abundance was profiled using the splicing-sensitive microarrays. The net-expression of all mRNA splice junctions was calculated and, after population normalization and averaging of the biological replicates, the junctions whose abundance was significantly affected in the knockdown samples (2 or more standard deviations from the population net-expression average) were grouped together. Reduced expression of hrp36, hrp38, hrp40 and hrp48 was confirmed by immunoblot analysis for each protein (Figure 1A). The level of knockdown, evaluated to be around or greater than 90% of the endogenous level, was similar between the triplicate samples and between the different hnRNPs tested (Figure 1B). In addition, the specificity of the knockdowns can be seen by the absence of appreciable changes in expression of the other three hnRNP proteins in each knockdown (Figure 1A). Finally, using hrp36 knockdown as a test-case, no major off-target effects were detected as the overall changes in net-expression between two different non-overlapping dsRNAs were very similar (Figure 1C, Pearson correlation coefficient = 0.952).
Figure 1.
Knockdown of expression of four members of the Drosophila hnRNP A/B family by RNAi in Drosophila S2 cells. A) Western blot analysis shows that hrp36, hrp38, hrp40 and hrp48 are efficiently knock down in triplicate samples compare to cells treated with a non-specific dsRNA (NS). Alpha-tubulin is used as control to confirm that similar levels of proteins were loaded. B) Titration of the protein sample treated with non-specific dsRNA is used to evaluate the knockdown efficiency. C) The overall changes in the net-expression value is similar for cells knockdown in the hrp36 expression with two different non-overlapping dsRNA. A splicing junction in one knockdown is plotted against the same splicing junction in the second knock down. The relative feature density is expressed as darker shade of yellow. The blue line is the fitted linear model used to calculate the R-value. The dashed red line corresponds to a perfect correlation.
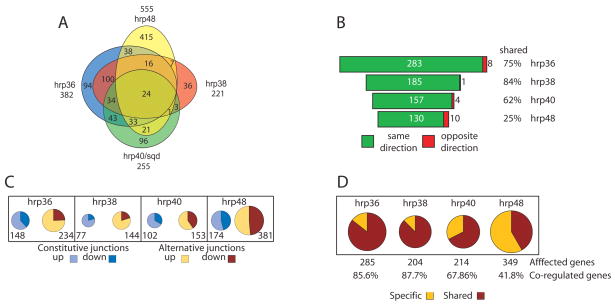
Using the splicing-sensitive microarrays, we found that the number of significantly affected splice junctions in the knock-downs of hrp36, hrp38, hrp40 and hrp48 were estimated to be 382, 221, 255 and 555 respectively, (Figure 2A) with a validation rate estimated by RT-PCR to be around 70% (Supplemental Figure 2). These numbers are similar to what we had previously found for RNAi knock-downs of the Drosophila SR proteins, dASF/SF2 and B52/SRp55 (Blanchette et al., 2005). We found that both alternatively and, to a lesser extent, constitutively spliced junctions were affected (Figure 2C) suggesting that either the current annotation does not account for all the different alternatively spliced mRNAs or that several cryptic splice sites are being used when the expression level of a given hnRNP protein is reduced.
Figure 2.
Genome-wide identification of splice junctions regulated by individual members of the Drosophila hnRNP A/B family. A) Venn diagram of the extent of overlap of the splice junctions regulated by each individual hnRNP protein. B) Representation of the co-regulated junctions for each splicing regulator demonstrating that most of the co-regulated junctions are affected in the same direction (green) or the opposite direction (red) together with the fraction of the junctions co-regulated for each individual hnRNP protein. C) Breakdown of the number of junctions found to be significantly affected in each individual RNAi knockdown. Constitutive junctions are shown in light blue (up) and dark blue (down). Alternative junctions are shown in light orange (up) or red (down). D) Number of genes with junctions regulated by a single hnRNP protein (specific-light yellow) together with the fraction of genes co-regulated by a second hnRNP protein (shared-red).
We next asked whether a significant number of regulated splicing events were under the control of more than one individual hrp protein. We found that hrp36 and hrp38 shared the vast majority of their regulated junctions with the other hrp proteins, (Figure 2A and 2B; 75% and 84% of the affected junctions respectively) while hrp48 shared the lowest number of junctions (Figure 2A and 2B; 140 junctions or 25% of the affected junctions). Interestingly, the vast majority of the junctions co-regulated by more than one hnRNP proteins (more than 93% in every case) were affected in the same direction (i.e., a junction going up in a given knock down also goes up in the other knock downs and vice versa; Figure 2B).
Using the current splicing-sensitive microarray, using a cut-off of 2 standard deviationds, the number of alternatively spliced transcripts regulated by hrp36, hrp38, hrp40 and hrp48 in Schneider Line 2 cells (S2) is estimated to be 285, 204, 214 and 349 genes, respectively (Figure 2D). Using FatiGO (Al-Shahrour et al., 2004) to assess over or under representation of gene ontology terms in the populations of genes regulated by the different hnRNP proteins, no significant functional clustering was detected in the genes regulated by hrp36, hrp38 and hrp40, while only a small number of genes involved in neuronal development showed a significant over-representation in the hrp48 affected genes (data not shown and Supplemental Table 1). This suggests that the Drosophila hnRNP A/B homologues regulate broad and unrelated sets of genes.
Different hnRNP members associate with different RNA subsets
Analysis of exon-junction utilization in different knockdown environments suggests that different members of the Drosophila hnRNP A/B family regulate alternative splicing of distinct, but overlapping, sets of target pre-mRNAs. One question raised by this observation is whether different proteins that share a target RNA bind to the same region or to different locations. We used antibodies specific to the four hnRNP proteins to immunopurify RNAs from nucleoplasmic ribonucleoprotein (RNP) particles containing each protein (Supplemental Figure 3) and determined their genomic locations on the pre-mRNAs isolated from each purification using a 35 nt tiling array. The RIP-Chip approach (RNA IP detected on DNA Chip) was first validated by confirming the presence of six previously known targets of hrp48 in the immunopurified RNA, as well as tracts of significantly overrepresented signals on the tiling array data (Supplemental Figure 4). In addition, 20 randomly selected RNA regions bound by hrp36 on the tiling array were selected and their level of enrichment in immunopurified RNA was compared to the starting RNP extract. The RNA abundance relative to three different highly expressed but unbound genes, RpS15, Hsc70-4 and Ef2b, was evaluated by real-time RT-PCR from two individually hrp36-immunopurified RNA samples and compared to two RNA samples isolated from the starting RNP extract. The real-time RT-PCR analysis shows a significant enrichment of 18 out of the 20 regions predicted to be bound by hrp36 in all three comparisons (Supplemental Figure 5, validation rate of 90%).
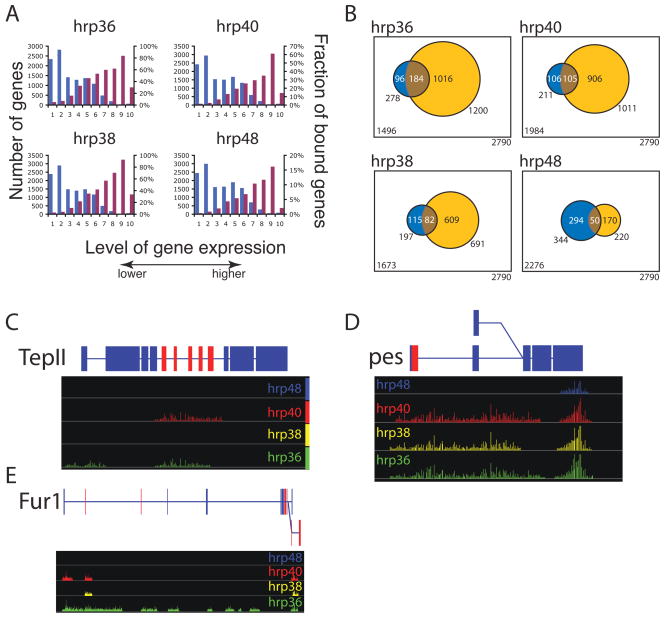
We found that hrp36, hrp38, hrp40 and hrp48 bind to RNAs that map to 5658, 2286, 4001 and 1359 regions of the Drosophila genome, respectively (Table 1). Within these bound regions, hrp36 binds to the largest number of genes (2410 different genes; Table 1), while hrp48 binds to only 341 different genes (Table 1). The number of genes bound by hrp38 and hrp40 was intermediate between these two extremes, 1219 and 1922 different genes, respectively. Interestingly, intronic sequences were mainly found associated with hrp36, hrp38 and hrp40 (54.1% for hrp36 and hrp38 and 50.9% for hrp40) while, in contrast, hrp48 was found to bind preferentially to exonic sequences (44.9%; Table 1). Intergenic sequences, although to a lesser extent, were also found associated with all four hnRNP proteins. Between 7.3% for hrp48 to 11.7% for hrp36 of the tract (Table 1) comes from intergenic sequences disconnected from nearby annotated genes and might represent un-annotated genes or, most likely, un-annotated remote 5′ or 3′ exons connected to distant genes (Manak et al., 2006). These observations, together with the fact that hrp48 shares the least number of regulated splice sites, raise the possibility that hrp48 has more distinct and specialized functions than the other three Drosophila hnRNP A/B family members.
Further, we determined the abundance of all expressed mRNAs in Drosophila S2 cells using Affymetrix GeneChip Drosophila Genome 2.0 arrays and the fraction of genes bound by a given hnRNP protein. If, as it is often assumed, that hnRNP proteins bind RNA non-specifically (Dreyfuss et al., 2002 and references therein), there should be a strong correlation between the representation of a specific mRNA in the immunopurified material and the abundance of that mRNA in the starting RNP fraction. Contrary to this notion, we found that the most highly expressed genes (Figure 3A, higher 10 percentile) are not as frequently bound as their less expressed counterparts (Figure 3A, percentiles 30–90). With the exception of genes that are not expressed or expressed at very low levels (Figure 3A, lower 30 percentiles) the fraction of genes bound by each individual hnRNP is very similar (Figure 3A). This observation strongly suggests that the four members of Drosophila hnRNP A/B family of proteins bind to distinct, but only partially overlapping populations of mRNAs in a selective manner.
Figure 3.
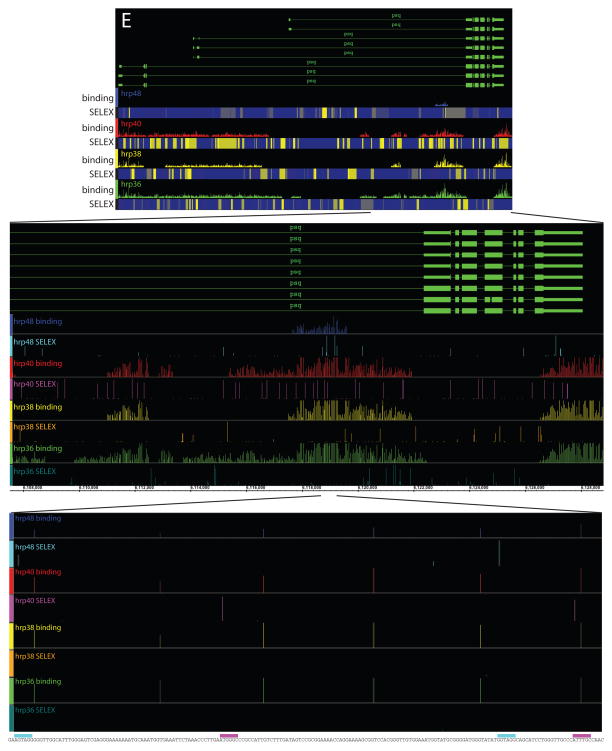
Genome-wide identification of the RNAs associated with the different hnRNP proteins in RNP complexes. A) Distribution of the level of gene expression measured by Affymetrix arrays measured in S2 cells (blue bar) and the fraction of RNA bound by a hnRNP protein within a given level of expression (purple bar). B) The overlap between the genes that were found regulated in the splice junction microarray (blue) and bound in the tiling array (yellow). The highly significant overlapping transcripts are represented in brown (Fisher exact test, p-value<2×10−5). D) TepII and pes, two representative genes bound by hnRNP proteins showing extensive coverage suggestive of the spreading model. C) Fur1 a representative gene bound by hnRNP proteins showing discrete intronic binding tracts for hrp40 suggestive of the looping model. Significantly over-represented signal in the IP samples over the signal measured from RNA extracted from the starting RNP extract are represented by bars along the genome with the eight of the bar corresponding to p-values (expressed as −Log (p)). Above is a representation of the gene structure, blue boxes are constitutive exons, red boxes are alternative exons, lines are introns. Pes has two alternative promoters while Fur1 as two alternative poly-A signals represented by the 2 levels in the gene structure.
Examination of individual genes provides diverse examples of differential binding by hnRNP proteins. For example, the alternatively spliced Fur1 pre-mRNA shows discrete binding for hrp36, hrp38 and hrp40, where some of the bound regions overlap while other transcript regions are specific to individual proteins (Figure 3E). A very different outcome is observed on the pre-mRNA encoding the alternatively spliced TepII and pes genes, where binding of the different hnRNP proteins are contiguous over multiple regions (Figure 3C and 3D). These and other examples support the specific nature of interaction between hnRNP A/B family members and individual pre-mRNAs in Drosophila. In addition, the strong bias of hrp36, hrp38 and hrp40 for intronic RNA is additional evidence of the specific nature of their RNA binding properties, introns are short lived, nuclear RNA species and they are not expected to be preferentially associated and recovered with proteins with low RNA binding specificity.
We next sought to determine whether binding of a given hnRNP protein to an RNA transcript correlates with the ability of that hnRNP protein to regulate alternative splicing of the bound pre-mRNA. We found an extensive overlap between genes with RNAs found associated with a given hnRNP protein on the tiling arrays and alternative pre-mRNA splicing regulated by the same protein on the splicing-sensitive microarrays (Exact Fisher test p-value<2×10−5; Figure 3B). This results support the notion that the specific binding of a given hnRNP protein to a pre-mRNA is likely to be associated with the control of alternative splicing of the bound pre-mRNA.
Binding site specificity of the different members of the Drosophila hnRNP A/B protein family
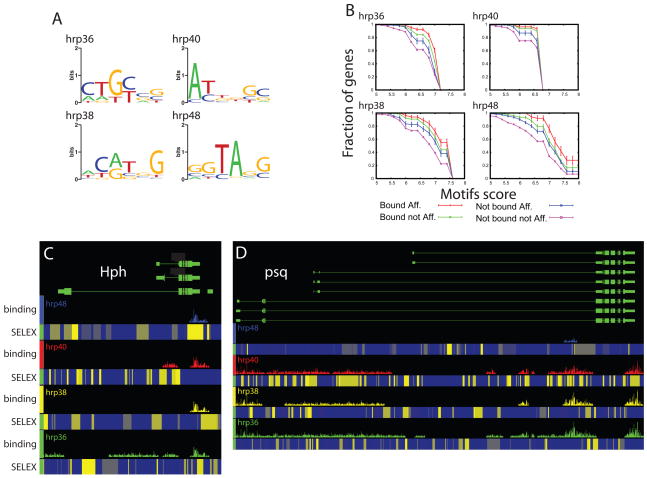
Our experiments show that 270 genes were regulated by more than one hnRNP protein, and also that 1888 RNAs were bound by more than one hnRNP protein. What remained unknown was whether different hnRNP proteins acting on the same target pre-mRNA interact with the same or different cis-acting RNA elements. To address the RNA binding specificities of the hrp proteins, we performed in vitro selection of binding sites (SELEX) on recombinant hrp36, hrp38, hrp40 and hrp48 proteins. Following eight rounds of selection, individual bound RNAs were cloned and sequenced. Sequences were aligned and the common motifs in them were identified using MEME (Bailey and Elkan, 1994; Figure 4A). The models of the most overrepresented sites derived for each of the RNA-binding proteins appear to be unrelated to each other (Figure 4A). Comparison of these models to the Drosophila transcriptome revealed a significant over-representation of each of these preferred binding sites in the population of genes regulated by the corresponding hnRNP protein, compared to the rest of the genes queried by the splicing-sensitive microarray analyses (Supplemental Figure 6). If we examined the subset of genes that were both found in a complex with a given hnRNP protein and exhibited differential splicing in the cell lines where the same protein was knocked-down, the extent of enrichment in the corresponding preferred binding site was even higher (Figure 4B, red lines). Moreover, in all cases tested, the second and third largest fraction of genes having high scores for the preferred SELEX binding motifs were the genes found to be bound only by the corresponding hnRNP protein and found to only exhibit differential splicing in the RNAi knock-down experiment, respectively (Figure 4B, green and blue lines respectively). Finally, in all four cases studied here, the populations of genes that were neither bound nor regulated by a specific hnRNP proteins had the lowest fraction of genes with high scoring in vitro-selected binding sites for the corresponding hnRNP protein (Figure 4B, purple lines). This series of observations provide further evidences that each hnRNP protein regulates alternative splicing through the recognition of specific and different binding sites within its target pre-mRNAs, and that multiple hnRNP proteins may simultaneously target the same pre-mRNA through distinct binding sites.
Figure 4.
HnRNP proteins bind to different and specific RNA motifs that are over-represented in the genes regulated and bound by the respective hnRNP proteins. A) Graphical representation of the preferred binding motif identified by SELEX for each individual hnRNP protein (Crooks et al., 2004). Height of each bar shows the information content at each position of the binding motif in bits (log-odds in base 2). B) Cumulative distribution of the presence of at least three occurrences of the SELEX motif in the genes identified as being regulated by the splice junction array or identified as bound by the tiling array. Error bars are determined analytically using the binomial distribution and correspond to 1 standard deviation: sqrt[np(1 − p)]/n, where n is the number of sequences searched and p is the observed probability of having sites of given score. C–D) Hph and psq, two representative genes showing that the binding tracts identified on the tiling arrays (colored bar tracts) are frequently associated with clusters of high affinity binding sites. A moving window using the SELEX motifs was used to calculate the score at each position along the fly genome. The motif score at a given position is represented as a heat map, blue low motif score-yellow high motif score. E) Magnification of Hph binding tracts showing a 20,000 nt long region (middle panel) to the nucleotide level showing the underlying genomic sequence (bottom of the window) with the presence of the SELEX motifs (colored bar above the sequence) corresponding to the hrp48 and hrp40 recognized motifs (cyan and magenta boxes respectively).
The preferred RNA binding motifs identified in the SELEX experiments can be used to search for putative binding sites within the RNA regions identified in the RIP-Chip analysis. In many cases, exemplified by the Hph and psq genes, one or more of the respective preferred binding motifs are indeed found in the regions covered by hnRNP proteins (Figure 4C and D). The gene regions identified on the tiling arrays following immunopurification of the hnRNP complexes are often found to contain clusters of the preferred SELEX motifs (Figure 4C, D and E for a nucleotide level blow up and Supplemental Figure 7). Among the genes bound by multiple hnRNP proteins, we noted that the high-scoring motifs for a given hnRNP protein are not always evenly distributed along the bound region. Frequently, the bound regions are found associated with clusters of the preferred binding motifs of co-associated hnRNP proteins as with Hph and psq (Figure 4C and D). These observations strongly suggest the involvement of cooperative binding among different hnRNPs to form the mature RNP particles (Domsic et al., 2003; Zhu et al., 2001). This cooperative arrangement of low affinity and high affinity binding sites for different proteins is reminiscent of the architecture of transcription factor-binding sites in eukaryotic transcriptional enhancers (Papatsenko et al., 2006; Zeitlinger et al., 2007; Zinzen et al., 2006).
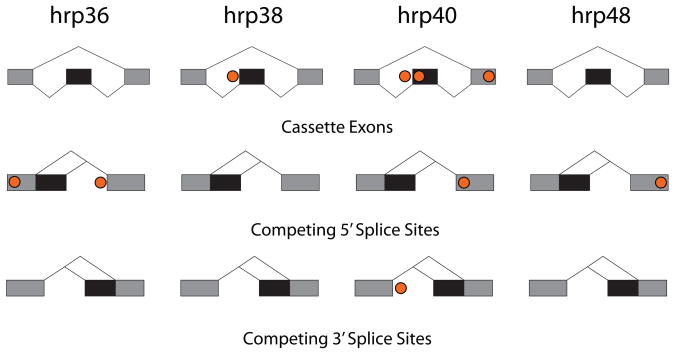
Knowing the target transcripts and specific splice junctions affected by RNAi to each of the four hnRNP proteins and the RNA binding site motifs from the SELEX experiments, we asked if there was enrichment of the SELEX motifs near the affected splice junctions. We have previously performed a similar analysis for the two SR proteins, B52/SRp55 and dASF/SF2 (Blanchette et al., 2005) and similar studies were done with mouse Nova-1/2 target transcripts (Ule et al., 2006). Here, the affected splice events were grouped according to splicing patterns (cassette exon, competing 5′ splice sites and competing 3′ splice sites). This analysis looked for the enrichment of preferred binding site scoring matrices within windows adjacent to affected splice events compared to splice events not affected by the hnRNP (see Experimental Procedures and Supplemental Information). Positions enriched with hnRNP binding sites are shown in Figure 5 (also Supp. Figure 8). These findings suggest a preferential binding site location for a given factor controlling a specific splice event, as seen previously (Aznarez et al., 2008; Ben-Dov et al., 2008; Blanchette et al., 2005; Castle et al., 2008; Licatalosi et al., 2008; Ule et al., 2006; Zhang et al., 2008). However, none of the enrichments were significant after using a conservative Bonferroni correction for >2,000 comparisons between affected splicing events and controls. Thus, while the target transcripts affected by RNAi against a given hrp protein appear to be enriched in the SELEX motifs for that factor (Fig. 4B), there does not seem to be a strict fixed location at which a given factor acts on a specific class of splicing events (Fig. 5). Other studies have shown a complex relationship between the location of splicing control elements and their activities (Goren et al., 2006; Wang et al., 2006) and this lack of a strict spacing of regulatory elements may reflect differences in the mechanisms of action of the Drosophila hnRNP A/B family of proteins (Cartegni et al., 2002) and the KH domain splicing factor, Nova-1 (Ule et al., 2006). This variability in the spacing of regulatory elements is also common in transcriptional enhancers and silencers (Levine and Tjian, 2003). Our bioinformatic analysis also does not take into account the possible role of weak or non-consensus binding sites, which could be important in the context of cooperative interactions with other splicing factors and might explain the lack of significant enrichment of strong binding sites within fixed windows near affected splice junctions.
Figure 5.
Locations of binding site enrichment for each hnRNP. Circles show positions that had an enrichment of SELEX motifs in affected splicing events (two-sided Wilcoxon ranked-sum test, p-value<0.01 without multiple trials correction for >2,000 trials) using any of several measures for enrichment (see Experimental Procedures and Supp. Figure 8 for details).
Control of alternative splicing by hnRNP and SR proteins
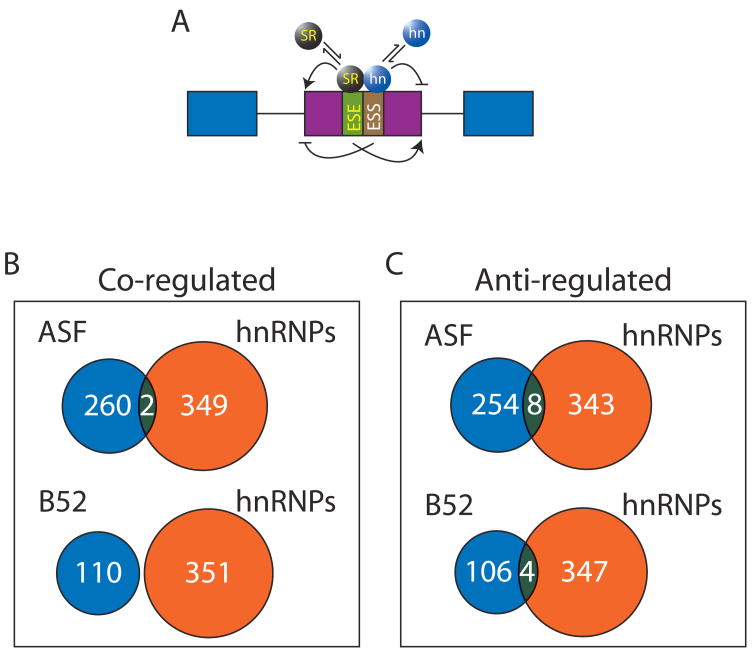
A model of antagonistic control of alternative splicing, mediated by the relative levels of hnRNP and SR proteins, has been proposed (Cáceres et al., 1994; Eperon et al., 2000; Mayeda et al., 1993; Mayeda and Krainer, 1992; Zhu et al., 2001; Figure 6A). We compared the results of our current analysis with the data generated from a previous study where the expression of two SR proteins, dASF/SF2 and dSRp55/B52, were knocked-down (Blanchette et al., 2005). The most important conclusion from our previous comparison was that the SR and the two different hnRNP protein knock-downs affected distinct sets of splice junctions. With a very few exceptions, a similar conclusion was reach when this previous analysis was extend to include the four drosophila hnRNP A/B homologues. Namely, that very few splice junctions were affected in the same and opposite directions (Figure 6B and C), suggesting that, at least in the Drosophila cell culture system with the two SR proteins examined here, an antagonistic relationship of SR and hnRNP proteins in alternative splicing is extremely rare. This issue needs to be addressed further in vivo in other model organisms especially in mammals.
Figure 6.
No significant antagonistic regulation is found between two SR proteins and four hnRNP A/B family proteins in a single defined cellular environment. A) It has been suggested that several genes are under the antagonistic control of SR and hnRNP proteins where the pattern of splice isoforms is dependent on the relative concentrations of SR and hnRNP proteins. B) Comparison of the genes that are found to be regulated in the same direction between hnRNPs (this analysis) and two different SR proteins (Blanchette et al., 2005). B) Comparison of the genes that are found to be regulated in opposite directions between hnRNPs (this analysis) and two different SR proteins (Blanchette et al., 2005).
Discussion
In this study, we report a genome-wide analysis of control of alternative splicing by four Drosophila homologues of the mammalian hnRNP A/B proteins, a family of proteins that has been implicated in a variety of cellular processes including telomere biogenesis, mRNA localization, translation and stability (reviewed in Krecic and Swanson, 1999). By interrogating each predicted splice site by specific molecular probe in an array format, we have found that the four different Drosophila hnRNP A/B protein homologues regulate alternative splicing of specific, but partially overlapping sets of pre-mRNAs. We also found that a majority of the targets of the hrp36, hrp38, and hrp40 proteins were co-regulated by more than one member of this protein family, while hrp48 regulated a larger and more distinct subset of pre-mRNAs. Moreover, RIP-Chip experiments indicate that the former three proteins bind predominantly intronic regions of pre-mRNA, while hrp48 predominantly binds to exonic sequences, in both discreet and more extensive transcript regions. The preferred RNA binding motifs for each individual protein, determined by the SELEX method, were different for each protein and the transcripts regulated and bound by each factor were found to be enriched in the corresponding binding sites.
HnRNP proteins and splicing silencer function: spreading or looping?
Different models have been proposed to explain the molecular mechanisms by which hnRNP proteins control alternative splicing at splicing silencer elements. Our RIP-Chip data show the binding distributions of the hrp proteins on target transcripts and can be used to address the models for how splicing silencers are controlled by hnRNP proteins. It had first been proposed that hnRNP A1 controls its own alternative pre-mRNA splicing by a mechanism involving protein-protein interactions between distantly bound A1 protein at high-affinity binding sites. These distant binding sites would bring the widely-separated splice sites in close proximity, while preventing utilization of the more proximal splice sites, presumably by looping-out of the alternative exon leading to exon skipping (Blanchette and Chabot, 1999). This model, involving protein-mediated looping, also predicted that hnRNP A1 binding sites should be able to act as splicing enhancers to promote intron definition when the intron is large, which was recently documented (Martinez-Contreras et al., 2006; Nasim et al., 2002). Consistent with the idea of hnRNP-mediated looping, several discrete binding regions that are distantly spaced and flank alternative exons can be found on specific pre-mRNAs like on the Fur1 pre-mRNA (Figure 3E), suggesting that the Drosophila hnRNP A/B members could regulate alternative splicing in vivo by a mechanism similar to the looping model defined by biochemical experiments in vitro. More recently, a different model has also been proposed to explain how mammalian hnRNP A1 regulates alternative splicing. It has been proposed that hnRNP A1 binding to exonic regions prevents binding of SR proteins to their cognate splicing enhancers by nucleating and spreading over extended regions of the pre-mRNA via cooperative RNA binding to weak adjacent sites which is also mediated by protein-protein interactions between hnRNP proteins (Domsic et al., 2003; Zhu et al., 2001). Consistent with the spreading mechanism, large regions of pre-mRNA were found to be associated with the four different Drosophila hnRNP proteins, as in the TepII and pes pre-mRNA (Figure 3C and D), suggesting that a similar nucleation mechanism might also be used by this family of splicing factors to control alternative splicing events. Interestingly, we also see that local smaller regions bound by one hnRNP protein can be found within a larger region covered by a different hnRNP protein as seen for FurI where discrete regions of hrp40 binding are found within a large portion of this pre-mRNA that is bound by hrp36. These observations suggest that both biochemical models might actually function together and provide the first in vivo evidence supporting both the spreading and looping models for splicing silencer function.
An RNA Splicing Code
Recently, several genome-wide surveys of the patterns of alternative splicing across different mammalian tissues have identified known or novel RNA sequence motifs associated with specific differential splicing events (Castle et al., 2008; Ule et al., 2006; Wang et al., 2008). It is now becoming clear that specific splicing regulators rely on an RNA code in their target transcripts and that this RNA code is responsible for establishing the cellular constellation of differentially spliced mRNAs (Aznarez et al., 2008; Castle et al., 2008; Licatalosi et al., 2008; Wang et al., 2008; Zhang et al., 2008). However, very few and only individual factors regulating alternative splicing have been globally linked to their cis-acting regulatory elements (Aznarez et al., 2008; Licatalosi et al., 2008; Zhang et al., 2008). In this analysis, we found that a large proportion of the pre-mRNAs regulated by either hrp36, hrp38 hrp40 or hrp48 were also regulated by at least one additional hnRNP protein. However, even though their protein sequences are similar (Supplemental Figure 1), our studies showed that the RNA motifs recognized by each hrp protein are distinct. Nonetheless, their binding locations observed across target transcripts are extensively overlapping, which raises the possibility that some hnRNP A/B family proteins may have redundant functions at individual splice junctions. Interestingly in Drosophila, single hrp36 or hrp38 mutants are viable, but combining mutations in both genes causes synthetic lethality (Haynes et al., 1991; Zu et al., 1996; Haynes S. personal communication), which is consistent with the largely overlapping molecular data sets of bound RNAs and regulated splice-junctions identified here. By contrast, individual hrp40, also known as squid (sqd), and hrp48 mutations are lethal (Hammond et al., 1997; Norvell et al., 1999).
It is also interesting to note that both hrp36 and hrp38 over-expression were able to affect the alternative splicing of dopa decarboxylase (Ddc) in vivo (Shen et al., 1995; Zu et al., 1998; Zu et al., 1996) and that no change in Ddc splicing pattern was observed in the hrp36 null mutant genetic background (Zu et al., 1996), possibly due to the redundant activity of the hrp38 protein. One alternative explanation for the large fraction of co-regulated targets between hrp36, hrp38 and hrp40 is the possibility that they might exert their action through a combinatorial mode of regulation. Several independent pre-mRNAs have been found to be under the control of a complex network of protein-protein and protein-RNA interactions and it is thought that a given splicing pattern is generated by the combination of these interactions (For reviews see Matlin et al., 2005; Smith and Valcarcel, 2000). For example, the skipping of a given alternative exon might first require binding of an hnRNP protein that, through protein-protein interactions, may mediate cooperative binding of a second hnRNP protein to lower affinity binding sites nearby, leading to the repression of splicing of the alternative exon. In this case, the proper control of alternative splicing would require the combination of these two events the absence of either hnRNP proteins would lead to the same phenotype. A combinatorial mode of regulation is consistent with the high frequency of shared binding regions observed in the data from the RIP-Chip analysis, as well as the extensive overlap between the alternative splicing patterns affected by individual hnRNP proteins.
By comparing the pre-mRNAs regulated by the four hrp proteins with data from a genome-wide analysis of the genes regulated by two SR proteins, dASF/SF2 and dB52/SRp55 (Blanchette et al., 2005), we have found that, at least for the SR proteins tested, an antagonistic regulation between SR and hnRNP proteins is not a major mode of regulation in Drosophila cell culture. Similarly, over-expression of hrp36 did not affect the overall localization of SR proteins on transcripts expressed in larval polytene nuclei (Zu et al., 1996), suggesting that dramatic changes in the SR/hnRNP ratio do not significantly affect the distribution of SR proteins on pre-mRNAs. However, it is possible that when the level of an hnRNP is reduced, for example in the case of an RNAi knockdown, that sites normally occupied by hnRNP proteins, are now freed up and become accessible to binding by SR proteins resulting in SR-dependent changes in alternative splicing profiles. In fact, an example of this type of situation has been observed in the Dscam pre-mRNA, where SR protein dependent ectopic inclusion of multiple exons can only be seen when the level of hrp36 is reduced by RNAi (Olson et al., 2007). Global analysis of pairwise RNAi knockdowns would provide a better understanding of these complex interactions among alternative splicing regulators.
Experimental Procedures
Cell culture and RNAi
Serum-free adapted D.mel2 cells (Gibco) were maintained at 25°C in Drosophila SFM. In vitro transcription, RNA purification and preparation of double-stranded RNA was done as described (Blanchette et al., 2005). RNAi knock-downs were performed by seeding 2×106 cells per well in a 6 well plate and by adding 10 μg of dsRNA. After 48 hours of incubation, a second round of 10 μg of dsRNA was added and the cells were allowed to grow for 48 more hours before being harvested and the RNA purified using the RNeasy kit following the manufacturer’s protocol (Qiagen).
Splicing sensitive microarray; design and hybridization
36 nucleotides exon-junction and exon-body probes were selected from all genes from the GadFly 4.2.1 Drosophila melanogaster genome annotation that contained more than one annotated transcript. A complete description of the design and the hybridization procedure and data analysis can be found in the supplementary materials and references there in.
Tiling array hybridization
Nuclear RNP fractions from S2 cells were prepared using a modified version the mammalian RNP protocol (See Supplementary Materials for more details; Pinol-Roma et al., 1990). The immuno-purification, labeling and hybridization were done as previously described (Olson et al., 2007). The statistical analysis of the bound regions was performed using the TiMAT analysis suite (http://bdtnp.lbl.gov/TiMAT/) essentially as described (Li et al., 2008).
RNA SELEX
In vitro selection of the RNA-preferred binding motif was performed using recombinant protein as described elsewhere (Amarasinghe et al., 2001; Fitzwater and Polisky, 1996). A detail description of the procedure can be found in supplementary materials.
Bioinformatic analysis of position specific motif enrichment
Specific alternative splicing events were inferred from the Drosophila melanogaster genome annotation and computationally searched for enrichment of binding sites in the hnRNP affected genes and compared to the rest of the genome for statistically significant over-representation. A detail description of the bioinformatic analysis can be found in the supplemental materials.
Accession codes
Microarray data have been deposited with to the NCBI GEO public database. Accession number XXXX (Waiting for it from GEO)
Supplementary Material
Acknowledgments
We are grateful to A. Mushegian, B. Chabot, K. Hansen and M. Levine for critical comments and suggestions on the manuscript. Many thanks to M. Adams for the production and purification of recombinant hrp48. Special thanks to R. Tjian, M. Biggin and G. Rubin for providing us early access to Drosophila whole genome tiling arrays. We would also like to thank Agilent Technologies for granting us access to their clean room scanning facility. This work was supported by a U.S. National Institutes of Health grant (R01GM61987). M.B. was the recipient of a Human Frontier Science Program long-term fellowship. A.N.B. is supported by an NSF Graduate Research Fellowship. S.E.B. and A.N.B. are also supported by National Institutes of Health grants R01GM071655 and U01HG004271.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Amarasinghe AK, MacDiarmid R, Adams MD, Rio DC. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA. 2001;7:1239–1253. doi: 10.1017/s1355838201010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Domsic JK, Mayeda A, Krainer AR, Stoltzfus CM. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J Virol. 2001;75:8487–8497. doi: 10.1128/JVI.75.18.8487-8497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu Rev Biochem. 2003 doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsic JK, Wang Y, Mayeda A, Krainer AR, Stoltzfus CM. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol. 2003;23:8762–8772. doi: 10.1128/MCB.23.23.8762-8772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences--The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Hammond LE, Rudner DZ, Kanaar R, Rio DC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Johnson D, Raychaudhuri G, Beyer AL. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991;19:25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Hutchison S, LeBel C, Blanchette M, Chabot B. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277:29745–29752. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Helfman DM, Krainer AR. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim FU, Hutchison S, Cordeau M, Chabot B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/s1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 1999;13:864–876. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S, Blanchette M, Park J, Savva Y, Yeo GW, Yeakley JM, Rio DC, Graveley BR. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Kislyuk A, Levine M, Dubchak I. Conservation patterns in different functional sequence categories of divergent Drosophila species. Genomics. 2006;88:431–442. doi: 10.1016/j.ygeno.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Swanson MS, Matunis MJ, Dreyfuss G. Purification and characterization of proteins of heterogeneous nuclear ribonucleoprotein complexes by affinity chromatography. Methods Enzymol. 1990;181:326–331. doi: 10.1016/0076-6879(90)81133-f. [DOI] [PubMed] [Google Scholar]
- Shen J, Zu K, Cass CL, Beyer AL, Hirsh J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc Natl Acad Sci U S A. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Zu K, Sikes ML, Beyer AL. Separable roles in vivo for the two RNA binding domains of Drosophila A1-hnRNP homolog. RNA. 1998;4:1585–1598. doi: 10.1017/s135583829898102x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu K, Sikes ML, Haynes SR, Beyer AL. Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol Biol Cell. 1996;7:1059–1073. doi: 10.1091/mbc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.