Abstract
The human deafness dystonia syndrome results from the mutation of a protein (DDP) of unknown function. We show now that DDP is a mitochondrial protein and similar to five small proteins (Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p) of the yeast mitochondrial intermembrane space. Tim9p, Tim10p, and Tim12p mediate the import of metabolite transporters from the cytoplasm into the mitochondrial inner membrane and interact structurally and functionally with Tim8p and Tim13p. DDP is most similar to Tim8p. Tim8p exists as a soluble 70-kDa complex with Tim13p and Tim9p, and deletion of Tim8p is synthetically lethal with a conditional mutation in Tim10p. The deafness dystonia syndrome thus is a novel type of mitochondrial disease that probably is caused by a defective mitochondrial protein-import system.
Most mitochondrial proteins are synthesized with a cleavable NH2-terminal targeting sequence and imported into mitochondria by a general import pathway composed of cytosolic chaperones and two heterooligomeric membrane complexes: a TOM (translocase of the outer membrane) complex in the outer membrane and a TIM complex in the inner membrane (1–4). Recently, another translocase of the mitochondrial inner membrane specific for the import and insertion of multispanning carrier proteins has been identified. This new TIM machinery includes soluble components, Tim9p and Tim10p, in the mitochondrial intermembrane space, and Tim12p, Tim22p, and Tim54p of the mitochondrial inner membrane (5–9). Tim9p and Tim10p are partner proteins in a 70-kDa complex that transfers the carriers from the outer membrane to the inner membrane. A 300-kDa complex consisting of Tim12p, Tim22p, Tim54p, and a fraction of the Tim9p and Tim10p mediates insertion of the carrier into the inner membrane.
Because the mitochondrion is essential for energy production in higher eukaryotes and many diseases already have been linked to errors in mitochondrial metabolism (10, 11), we reasoned that mitochondrial protein import defects potentially could lead to disease. The human deafness dystonia syndrome (MTS/DFN-1) is a recessive, X-linked neurodegenerative disorder characterized by progressive sensorineural deafness, cortical blindness, dystonia, dysphagia, and paranoia (12). It is caused by truncation or deletion of an 11-kDa protein (referred to as DDP1) whose intracellular location and function are unknown (13). DDP1 is closely related to the ORF YJR135w-a in Saccharomyces cerevisiae (14).
By searching various sequence databases, we now report that DDP and YGR135w-a (referred to as Tim8p) are similar to Tim9p, Tim10p, and Tim12p, which mediate the import of mitochondrial carrier proteins. In addition, we have identified another similar protein in S. cerevisiae (YGR181w, referred to as Tim13p) and another in humans termed DDP2. DDP1, Tim8p, and Tim13p are localized to the mitochondrial intermembrane space, and the yeast homolog of DDP1, Tim8p, interacts with the import system for multispanning inner membrane proteins. The deafness dystonia syndrome thus is a mitochondrial disease that may be caused by a defective protein-import machinery.
MATERIALS AND METHODS
Plasmids and Strain Construction.
DDP1 was amplified by PCR with Pfu polymerase (Stratagene) from a first-strand liver cDNA pool using primers prHD1 5′-gcccggatcccgccctgggatggattcctcc-3′ (complementary to codons 1–4) and prHD2 5′-gcccgtcgactaccttccttttccaaagaggt-3′ (complementary to codons 95–97). The amplified 300-bp fragment was cleaved with BamHI and SalI and cloned into pSP65 (Promega), sequenced, and used as the template for further plasmid construction. A gene encoding DDP1 C-terminally tagged with a hemagglutinin epitope (DDP1HA) was constructed with the primer prHD1 and a primer coding for the 12-aa epitope of the hemagglutinin tag (CYPYDVPDYASL) by PCR with Pfu polymerase. DDP1HA was subcloned into plasmids pGK1 (15) for expression in yeast and pCDNA3 (Invitrogen) for expression in mammalian cells. To construct yeast strains lacking Tim8p and/or Tim13p, one of two ORFs of TIM8 and TIM13 in a diploid yeast strain was replaced completely with the URA3 and kanr markers (16), respectively, and haploid segregants carrying the replacement markers were selected. The double disruptant was constructed by mating the single disruptants and sporulating the resulting diploids. For synthetic lethal studies, a tim10–1 strain was used in which TIM10 was replaced with HIS3 and the temperature-sensitive (ts) tim10–1 allele was integrated at the leu2 locus (5, 6); this genotype is designated as Δtim10∷HIS3 tim10–1:LEU2. The ts tim10–1 strain was mated to the strain lacking Tim8p (Δtim8∷URA3) followed by sporulation and tetrad analysis. Viability of dissected spores was tested at 25°C, and auxotrophic markers were screened on the appropriate medium. For plasmid-loss experiments, two ts tim10–1 strains that either contained TIM8 or lacked TIM8 (Δtim8∷URA3) were constructed containing the wild-type TIM10 gene on a centromeric plasmid ([pTIM10:TRP1 CEN]). Standard procedures were used for yeast genetics and for manipulations of yeast (17) and Escherichia coli.
Expression in Mammalian Cells.
The pCDNA3-DDP1HA construct was transiently transfected into cultured COS7 cells by using lipofectin (Life Technologies, Gaithersburg, MD) according to the manufacturer’s procedure. After 36 hr, 1 μM Mitotracker (Molecular Probes) was added, the cells were fixed with 3% paraformaldehyde (18), excess paraformaldehyde was quenched with ammonium chlorite, and cells were permeabilized with 0.1% Triton X-100. For immunofluorescence, cells were incubated for 1–2 hr with monoclonal 12CA5 antibody (1:1,000, Babco, Richmond, CA) against the hemagglutinin (HA) tag in 1% BSA-PBS. Cells were washed three times with 1% BSA-PBS and incubated with fluorescein isothiocyanate-conjugated rabbit anti-mouse IgG (1:200, Organon Teknika–Cappel) for 30 min, mounted with Mowiol (Hoechst Pharmaceuticals) mounting medium, and examined in a Zeiss Axiophot microscope.
Biochemical Manipulations.
Standard procedures were used for purification of mitochondria (19), import of radiolabeled proteins into isolated yeast mitochondria and immunoprecipitations (5, 6), and blue-native gel electrophoresis (20).
RESULTS
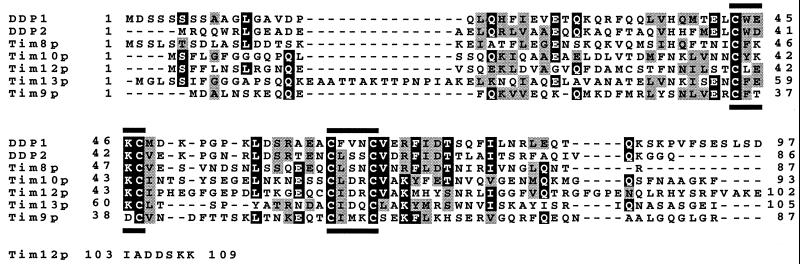
Previous work has shown that import of multispanning proteins from the cytoplasm into the mitochondrial inner membrane is mediated by three related proteins termed Tim9p, Tim10p, and Tim12p (5, 6). By searching the S. cerevisiae genome database for additional members of this protein family, we identified two novel homologs, YJR135w-a and YGR181w, that we termed Tim8p and Tim13p, respectively (Fig. 1). Tim8p and Tim13p also were discovered independently as mitochondrial intermembrane space proteins that utilized Tom5p for import (M. Ryan and N. Pfanner, personal communication). In addition, two sequences encoding similar proteins, DDP (ref. 13; now renamed DDP1) and DDP2, were identified in the human expressed sequence tag database (ref. 21; last searched October 1998). The sequence of the entire ORF was assembled from related cDNAs represented in the Unigene database (21). Previously, loss of the functional DDP1 protein was shown to cause a recessive, X-linked neurodegenerative syndrome (13). All proteins in this family share a conserved “twin CX3C” motif (ref. 6; Fig. 1).
Figure 1.
Members of the Tim family in the mitochondrial intermembrane space share a “twin CX3C” motif. Sequence comparison of DDP1, DDP2, Tim8p, Tim10p, Tim12p, Tim13p, and Tim9p. Black boxes, identical amino acids; shaded boxes, similar amino acids; black bars, “CX3C” motif.
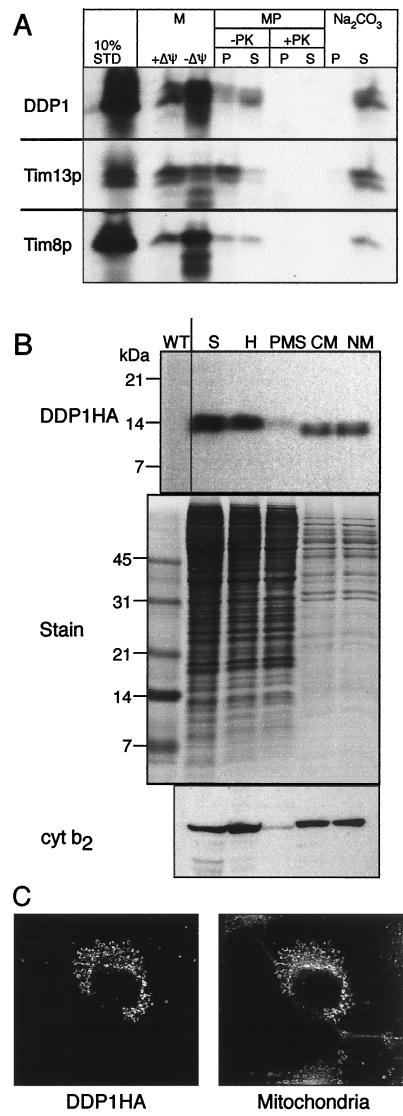
To address whether Tim8p, Tim13p, and DDP1 might be functionally related to Tim9p, Tim10p, and Tim12p, we first determined their subcellular localization. Tim9p, Tim10p, and Tim12p are located in the yeast mitochondrial intermembrane space (5, 6, 9). The same is true of DDP1. When DDP1 was transcribed and translated in vitro in the presence of [35S]methionine, it yielded a radioactive protein of the expected size that was imported into the intermembrane space of isolated yeast mitochondria (Fig. 2A). Import did not require a potential across the inner membrane, suggesting that it bypassed the two protein insertion systems (22) in that membrane. When mitochondria that had imported DDP1 were subjected to graded osmotic shock to rupture the outer membrane, DDP1 became protease-accessible in parallel with cytochrome b2, a marker for the soluble intermembrane space (data not shown). When DDP1 epitope tagged at its C terminus was expressed in yeast cells, it copurified with mitochondrial marker cytochrome b2 (Fig. 2B); when it was expressed transiently in cultured monkey cells and visualized by immunofluorescence, it colocalized with the mitochondria-specific stain Mitotracker (Fig. 2C) and with mitochondria-targeted preornithine transcarbamylase fused to green fluorescent protein (ref. 23; data not shown).
Figure 2.
DDP1 is located in the mitochondrial intermembrane space. (A) Import into isolated yeast mitochondria. Radiolabeled Tim8p, Tim13p, and DDP1 were synthesized in vitro and incubated for 10 min at 25°C with wild-type yeast mitochondria in the presence (+) or absence (−) of a membrane potential (Δψ). Mitochondria (M) were treated with trypsin to digest nonimported precursor and then with soybean trypsin inhibitor, and finally were analyzed by SDS/PAGE and fluorography. Equal aliquots of the mitochondria that had imported precursor in the presence of a membrane potential were subjected further to osmotic shock or alkali extraction (5, 6). Osmotic shock (that selectively ruptures the outer membrane and generates mitoplasts, MP) was performed in the absence or presence of 50 μg/ml proteinase K (PK). After addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), samples were centrifuged at 14,000 × g for 10 min to separate pellet (P) and supernatant (S). For alkali extraction (Na2CO3), mitochondria were incubated with 100 mM Na2CO3 for 30 min, followed by centrifugation at 100,000 × g for 15 min to separate pellet (P) and supernatant (S). 10% STD, 10% of the radioactive precursor present in each assay. (B) Expression in yeast cells. A yeast transformant expressing DDP1 with a C-terminal HA tag from a multicopy plasmid was grown in semisynthetic lactate medium at 30°C, converted to spheroplasts (S), and fractionated into total homogenate (H), postmitochondrial supernatant (PMS), crude mitochondria (CM), and mitochondria purified on a Nycodenz gradient (NM). An equal amount of each fraction was analyzed by SDS/PAGE and immunoblotting with an mAb against HA followed by rabbit anti-mouse antibody or with a rabbit antiserum against cytochrome b2 (cyt b2). The immune complexes then were decorated with [125I]protein A. As a control, a Coomassie blue-stained gel of the fractions is included (Stain). WT, mitochondria from a strain not transformed with the tagged DDP1 gene. (C) Expression in monkey cells. DDP1 was transiently expressed in cultured COS7 cells under the control of the cytomegalovirus promoter. After staining, the cells were examined by indirect immunofluorescence (ref. 18; primary antibody for DDP1HA, monoclonal 12CA5 antibody; secondary antibody, FITC-conjugated rabbit anti-mouse) as well as by staining with the mitochondria-specific fluorescent dye Mitotracker (Mitochondria).
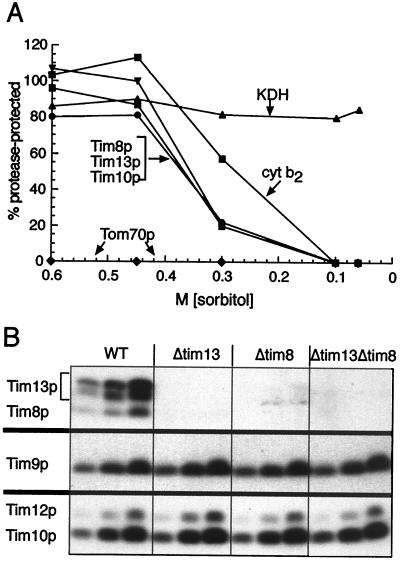
What is the function of DDP1? To address this question, we studied the yeast homolog of DDP1, Tim8p. Tim8p was not essential for viability and its deletion did not significantly affect growth on different carbon sources or at different temperatures. The same was true of Tim13p, the second novel Tim protein (data not shown). Both proteins were located in the mitochondrial intermembrane space because they became protease-accessible after the outer membrane was disrupted by osmotic shock (Fig. 3A); the same behavior was observed with the intermembrane space markers Tim10p and cytochrome b2. Both Tim8p and Tim13p also were imported in vitro into the mitochondrial intermembrane space (Fig. 2A).
Figure 3.
Tim8p and Tim13p are located in the intermembrane space and stabilize each other. (A) Submitochondrial localization. Isolated yeast mitochondria were incubated in 20 mM Hepes-KOH, pH 7.4, 100 μg/ml proteinase K, and the indicated sorbitol concentrations on ice for 30 min. After addition of 1 mM PMSF and centrifugation, the pellet was analyzed by SDS/PAGE and immunoblotting for Tim8p, Tim10p, Tim13p, cytochrome b2 (cyt b2; intermembrane space marker), α-ketoglutarate dehydrogenase (KDH; matrix marker), and Tom70p (outer membrane marker). Antigen protected from protease was quantified by densitometry and expressed as a percentage of the antigen measured in the absence of protease. (B) Deletion of Tim8p causes loss of Tim13p, and vice versa. The parental wild-type strain (WT) and strains deleted for TIM13 (Δtim13), TIM8 (Δtim8), or both TIM13 and TIM8 (Δtim13Δtim8) were grown in lactate medium for 16 hr at 30°C. Three different amounts (50, 100, and 200 μg protein) of purified mitochondria were analyzed by SDS/PAGE and immunoblotting, with monospecific rabbit antisera for the proteins indicated on the left and (data not shown) for Tim22p, Tim54p, the ADP/ATP carrier, cytochrome b2, and porin. Blots were decorated with [125I]protein A and autoradiographed.
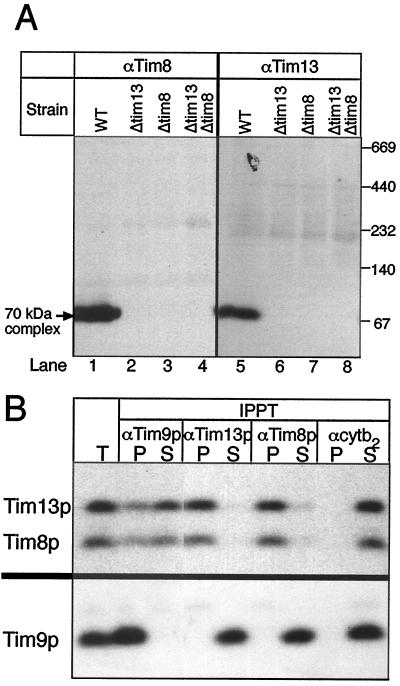
Cells deleted for either Tim8p or Tim13p had essentially normal steady-state levels of outer membrane porin, of the intermembrane space protein cytochrome b2, of Tim proteins 9, 10, 12, 22, and 54, and of mitochondrial metabolite carriers (Fig. 3B; data not shown). Deletion of Tim8p caused loss of Tim13p and vice versa, suggesting that the two proteins stabilize each other by forming a complex (Fig. 3B). This complex was visualized directly as a 70-kDa species by analyzing solubilized mitochondria by nondenaturing gel electrophoresis (20) followed by immunoblotting for Tim8p and Tim13p (Fig. 4A, lanes 1 and 5). The mobility of this complex was indistinguishable from that of the previously described (Tim9p)3(Tim10p)3 complex (6). Tim8p and Tim13p were not detected in the 300-kDa membrane complex that contains all of Tim12p, Tim22p, Tim54p, and a small fraction of Tim9p and Tim10p (6).
Figure 4.
Tim8p, Tim13p, and a fraction of Tim9p form a 70-kDa complex in the intermembrane space. (A) Proteins solubilized from purified yeast mitochondria with 0.16% n-dodecylmaltoside were subjected to “blue native gel” electrophoresis (ref. 20; 6–16% acrylamide) and analyzed for Tim8p and Tim13p by immunoblotting and decoration with [125I]protein A. WT, parental strain; Δtim13, TIM13 deleted; Δtim8, TIM8 deleted; Δtim13Δtim8, TIM13 and TIM8 deleted. All strains were haploid. (B) A soluble intermembrane space fraction was incubated at 4°C for 2 hr with protein A-Sepharose that had been coupled to IgGs against Tim9p (αTim9p), Tim13p (αTim13p), Tim8p (αTim8p), or cytochrome b2 (αcytb2). After centrifugation, aliquots (equivalent to 200 μg mitochondria) of unbound (S) and bound (P) proteins were analyzed by SDS/PAGE and immunoblotting for Tim13p and Tim8p (Upper) or for Tim9p (Lower). IPPT, immunoprecipitation; T, total mitochondrial intermembrane space fraction analyzed without immunoprecipitation.
To define the interactions among Tim8p, Tim9p, Tim10p, and Tim13p, a soluble intermembrane space fraction was subjected to immunoprecipitations with monospecific antibodies against each protein. As expected, each antibody quantitatively precipitated its cognate antigen (Fig. 4B; see also ref. 6). In addition, antibody against Tim9p coprecipitated 20–40% of Tim8p, Tim10p, and Tim13p; antibody against Tim8p coprecipitated Tim13p, but virtually no Tim9p and no Tim10p; antibody against Tim13p coprecipitated Tim8p, virtually no Tim9p, and no Tim10p; and antibody against Tim10p coprecipitated most of Tim9p, but no Tim8p or Tim13p. Antibody against cytochrome b2, an abundant intermembrane space protein, did not coprecipitate any Tim protein, confirming the specificity of the immunoprecipitations. These results and those reported earlier (5, 6, 9) suggest that the yeast mitochondrial intermembrane space contains at least two distinct 70-kDa Tim complexes: the (Tim9p)3(Tim10p)3 complex, representing the bulk of Tim9p and Tim10p (6, 9), and a Tim8p/Tim9p/Tim13p complex that represents all of Tim8p and Tim13p and a small fraction of Tim9p. Quantitative immunoblotting (data not shown) confirmed that Tim9p and Tim10p are 10 to 20 times more abundant (120–140 pmol of each/mg mitochondrial protein) than either Tim8p or Tim13p (5–15 pmol of each/mg protein). In immunoprecipitation experiments, the small percentage of Tim9p associated with Tim8p and Tim13p probably is difficult to detect against the large excess of Tim9p associated with Tim10p. As reported earlier (5, 9), Tim12p is not present in the soluble intermembrane space, but bound to the outer face of the inner membrane.
We purified the Tim8p/Tim9p/Tim13p complex to about 90% purity from the soluble intermembrane space fraction by ion-exchange chromatography on MonoS and MonoQ columns. Peptide sequencing of the final preparation revealed Tim8p, Tim9p, and Tim13p as the only near-stoichiometric proteins (data not shown). After each purification step, Tim8p, Tim13p, and a fraction of Tim9p migrated as a single 70-kDa species in nondenaturing gel electrophoresis.
Because Tim8p and Tim13p form a complex with Tim9p, they interact structurally with the mitochondrial import system for metabolite carriers. But Tim8p also interacts with this import system functionally because a deletion of TIM8 was synthetically lethal with the ts tim10–1 mutation. Synthetic lethality was shown by the absence of viable haploid Δtim8,tim10–1 meiotic segregants in appropriate crosses (Table 1) and by the inability of Δtim8Δtim10–1 cells carrying a plasmid-borne wild-type TIM10 gene to lose this gene during mitotic growth at 25°C (Table 2). Because deletion of Tim8p causes loss of Tim13p (Figs. 3B and 4A), we suspect that the tim10–1 mutation also is synthetically lethal with a Tim13p deletion. However, this has not yet been shown directly. A TIM8 disruption was not synthetically lethal with the ts tim12–1 mutation, suggesting that the observed genetic interaction between Tim8p and Tim10p is specific.
Table 1.
Deletion of TIM8 is synthetically lethal with the temperature-sensitive tim10-1 mutation: Segregation analysis*
| Genotype | Viability† | Frequency‡ |
|---|---|---|
| TIM10 leu2 TIM8 | + | 0.115 |
| TIM10 tim10-1:LEU2 TIM8 | + | 0.125 |
| TIM10 leu2 Δtim8∷URA3 | + | 0.135 |
| Δtim10∷HIS3 leu2 TIM8 | − | 0.131 |
| TIM10 tim10-1:LEU2 Δtim8∷URA3 | + | 0.135 |
| Δtim10∷HIS3 leu2 Δtim8∷URA3 | − | 0.115 |
| Δtim10∷HIS3 tim10-1:LEU2 TIM8 | + | 0.128 |
| Δtim10∷HIS3 tim10-1:LEU2 Δtim8∷URA3§ | − | 0.115 |
The diploid strain TIM10/Δtim10∷HIS3 TIM8/Δtim8∷URA3 leu2/tim10-1:LEU2 was sporulated and 76 tetrads were analyzed for viability and segregation of LEU2, TRP1, and URA3 markers. The genotype of the inviable spores was deduced assuming a 2:2 segregation of auxotrophic markers.
Growth on rich-glucose medium: +, viable; −, lethal.
Expected frequency of independent segregation of each marker is 0.125, P < 0.01 (χ2 test).
Progeny with this genotype contain only the temperature-sensitive tim10-1 allele and URA3 allele (replacing TIM8) and were inviable, indicating that disruption of TIM8 is synthetically lethal with the tim10-1 allele.
Table 2.
Deletion of TIM8 is synthetically lethal with the temperature-sensitive tim10-1 mutation: Plasmid loss analysis*
| Genotype | TIM8 locus | Plasmid loss† |
|---|---|---|
| Δtim10∷HIS3 tim10-1:LEU2 [pTIM10:TRP1 CEN] | Wild type | 38% |
| Δtim10∷HIS3 tim10-1:LEU2 Δtim8∷URA3[pTIM10:TRP1 CEN] | Null | 0% |
Strains were grown overnight in liquid rich-glucose medium at 25°C to saturation and then plated at 25°C for single colonies on rich-glucose medium. After 2 days, colonies were replica-plated to rich-glucose medium and minimal medium lacking tryptophan and scored for growth after 2 days.
Three independent isolates of each strain were tested; approximately 800 colonies were scored.
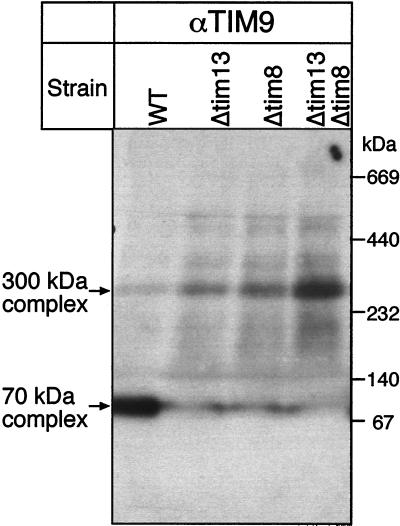
We studied the role of Tim8p and Tim13p in protein import in greater detail. Mitochondria isolated from the Tim8p or Tim13p deletion strains imported radiolabeled adenine nucleotide carrier or phosphate carrier at normal rates in vitro. Neither Tim8p nor Tim13p could be crosslinked to a partly translocated adenine nucleotide carrier in isolated mitochondria (data not shown) whereas corresponding crosslinks of the partly translocated precursor to Tim9p, Tim10p, and Tim12p were readily detected (5, 6). Tim8p and Tim13p have a different role in the general import pathway for metabolite carriers, albeit a role that is not essential. What is the function of Tim8p and Tim13p? Deletion of Tim8p or Tim13p greatly diminished the amount of Tim9p in the 70-kDa complex and increased its abundance in the 300-kDa membrane complex (Fig. 5). Because a similar, but smaller effect was noted for Tim10p, partitioning of the small Tim proteins between the 70- and 300-kDa complexes appears to be dynamic. Our results are consistent with the possibility that Tim8p and Tim13p regulate this partitioning. Biosynthesis of the different Tim complexes in the intermembrane space, the stability of the complexes in vivo, and the role of Tim8p and Tim13p in these dynamic processes require further study.
Figure 5.
Deletion of TIM8 or TIM13 causes redistribution of Tim9p from the 70-kDa complex to the 300-kDa complex. Proteins solubilized from purified yeast mitochondria with 0.16% n-dodecylmaltoside were subjected to “blue native gel” electrophoresis (ref. 20; 6–16% acrylamide) and analyzed for Tim9p by immunoblotting and decoration with [125I]protein A. WT, parental strain; Δtim13, TIM13 deleted; Δtim8, TIM8 deleted; Δtim13Δtim8, TIM13 and TIM8 deleted.
DISCUSSION
We have identified two additional members of a novel protein family in the yeast mitochondrial intermembrane space, bringing the total to five proteins. Unlike the previously identified members, the two newest members, Tim8p and Tim13p, are not essential for viability. However, they form a 70-kDa complex with a fraction of Tim9p, which is an essential component of the carrier import pathway. Tim8p and Tim13p interact structurally and functionally with Tim9p and Tim10p because the Tim8p/Tim9p/Tim13p complex is required for viability in the ts tim10–1 mutant and because Tim9p repartitions to the 300-kDa complex when Tim8p or Tim13p is deleted. Thus, although Tim8p and Tim13p are not directly required for import of carrier proteins, they seem to regulate the formation of the Tim9p/Tim10p complex and may influence the partitioning of Tim9p and Tim10p between the intermembrane space and inner membrane.
Our results show that human DDP1, like its yeast homolog Tim8p, is located in the mitochondrial intermembrane space. Most likely, DDP1 interacts with several components of the mammalian mitochondrial import system for metabolite carriers. Why does a loss of DDP1/Tim8p have dramatic consequences for mammals, but not for yeast? The DDP1 transcript is ubiquitously expressed in human tissues (13), indicating that DDP1 is important for basic cellular processes; loss of DDP1 may manifest itself selectively in different tissue types because the energy requirements of tissues may differ, particularly during development. DDP1 may be important for import of a subset of metabolite carriers that either are not present in yeast or have a crucial role in neural development of mammals. Alternatively, yeast Tim8p may be essential under specific growth conditions that are yet to be identified or may function in pathways other than protein import. Although this question remains open, the present results show clearly that the human deafness dystonia syndrome is a novel type of mitochondrial disease that most likely reflects a defect in mitochondrial protein import.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (to G.S. and Martin Spiess), the Human Frontier Science Program Organization (to G.S.), and the European Economic Community (to G.S.), the Damon Runyon-Walter Winchell Cancer Research Foundation and the U.S. National Science Foundation (to C.M.K.), and the U.S. National Science Foundation MCB-9752974 and the Guggenheim Foundation (to S.M.). We thank Dr. Radek Skoda for providing the first-strand cDNA pool from liver, Dr. Natasha Kralli for the yeast expression vector pGK1, and Dr. Masataki Mori for the plasmid encoding preornithine transcarbamylase C-terminally fused to green fluorescent protein.
ABBREVIATIONS
- DDP
deafness dystonia polypeptide
- HA
hemagglutinin
- TIM or Tim
translocase of the mitochondrial inner membrane
- TOM
translocase of the outer membrane
- ts
temperature sensitive
Footnotes
A Commentary on this article begins on page 1817.
References
- 1.Ryan K R, Jensen R E. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 2.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 3.Neupert W. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 4.Pfanner N. Curr Biol. 1998;8:R262–R265. doi: 10.1016/s0960-9822(98)70168-x. [DOI] [PubMed] [Google Scholar]
- 5.Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 6.Koehler C M, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerscher O, Holder J, Srinivasan M, Leung R S, Jensen R E. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirrenberg C, Bauer M F, Guiard B, Neupert W, Brunner M. Nature (London) 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 9.Sirrenberg C, Endres M, Folsch H, Stuart R A, Neupert W, Brunner M. Nature (London) 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 10.Schapira A H. Biochim Biophys Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 11.Wallace D C, Shoffner J M, Trounce I, Brown M D, Ballinger S W, Corral-Debrinski M, Horton T, Jun A S, Lott M T. Biochim Biophys Acta. 1995;1271:141–151. doi: 10.1016/0925-4439(95)00021-u. [DOI] [PubMed] [Google Scholar]
- 12.Tranebjaerg L, Schwartz C, Eriksen H, Andreasson S, Ponjavic V, Dahl A, Stevenson R E, May M, Arena F, Barker D, et al. J Med Genet. 1995;32:257–263. doi: 10.1136/jmg.32.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, May M, Tranebjaerg L, Kendall E, Fontan G, Jackson J, Subramony S H, Arena F, Lubs H, Smith S, et al. Nat Genet. 1996;14:177–180. doi: 10.1038/ng1096-177. [DOI] [PubMed] [Google Scholar]
- 14.Foury F. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 15.Schena M, Picard D, Yamamoto K R. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 16.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 18.Wahlberg J M, Geffen I, Reymond F, Simmen T, Spiess M. J Cell Biol. 1995;130:285–297. doi: 10.1083/jcb.130.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glick B S, Pon L. Methods Enzymol. 1995;260:213–233. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 20.Schägger H, Cramer W A, von Jagow G. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 21.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 22.Pfanner N, Meijer M. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- 23.Yano M, Kanazawa M, Terada K, Takeya M, Hoogenraad N, Mori M. J Biol Chem. 1998;273:26844–26851. doi: 10.1074/jbc.273.41.26844. [DOI] [PubMed] [Google Scholar]