Abstract
In this study, we report for the first time that both t-SNAREs and v-SNARE and their complexes in buffered suspension, exhibit defined peaks at CD signals of 208 and 222 nm wavelengths, consistent with a higher degree of helical secondary structure. Surprisingly, when incorporated in lipid membrane, both SNAREs and their complexes exhibit reduced folding. In presence of NSF–ATP, the SNARE complex disassembles, as reflected from the CD signals demonstrating elimination of α-helices within the structure.
1. Introduction
Understanding biochemical processes requires the determination of how participating molecules interact. This process is even more critical when such interactions are between membrane proteins required for membrane fusion in cells. A general understanding of membrane fusion in cells has been made possible following discovery of an N-ethylmaleimide-sensitive factor (NSF) [1] and SNARE proteins [2–4], and the mechanism of their participation [5–11]. Target membrane proteins at the cell plasma membrane SNAP-25 and syntaxin termed t-SNAREs, and secretory vesicle-associated membrane protein VAMP or v-SNARE, are part of the conserved protein complex involved in fusion of opposing cellular membranes. VAMP and syntaxin are both integral membrane proteins, whereas the soluble SNAP-25 protein associates with syntaxin. Therefore, the key to our understanding of SNARE-induced membrane fusion requires determination of the atomic arrangement and interaction between membrane-associated v- and t- SNAREs. Ideally, the atomic coordinates of membrane-associated SNARE complex using X-ray crystallography would help elucidate the chemistry of SNARE-induced membrane fusion in cells. So far such structural details at the atomic level of membrane-associated t-/v-SNARE complex have not been realized, primarily due to solubility problems of membrane-associated SNAREs and due to the fact that v-SNARE and t-SNAREs need to reside in opposing membranes when they meet to be able to assemble in a physiologically relevant conformation [6,8,9]. The remaining option has been the use of nuclear magnetic resonance spectroscopy (NMR), which too has been unsuccessful due to the size of the t-/v-SNARE complex being larger than current NMR capabilities. Regardless of these setbacks, AFM force spectroscopy has provided, at nm resolution, an understanding of the structure, assembly, and disassembly of membrane- associated t-/v-SNARE complex, in physiological buffered solution [6,8,9,11]. The structure and arrangement of SNARE-associated with lipid bilayer were first determined using AFM [6]. A bilayer electrophysiological setup allowed measurements of membrane conductance and capacitance during fusion of v- SNARE-reconstituted liposomes with t-SNARE-reconstituted membrane, and vice versa. Results from these studies demonstrated that t-SNAREs and v-SNARE when present in opposing membranes interact and assemble in a circular array (ring complexes), and form conducting channels in presence of calcium [6]. The interaction between t-SNAREs and v-SNARE proteins to form such conducting channel is strictly dependent on the association of SNAREs in opposing bilayers. In the absence of membrane, either v-SNARE or t- SNAREs fail to appropriately interact and form the ring complex, and to establish continuity between the opposing bilayers [6].
Four intertwined α-helices have previously been reported in the crystal structure of a truncated SNARE complex, formed using vand t-SNAREs lacking their membrane-spanning domain [12]. Since membrane association of SNAREs is critical to their assembly and function, the molecular conformations in the assembly and disassembly of full length SNAREs both in buffered suspension and in membrane, was the focus of the current study using CD spectroscopy. Our results demonstrate that the α-helices both within v-SNARE and t-SNAREs, and their complex, are significantly less pronounced when membrane-associated than in suspension. Furthermore, exposure of the t-/v-SNARE–NSF complex to ATP results in dissociation of α-helices present within the SNARE complex.
2. Materials and methods
2.1. Protein purification
N-Teminal 6xHis-tag constructs for SNAP25 and NSF, C-terminal 6xHis-tag constructs for Syntaxin 1A and VAMP2 were generated. All four proteins were expressed with 6xHis at full length in E. coli (BL21DE3) and isolated by Ni–NTA (nickel-nitrilotriacetic acid) affinity chromatography (Qiagen, Valencia, CA). Protein concentration was determined by BCA assay.
2.2. Preparation of proteoliposomes
All lipids were obtained from Avanti Polar Lipids (Alabaster, AL). A 5 mM lipid stock solution was prepared by mixing DOPC (1,2-dioleoyl phosphatidylcholine): DOPS (1,2-dioleoyl phosphatidylserine) in 70:30 mol/mol ratios in glass test tubes. The lipid mixture was dried under gentle stream of nitrogen and resuspended in decane. Lipids were suspended in 5 mM sodium phosphate buffer, pH 7.5, by vortexing for 5 min at room temperature. Unilamellar vesicles were formed following five times sonications for 2 min/sonication, followed by passing the resultant liposomes through a 200 nm pore size membrane using an extruder. Typically, vesicles ranging in size from 180 to 220 nm in diameter were obtained as assessed by atomic force microscopy (AFM) and photon correlation spectroscopy (PCS) (data not shown). Proteoliposomes were prepared by gently mixing either t-SNARE complex (equal amounts of Syntaxin 1-His6 and His6-SNAP-25, final concentration 10 μM) or VAMP2–His6 (final concentration 10 μM) with liposomes [6,7], followed by three freeze/thaw cycles to enhance protein reconstitution at the vesicles membrane. PCS was performed for the measurement of proteoliposome size [9,11], using a Zetasizer Nano ZS, (Malvern Instruments, UK).
2.3. Circular dichroism (CD) spectroscopy
Overall secondary structural content of SNAREs and their complexes, both in suspension and membrane-associated, were determined by CD spectroscopy using an Olis DSM 17 spectrometer. Data were acquired at 25 °C with a 0.01 cm path length quartz cuvette (Helma). Spectra were collected over a wavelength range of 185–260 nm using a 1-nm step spacing. In each experiment, 30 scans were averaged per sample for enhanced signal to noise, and data were acquired on duplicate independent samples to ensure reproducibility. SNAREs and their complexes, both in suspension and membrane-associated, were analyzed for the following samples: v-SNARE, t-SNAREs, v-SNARE + t-SNAREs, v-SNARE + t- SNAREs + N-ethylmaleimide sensitive factor (NSF) and v-SNARE + t-SNAREs + NSF + 2.5 mM ATP. All samples had final protein concentrations of 10 μM in 5 mM sodium phosphate buffer at pH 7.5 and were baseline subtracted to eliminate buffer (or liposome in buffer) signal. Data were analyzed using the Globalworks software (Olis), which incorporates a smoothing function [13] and fit using the CONTINLL algorithm [14].
2.4. Atomic force microscopy
Atomic force microscopy was performed on mica surface in buffer as previously reported [9]. The same samples used in CD spectra analysis were imaged using the Nanoscope IIIa AFM (Digital Instruments, Santa Barbara, CA). Images were obtained in the ‘tapping’ mode in fluid, using silicon nitride tip with a spring constant of 0.06 N m−1, and an imaging force of <200 pN. Images were obtained at line frequencies of 1.5 Hz, with 512 lines per image, and constant image gains. All images were analyzed by the nanoscope IIIa 4.43r8 software, supplied by Digital Instruments.
3. Results and discussion
It is well established that t- and v-SNAREs in opposing bilayers form circular arrays in the presence of calcium to form conducting channels [6]. The purified SNARE proteins are observed to interact in suspension but do not retain the capacity to form rosettes or create conducting channels [6]. Therefore, it is logical to hypothesize that these proteins retain different structural arrangements when associated with lipid than in buffered suspension. The present study directly tests this hypothesis using CD spectroscopy and AFM.
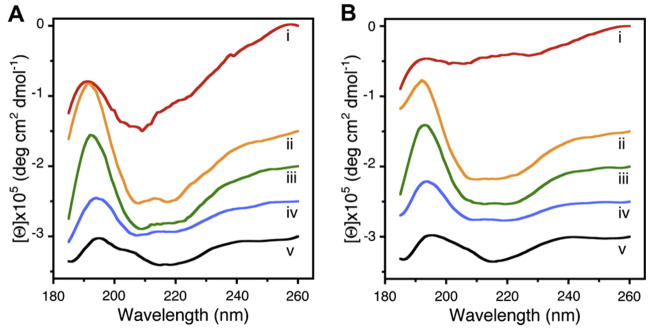
The overall secondary structural content of full-length neuronal v-SNARE and t-SNAREs, and the t-/v-SNARE complex, both in suspension and membrane-associated, were determined by CD spectroscopy using an Olis DSM 17 spectrometer. Circular dichroism spectroscopy reveals that v-SNARE in buffered suspension (Fig. 1Ai), when incorporated into liposomes (Fig. 1Bi), exhibit reduced folding (Table 1). This loss of secondary structure following incorporation of full-length v-SNARE in membrane may be a result of self-association of the hydrophobic regions of the protein in absence of membrane.
Fig. 1.
Circular dichroism data reflecting structural changes to SNAREs, both in suspension and in association with membrane. Structural changes, following the assembly and disassembly of the t-/v-SNARE complex is further shown. (A) CD spectra of purified full-length SNARE proteins in suspension and (B) in membrane-associated; their assembly and (NSF–ATP)-induced disassembly is demonstrated. (i) v-SNARE; (ii) t-SNAREs; (iii) t-/v-SNARE complex; (iv) t-/v-SNARE + NSF and (v) t-/v-SNARE + NSF + 2.5 mM ATP, is shown. CD spectra were recorded at 25 °C in 5 mM sodium phosphate buffer (pH 7.5), at a protein concentration of 10 μM. In each experiment, 30 scans were averaged per sample for enhanced signal to noise, and data were acquired on duplicate independent samples to ensure reproducibility.
Table 1.
Secondary structural fit parameters of SNARE complex formation and dissociation
| Proteinb | Suspension (100 × fa) |
Membrane-associated (100 × f) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| α | β | O | U | Fitc | α | β | O | U | Fit | |
| v-SNARE | 4 | 36 | 18 | 43 | 0.19 | 0 | 30 | 32 | 38 | 0.21 |
| t-SNAREs | 66 | 34 | 0 | 0 | 0.02 | 20 | 15 | 21 | 44 | 0.84 |
| v-/t-SNAREs | 48 | 52 | 0 | 0 | 0.02 | 20 | 19 | 56 | 5 | 0.38 |
| v-/t-SNAREs+NSF | 20 | 25 | 0 | 55 | 0.07 | 18 | 6 | 8 | 68 | 0.2 |
| v-/t-SNAREs+NSF+ATP | 3 | 39 | 18 | 40 | 0.22 | 1 | 27 | 34 | 38 | 0.23 |
Abbreviations used: f, fraction of residues is a given conformational class; α, α-helix; β, β-sheet; O, other (sum of turns, distorted helix, distorted sheet); U, unordered.
Protein constructs: v-SNARE (VAMP2); t-SNAREs (SNAP-25 + syntaxin 1 A); NSF, N-ethylmaleimide sensitive factor; ATP, adenosine triphosphate.
Fit: goodness of fit parameter expressed as normalized spectral fit standard deviation (nm).
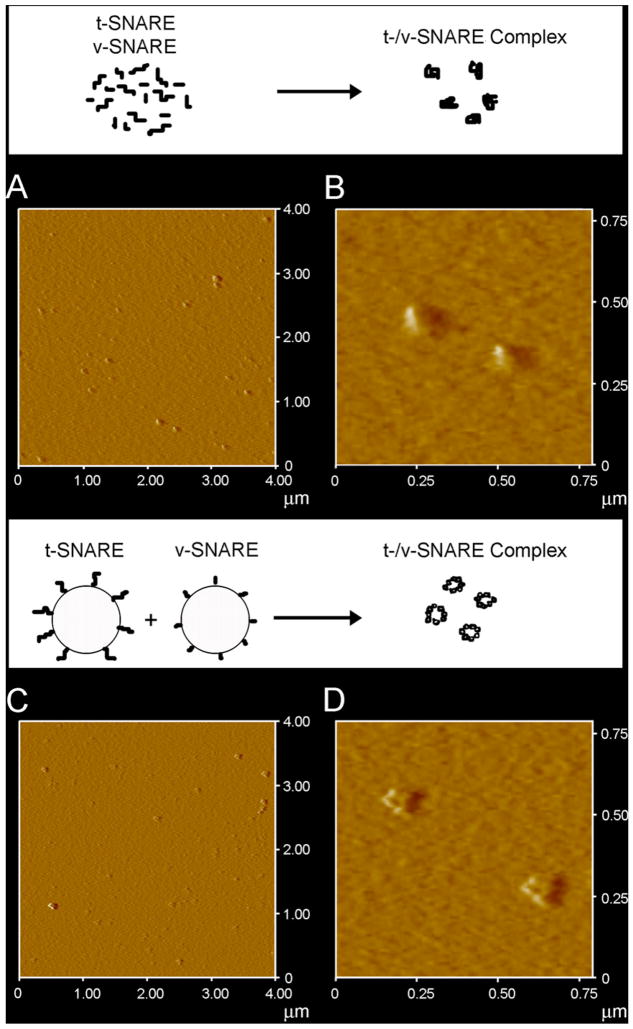
When incorporated into liposomes, v-SNARE may freely unfold without the artifactual induction of secondary structure, as reflective of the lack in CD signals at 208 and 222 nm, distinct for α-helical content. The t-SNAREs (Fig. 1Aii, Bii), shows clearly defined peaks at both these wavelengths, consistent with a higher degree of helical secondary structures formed both in buffered suspension and in membrane, at ca. 66 and 20%, respectively (Table 1). Again, the membrane-associated SNARE exhibits less helical content than when in suspension. Similarly, there appears to be a dramatic difference in the CD signal observed in t-/v-SNARE complexes in suspension, and those complexes that are formed when membrane-associated SNAREs interact (Fig. 1Aiii and Biii). Interestingly, we do not observe an increase of secondary structure upon complex formation. Rather, the CD spectra of the complexes are identical to a combination of individual spectra. Moreover, membrane associated t-/v-SNAREs are less folded than the purified SNARE complex. This data supports the hypothesis that lipid is required for proper arrangement of the SNARE proteins in membrane fusion. Addition of NSF to the t-/v-SNARE complex results in an increase in the unordered fraction (Fig. 1Aiv, Biv and Table 1), which may be attributed to an overall disordered secondary structure of the NSF, and not necessarily unfolding of the t-/v-SNARE complex. In contrast, activation of NSF by the addition of ATP almost completely abolishes all α-helical content within the multi-protein complex (Fig. 1Av, Bv). This is the first report of a direct observation of the helical unfolding of the SNARE complex using CD spectroscopy under physiologically relevant conditions (i.e. in membrane-associated SNAREs), and confirms earlier AFM reports on NSF–ATP induced t-/v-SNARE complex disassembly [9,11]. In further agreement with previously reported studies using the AFM [9], the consequence of ATP addition to the t-/v-SNARE–NSF complex is disassembly, regardless of whether the t-/v-SNARE+NSF complex is membrane-associated or in buffered suspension. In earlier AFM studies [8,9], 0.16–0.2 μg/Ml of SNARE proteins were used, as opposed to the 800–1000 μg/Ml protein concentration required for the current CD studies. To determine if t-SNARE and v- SNARE interact differently at higher protein concentrations, both membrane-associated and in-suspension v- and t-SNARE complexes used in CD studies, were imaged using the AFM. In confirmation to previously reported AFM studies [8,9], results from the current study (Fig. 2) demonstrate the formation of t-/v-SNARE ring complexes, only when t-SNARE-liposomes are exposed to v- SNARE-liposomes. Hence, higher SNARE protein concentrations are without influence on the membrane-directed self-assembly of the SNARE complex.
Fig. 2.
AFM micrographs of t-/v-SNARE complex formed in buffered suspension (A, B) and when membrane-associated t-SNAREs and v-SNARE interact (C, D). (A) and (C) are low resolution images; (B) and (D) are high resolution images of the t-/v-SNARE complexes formed.
In summary, we report for the first time that v-SNARE in suspension, when incorporated into liposomes, exhibits reduced folding. Similarly, t-SNAREs which exhibit clearly defined peaks at CD signals of 208 and 222 nm wavelengths, consistent with a higher degree of helical secondary structure in both the soluble and liposome- associated forms, exhibit reduced folding when membrane-associated. ATP-induced activation of NSF bound to the t-/v-SNARE complex, results in disassembly of the SNARE complex, eliminating all α-helices within the structure. In addition, these studies are a further confirmation of earlier reports [9] that NSF–ATP is sufficient for the disassembly of the t-/v-SNARE complex.
Acknowledgments
Financial support from NSF CBET-0730768, NIH NS-39918 and Wayne State University Research Enhancement Program (B.P.J.), and NIH DK068139 (T.L.S.), is gratefully acknowledged.
Footnotes
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- 1.Malhotra V, Orci L, Glick BK, Block MR, Rothman JE. Cell. 1988;54:221. doi: 10.1016/0092-8674(88)90554-5. [DOI] [PubMed] [Google Scholar]
- 2.Oyler GA, Higgins GA, Hart RA, Battenbarg M, Bloom FE, Wilson MC. J Cell Biol. 1989;109:3039. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett K, Calakos N, Scheller RH. Science. 1992;257:255. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 4.Trimble WS, Cowam DM, Scheller RH. P Natl Acad Sci USA. 1988;85:4538. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho WJ, Jeremic A, Jena BP. J Cell Mol Med. 2005;9:380. doi: 10.1111/j.1582-4934.2005.tb00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SJ, Kelly M, Rognlien KT, Cho JA, Horber JKH, Jena BP. Biophys J. 2002;83:2522. doi: 10.1016/s0006-3495(02)75263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeremic A, Kelly M, Cho J, Cho SJ, Horber JKH, Jena BP. Cell Biol Int. 2004;28:19. doi: 10.1016/j.cellbi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Cho WJ, Jeremic A, Jena BP. J Am Chem Soc. 2005;127:10156. doi: 10.1021/ja052442m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeremic A, Quinn AS, Cho WJ, Taatjes DJ, Jena BP. J Am Chem Soc. 2006;128:26. doi: 10.1021/ja056286v. [DOI] [PubMed] [Google Scholar]
- 10.Jeremic A, Cho WJ, Jena BP. J Biol Phys Chem. 2004;4:139. [Google Scholar]
- 11.Cho WJ, Jena BP. J Biomed Nanotechnol. 2007;3:209. [Google Scholar]
- 12.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Nature. 1998;395:347. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 13.Gorry PA. Anal Chem. 1990;62:570. [Google Scholar]
- 14.Provencher SW, Glöckner J. Biochemistry. 1981;20:33. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]