Abstract
AIM: To elucidate the possible difference in two promoter polymorphisms of the transforming growth factor-β1 (TGF-β1) gene (-800G > A, -509C > T) between ulcerative colitis (UC) patients and normal subjects.
METHODS: A total of 155 patients with established ulcerative colitis and 139 normal subjects were selected as controls. Two single nucleotide polymorphisms within the promoter region of TGF-β1 gene (-509C > T and -800G > A) were genotyped using PCR-RFLP.
RESULTS: There was a statistically significant difference in genotype and allele frequency distributions between UC patients and controls for the -800G > A polymorphism of the TGF-β1 gene (P < 0.05). The frequency of the TGF-β1 gene polymorphism at position -800 showed that the AA genotype and the allele A frequencies significantly differed between the patients and healthy controls (P < 0.05). At position -509, there was no statically significant difference in genotype and allele frequency between the patients and control subjects.
CONCLUSION: The results of our study indicate that there is a significant difference in both allele and genotype frequency at position -800G > A of TGF-β1 gene promoter between Iranian patients with UC and normal subjects.
Keywords: Transforming growth factor-β1, Ulcerative colitis, Promoter, Polymorphism, Iran
INTRODUCTION
Transforming growth factor-β1 is a cytokine produced by both immune and non immune cells, and it exhibits a broad range of functions. TGF-β1 controls the differentiation, proliferation, and state of activation of all immune cells, wound healing, angiogenesis and is implicated in immune abnormalities linked to cancer, autoimmunity, opportunistic infections, and fibrotic complications[1–5]. It has chemotactic properties and may stimulate cells to produce cytokines such as IL-1, IL6, and TNF-α at the sites of inflammation[1,6]. It is interesting that TGF-β1 plays an important role in promoting and activating extracellular matrix and bone remodeling. Paradoxically it is also involved in several immune suppressive mechanisms particularly as an inhibitor of intestinal epithelial proliferation[7–9]. It is proposed that the TGF-β1 production is under genetic control[7,10]. In the human TGF-β1 gene, which is located on chromosome 19q13 [11–15], eight polymorphisms are presently known. Three of them are localized in the promoter region at positions -988C > A, -800G > A and -509C > T from the first transcribed nucleotide[13].
Polymorphism of TGF-β1 has been investigated in several diseases. For instance, our own investigation indicates that polymorphism of this cytokine is not associated with the repeated spontaneous abortion[16,17]. Polymorphism of TGF-β1 and the risk of hepatocellular carcinoma have been investigated in patients with chronic hepatitis B virus infection[18,19]. In patients with liver cirrhosis, an association has been found between codon 10 and morphology of hepatocellular carcinoma in Korean population[20].
Inflammatory bowel disease is thought to result from inappropriate and ongoing activation of the mucosal immune system due to the presence of normal luminal flora. This aberrant response is most likely facilitated by defects in both the barrier function of the intestinal epithelium and the mucosal immune system[21,22]. As only two previous studies are available on the association of the -509 C > T TGF-β1 polymorphism with inflammatory bowel disease, especially Crohn's disease[23,24], we investigated whether these two polymorphisms (-509, -800) are associated in south Iranian patients with established UC.
MATERIALS AND METHODS
Subjects
The subjects in this study were comprised of 155 unrelated patients with UC (69 males and 86 females, aged 23-51 years, mean 36.4 years) attending the Department of Gastroenterology, Namazi and Saadi Academic Hospitals of Shiraz Medical University. Disease duration was at least 2 years. Patients were asked about acute phases of the disease after diagnosis. Every patient chart was revisited to complete all available information. A total of 139 age- and sex- matched health volunteers with no history of chronic bloody diarrhea and abdominal pain (50 males, 89 females) served as controls (aged 18-61 years, mean 35.5 years). Diagnosis of UC was established on the basis of conventional endoscopic and histological criteria. Since UC is a dynamic disease and patients can fluctuate into a different phenotype during the course of the disease, we analyzed the clinical records of our patients at two time points: when patients visited the hospital and for the first time he/she was followed up (mean time after diagnosis of 1.5 years). All patients were sub-classified according to gender, age of onset, localization of the disease, need for steroid therapy, and emergent surgical treatment. In UC patients, the localization of gut involvement was defined as proctitis, left-sided (up to the splenic flexure) or pancolitis (beyond the splenic flexure).
DNA extraction and TGF-β1 genotyping
Venous blood was collected into EDTA-coated tubes. DNA was extracted from whole blood using the salting-out method. The following primers (MOLBIOL, Germany) were used for amplification of promoter regions -509 and -800. The sequences of PCR primers for the -509C/T and -800G/A polymorphisms are 5’-CAGTAAATGTATGGGGTCGCAG-3’ (forward) and 5’-GGTGTCAGTGGGAGGAGGG-3’ (reverse), and 5’-ACAGTTGGCACGGGCTTTCG-3’ (forward) and 5’-TCAACACCCTGCGACCCCAT-3’ (reverse) respectively. The PCR product sizes from these primers were 153 bp and 388 bp, respectively. PCR was performed in a total volume of 20 μL containing 100 ng genomic DNA, 20 pmol/L of each primer, 0.2 mmol/L dNTPs, 20 mmol/L Tris-HCl (pH 8.8), 10 mmol/L MgCl2, and 1 unit of Taq polymerase (New England BioLabs Ipswich, MA). The PCR cycle conditions consisted of an initial denaturation at 94°C for 3 min followed by 35 cycles at 94°C for 60 s, at 61°C for 60 s, at 72°C for 60 s for -509 C/T, and at 94°C for 45 s, at 61°C for 45 s, at 72°C for 45 s for -800G/A, and a final elongation at 72°C for 5 min.
Genotyping was performed by restriction fragment length polymorphism (RFLP) analysis. The following restriction enzymes (Fermantes, Lithuania) were used for the digestion of amplified PCR products. For digestion of PCR products containing positions -800, NmuCI and -509, Eco811 was applied. The digested PCR products were resolved on 2% agarose gel and stained with ethidium bromide for visualization under UV light.
Statistical analysis
Allele frequencies at each polymorphic site were calculated by allele counting method. Deviation of the genotype counts from the Hardy-Weinberg equilibrium was tested using χ2 test with 1 degree of freedom. Differences in the allele frequencies and genotype distributions between the patients with UC and the controls were analyzed by χ2 or Fisher's exact tests when necessary.
RESULTS
Position -800 (G/A)
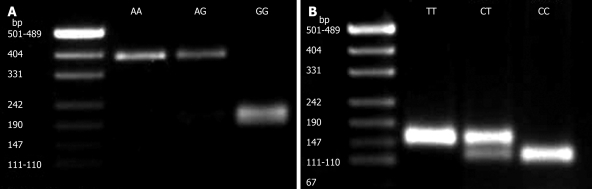
In this investigation, the change at position -800G > A of the TGF-β1 gene was studied using PCR-RFLP in 155 cases of UC and 139 normal Iranian subjects. At this position, homozygote GG was found in 125 (80.6%) UC patients and 104 (74.8%) normal subjects, heterozygote GA was observed in 29 (18.8%) UC patients and 27 (19.4%) normal subjects, while homozygote AA was demonstrated in only one of the patients, homozygote AA was shown in eight normal individuals. There was a statistically significant difference in genotype and allele frequency distributions between UC patients and controls for the -800G > A polymorphism of the TGF-β1 gene (P < 0.05) (Table 1). A typical genotyping at this position is represented in Figure 1A.
Table 1.
Distribution of TGF-β1 genotype and alleles in UC patients and controls
| TGF-β1 genotype and alleles | UC patients n = 155 (%) | Controls n =139 (%) | P value |
| Genotype -800 | < 0.05 | ||
| GG | 125 (80.6) | 104 (74.8) | |
| GA | 29 (18.8) | 27 (19.4) | |
| AA | 1 (0.6) | 8 (5.8) | |
| Allele frequency | < 0.05 | ||
| G | 279 (90) | 235 (84.5) | |
| A | 31 (10) | 43 (15.5) | |
| Genotype-509 | 0.3 | ||
| CC | 57 (36.8) | 56 (40.3) | |
| CT | 65 (41.9) | 63 (45.3) | |
| TT | 33 (21.3) | 20 (14.4) | |
| Allele frequency | 0.19 | ||
| C | 179 (57.75) | 175 (62.9) | |
| T | 131 (42.25) | 103 (37.1) |
Figure 1.
A: PCR-RFLP genotyping at position -800 (G/A). PCR product (388 bp) was digested with NmuCI restriction enzyme. From left to right: Lane 1: DNA size marker; lane 2: Homozygote AA; lane 3: Heterozygote AG; lane 4: Homozygote GG; B: PCR-RFLP genotyping at position −509 (C/T). PCR product (153 bp) was digested with Eco811 restriction enzyme. Lane 1: DNA size marker; lane 2: Homozygote TT; lane 3: Heterozygote CT; lane 4: Homozygote CC.
Position -509 (G/A)
In addition, the genotype at position -509C > T of the TGF-β1 gene was studied in the same study groups. At this position, homozygote CC was found in 57 (36.8%) UC patients and 56 (40.3%) normal subjects, heterozygote CT was observed in 65 (41.9%) UC patients and 63 (45.3%) normal subjects, homozygote TT was demonstrated in 33 (21.3%) UC patients and 20 (14.4%) normal subjects. There was no statistically significant difference in genotype distribution and allele frequency at this position between UC patients and controls. The distribution of TGF-β1 genotype and allele frequency in UC patients and controls are summarized in Table 1. A typical genotyping at position -509C > T is represented in Figure 1B. A Hardy-Weinberg equilibrium test was performed for the two polymorphisms.
Relation between other factors and polymorphisms
In our study, no significant relation was found between the two polymorphisms and factors such as gender, age of onset, cumulative dose of steroids, need for steroid therapy, localization of the disease, and need for emergent surgical treatment.
DISCUSSION
TGF-β1 is an immunoregulatory and immunosuppressive cytokine. Its role and function have been extensively investigated in inflammatory type of diseases[14]. It has been known that TGF-β1 has various biological and immunological functions, such as decreasing expression of MHC-II, reducing synthesis of TH1/TH2 cytokine and suppressing the activation of both T and B lymphocytes. Plasma levels of TGF-β1 have been detected in several inflammatory and malignant diseases and the consensus is that the synthesis of this immunosuppressive cytokine is under the control of genetic factors[3,6,11]. TGF-β1 gene is located on chromosome 19 and contains 7 exons.
Several polymorphisms have been reported in TGF-β1 gene at positions -988, -800 and -509 located in the gene promoter[13–21]. Garcia-Gonzalez and co-workers showed that codon 10 and 25 TGF-β1 polymorphisms in Dutch IBD patients and healthy controls do not participate in defining the susceptibility to and tile nature of the clinical course in IBD[24]. Polonikov AV et al[28] reported that TGF-β1 gene polymorphism plays a role in the pathogenesis of gastric ulcer disease. Su Zg and colleagues[16] reported that allele A at position -800 and allele T at position -509 of TGF-β1 gene are increased in Chinese population with chronic obstructive pulmonary disease. Schulte CM[23] showed that IBD susceptibility is not associated with genetic variations in TGF-β1 promoter polymorphism.
In addition to the reports on genetic variations in TGF-β1 polymorphism in IBD, several studies showed that serum level of TGF-β1[17] and mRNA expression[18] are significantly increased in IBD patients.
In the present study, the genotype distribution and allele frequencies of polymorphisms at position -509C > T were not significantly different between UC patients and controls, which is consistent with the previous reports[24,30]. However, at position -800G > A both allele and genotype frequencies were significantly different between UC patients and controls. It has been reported that -800G > A substitution is thought to disrupt a consensus half-site for binding to the nuclear transcription factor (CRE-binding protein), consequently contributing to a lower production of total TGF-β1 in the circulation[7,26].
In our study, AA genotype was reduced in patients with IBD compared with controls. A similar finding was seen in UC patients and controls when the frequency of allele A was analyzed. These findings indicate that individuals bearing allele A and genotype AA are less susceptible to IBD than those lacking allele A and genotype AA.
To the best of our knowledge, polymorphism at position -800 of TGF-β1 gene promoter has not yet been reported in IBD patients. However, both plasma level of TGF-β1 and mRNA expression should be further studied and compared in IBD patients carrying genotype AA at this position.
There are a number of other polymorphisms with known influence on different TGF-β1 expressions[3,4]. To study the role of TGF-β1 haplotype in the pathogenesis of IBD, analysis of other known TGF-β1 gene polymorphisms will strength our current observation.
In conclusion, there is no significant difference in TGF-β1 polymorphisms at position -509 C > T and allele A is significantly associated with genotype AA at position -800 of the promoter region between southern Iranian UC patients and healthy individuals.
COMMENTS
Background
Inflammatory bowel disease (IBD) is a common disorder in our population with an increasing incidence but no clear cut etiology. Cytokines and their receptors, immunogenetics and host -related factors are all involved in the disease susceptibility. TGF-β1 is produced by regulatory T lymphocytes with a profound suppressive effect on the induction phase of immune response. Plasma levels of TGF-β1 and its local synthesis, in addition to the contribution of genes encoding TGF-β1 have widely been investigated in IBD patients with no convincing and solid ground so far.
Research frontiers
Investigation of cytokine gene polymorphism in health and disease conditions has provided valuable information on the incidence of autoimmune diseases in different ethnics. Current data on TGF-β1 gene polymorphism in IBD are controversial due to the heterogeneity in clinical presentations of the disease and different methods. There is no evidence that TGF-β1 gene polymorphism is associated with IBD in European Caucasian population. Here we report the impact of allele A and genotype AA at -800 of TGF-β1 promoter region and susceptibility to Iranian patients with IBD.
Publications
Schults et al and Garcia Gonzalez reported that polymorphism at promoter region has no association with IBD in European individuals. Our data give another view on the significance of polymorphism of TGF-β1 promoter region in IBD patients.
Innovations and breakthroughs
Our findings provide a window for investigation on the mRNA and local secretion of TGF-β1 in IBD patients carrying allele A and genotype AA of TGF-β1 at position -800 of promoter region.
Application
By extending this investigation in a larger sample size within other ethnic groups or in a broader study in a format of meta-analysis, a genetic marker for screening of individual susceptible to IBD can be expected.
Peer review
This is a very interesting and original paper that merits to be fully published without any substantial modifications.It is also in good english written.
Supported by a grant from Shiraz University of Medical Sciences, No. 83-2363, and by a grant from Shiraz Institute for Cancer Research, ICR-8296
S- Editor Liu Y L- Editor Wang XL E- Editor Ma WH
References
- 1.Gewaltig J, Mangasser-Stephan K, Gartung C, Biesterfeld S, Gressner AM. Association of polymorphisms of the transforming growth factor-beta1 gene with the rate of progression of HCV-induced liver fibrosis. Clin Chim Acta. 2002;316:83–94. doi: 10.1016/s0009-8981(01)00738-0. [DOI] [PubMed] [Google Scholar]
- 2.Dieckgraefe BK, Stenson WF, Alpers DH. Gastrointestinal epithelial response to injury. Curr Opin Gastroenterol. 1996;12:109–114. [Google Scholar]
- 3.Kang Y, Prentice MA, Mariano JM, Davarya S, Linnoila RI, Moody TW, Wakefield LM, Jakowlew SB. Transforming growth factor-beta 1 and its receptors in human lung cancer and mouse lung carcinogenesis. Exp Lung Res. 2000;26:685–707. doi: 10.1080/01902140150216765. [DOI] [PubMed] [Google Scholar]
- 4.Lee-Chen GJ, Liu KP, Lai YC, Juang HS, Huang SY, Lin CY. Significance of the tissue kallikrein promoter and transforming growth factor-beta1 polymorphisms with renal progression in children with vesicoureteral reflux. Kidney Int. 2004;65:1467–1472. doi: 10.1111/j.1523-1755.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 5.Khalil MS, El Nahas AM, Blakemore AI. Transforming growth factor-beta1 SNPs: genetic and phenotypic correlations in progressive kidney insufficiency. Nephron Exp Nephrol. 2005;101:e31–e41. doi: 10.1159/000086227. [DOI] [PubMed] [Google Scholar]
- 6.Tag CG, Mengsteab S, Hellerbrand C, Lammert F, Gressner AM, Weiskirchen R. Analysis of the transforming growth factor-beta1 (TGF-beta1) codon 25 gene polymorphism by LightCycler-analysis in patients with chronic hepatitis C infection. Cytokine. 2003;24:173–181. doi: 10.1016/j.cyto.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, Carter ND, Spector TD. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8:93–97. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- 8.Hahm KB, Im YH, Lee C, Parks WT, Bang YJ, Green JE, Kim SJ. Loss of TGF-beta signaling contributes to autoimmune pancreatitis. J Clin Invest. 2000;105:1057–1065. doi: 10.1172/JCI8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo BM, Yeo M, Oh TY, Choi JH, Kim WW, Kim JH, Cho SW, Kim SJ, Hahm KB. Amelioration of pancreatic fibrosis in mice with defective TGF-beta signaling. Pancreas. 2005;30:e71–e79. doi: 10.1097/01.mpa.0000157388.54016.0a. [DOI] [PubMed] [Google Scholar]
- 10.Shah R, Hurley CK, Posch PE. A molecular mechanism for the differential regulation of TGF-beta1 expression due to the common SNP -509C-T (c. -1347C>T) Hum Genet. 2006;120:461–469. doi: 10.1007/s00439-006-0194-1. [DOI] [PubMed] [Google Scholar]
- 11.Mattey DL, Nixon N, Dawes PT, Kerr J. Association of polymorphism in the transforming growth factor {beta}1 gene with disease outcome and mortality in rheumatoid arthritis. Ann Rheum Dis. 2005;64:1190–1194. doi: 10.1136/ard.2004.031674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos Santos MC, Campos MI, Souza AP, Scarel-Caminaga RM, Mazzonetto R, Line SR. Analysis of the transforming growth factor-beta 1 gene promoter polymorphisms in early osseointegrated implant failure. Implant Dent. 2004;13:262–269. doi: 10.1097/01.id.0000140463.10204.87. [DOI] [PubMed] [Google Scholar]
- 13.Amani D, Dehaghani AS, Zolghadri J, Ravangard F, Niikawa N, Yoshiura K, Ghaderi A. Lack of association between the TGF-beta1 gene polymorphisms and recurrent spontaneous abortion. J Reprod Immunol. 2005;68:91–103. doi: 10.1016/j.jri.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Hahm KB, Im YH, Parks TW, Park SH, Markowitz S, Jung HY, Green J, Kim SJ. Loss of transforming growth factor beta signalling in the intestine contributes to tissue injury in inflammatory bowel disease. Gut. 2001;49:190–198. doi: 10.1136/gut.49.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahm KB, Lee KM, Kim YB, Hong WS, Lee WH, Han SU, Kim MW, Ahn BO, Oh TY, Lee MH, et al. Conditional loss of TGF-beta signalling leads to increased susceptibility to gastrointestinal carcinogenesis in mice. Aliment Pharmacol Ther. 2002;16 Suppl 2:115–127. doi: 10.1046/j.1365-2036.16.s2.3.x. [DOI] [PubMed] [Google Scholar]
- 16.Amani D, Zolghadri J, Dehaghani AS, Pezeshki AM, Ghaderi A. The promoter region (-800, -509) polymorphisms of transforming growth factor-beta1 (TGF-beta1) gene and recurrent spontaneous abortion. J Reprod Immunol. 2004;62:159–166. doi: 10.1016/j.jri.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 17.von Linsingen R, Bompeixe EP, Bicalho Mda G. A case-control study in IL6 and TGFB1 gene polymorphisms and recurrent spontaneous abortion in southern Brazilian patients. Am J Reprod Immunol. 2005;53:94–99. doi: 10.1111/j.1600-0897.2005.00250.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim YJ, Lee HS, Im JP, Min BH, Kim HD, Jeong JB, Yoon JH, Kim CY, Kim MS, Kim JY, et al. Association of transforming growth factor-beta1 gene polymorphisms with a hepatocellular carcinoma risk in patients with chronic hepatitis B virus infection. Exp Mol Med. 2003;35:196–202. doi: 10.1038/emm.2003.27. [DOI] [PubMed] [Google Scholar]
- 19.Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505–510. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Kwon OS, Song SH, Ju KT, Chung MG, Park DK, Kim SS, Kim YS, Koo YS, Kim YK, Choi DJ, et al. Polymorphism in codons 10 and 25 of the transforming growth factor-beta1 gene in Korean population and in patients with liver cirrhosis and hepatocellular carcinoma. Korean J Gastroenterol. 2003;42:212–219. [PubMed] [Google Scholar]
- 21.Podolsky DK. Mucosal immunity and inflammation. V. Innate mechanisms of mucosal defense and repair: the best offense is a good defense. Am J Physiol. 1999;277:G495–G499. doi: 10.1152/ajpgi.1999.277.3.G495. [DOI] [PubMed] [Google Scholar]
- 22.Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson IV. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation. 1998;66:1014–1020. doi: 10.1097/00007890-199810270-00009. [DOI] [PubMed] [Google Scholar]
- 23.Schulte CM, Goebell H, Rtiher HD, Schulte KM. C-509T polymorphism in the TGFB1 gene promoter: impact on Crohn's disease susceptibility and clinical course? Immunogenetics. 2001;53:178–182. doi: 10.1007/s002510100309. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Gonzalez MA, Crusius JB, Strunk MH, Bouma G, Perez-Centeno CM, Pals G, Meuwissen SG, Pena AS. TGFB1 gene polymorphisms and inflammatory bowel disease. Immunogenetics. 2000;51:869–872. doi: 10.1007/s002510000211. [DOI] [PubMed] [Google Scholar]
- 25.Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, Fiocchi C, Strober W. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn's disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157:1261–1270. [PubMed] [Google Scholar]
- 26.Syrris P, Carter ND, Metcalfe JC, Kemp PR, Grainger DJ, Kaski JC, Crossman DC, Francis SE, Gunn J, Jeffery S, et al. Transforming growth factor-beta1 gene polymorphisms and coronary artery disease. Clin Sci. 1998;95:659–667. doi: 10.1042/cs0950659. [DOI] [PubMed] [Google Scholar]
- 27.Polonikov AV, Ivanov VP, Belugin DA, Khoroshaya IV, Kolchanova IO, Solodilova MA, Tutochkina MP, Stepchenko AA. Analysis of common transforming growth factor beta-1 gene polymorphisms in gastric and duodenal ulcer disease: pilot study. J Gastroenterol Hepatol. 2007;22:555–564. doi: 10.1111/j.1440-1746.2006.04542.x. [DOI] [PubMed] [Google Scholar]
- 28.Su ZG, Wen FQ, Feng YL, Xiao M, Wu XL. Transforming growth factor-beta1 gene polymorphisms associated with chronic obstructive pulmonary disease in Chinese population. Acta Pharmacol Sin. 2005;26:714–720. [PubMed] [Google Scholar]
- 29.Wiercińska-Drapalo A, Flisiak R, Prokopowicz D. Effect of ulcerative colitis activity on plasma concentration of transforming growth factor beta1. Cytokine. 2001;14:343–346. doi: 10.1006/cyto.2001.0901. [DOI] [PubMed] [Google Scholar]
- 30.Wang YF, Wei B, Ouyang Q. Expression with TGFbeta1 in the patients with ulcerative colitis. Sichuan Da Xue Xue Bao Yi Xue Ban. 2005 Mar;36:204–206. [PubMed] [Google Scholar]