Abstract
A recombinant Aspergillus niger strain expressing the Hypocrea jecorina endoglucanase Cel7B was grown on spent hydrolysates (stillage) from sugarcane bagasse and spruce wood. The spent hydrolysates served as excellent growth media for the Cel7B-producing strain, A. niger D15[egI], which displayed higher endoglucanase activities in the spent hydrolysates than in standard medium with a comparable monosaccharide content (e.g., 2,100 nkat/ml in spent bagasse hydrolysate compared to 480 nkat/ml in standard glucose-based medium). In addition, A. niger D15[egI] was also able to consume or convert other lignocellulose-derived compounds, such as acetic acid, furan aldehydes, and phenolic compounds, which are recognized as inhibitors of yeast during ethanolic fermentation. The results indicate that enzymes can be produced from the stillage stream as a high-value coproduct in second-generation bioethanol plants in a way that also facilitates recirculation of process water.
Energy security, petroleum depletion, and global warming have been the main driving forces for the development of renewable fuels that can replace petroleum-derived fuels, such as gasoline and diesel. Bioethanol is currently the most commonly used renewable automobile fuel. It is largely produced by fermentation of sugar- or starch-containing feedstocks, such as cane sugar, corn, and wheat. However, the supply of these crops is relatively limited and many of them can be considered human food resources. Lignocellulose is a more-abundant and less-expensive raw material with the potential to give a high net energy gain (11, 17).
In the production of bioethanol from lignocellulosic materials, hydrolytic enzymes, such as cellulases and cellobiases, can be used to convert the lignocellulosic polysaccharides to monosaccharides. Microorganisms can be used to ferment the monosaccharides to ethanol. The yeast Saccharomyces cerevisiae is one of the most suitable microorganisms for ethanol production and is favored in industrial processes. However, S. cerevisiae only metabolizes hexose sugars. Many lignocellulosic materials consist of a significant proportion of xylan and arabinan, which give rise to pentose sugars. The cost of enzymes for the hydrolysis of polysaccharides and the inability of S. cerevisiae to utilize pentose sugars have been pointed out as two bottlenecks for commercialization of cellulosic ethanol production (9, 25). Considerable research efforts have therefore been focused on reducing the enzyme cost by producing more-efficient enzymes from cheaper growth media (25). Other efforts have been focused on different approaches to convert pentose sugars to ethanol by using recombinant microorganisms (3, 10).
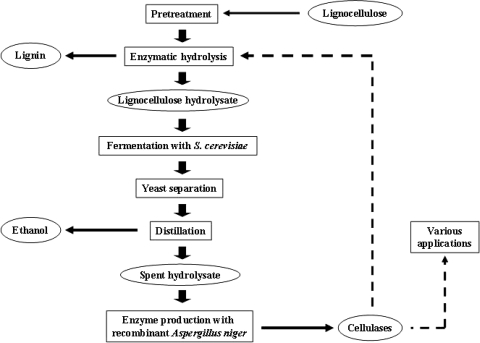
A novel approach to reduce the enzyme cost and to optimally utilize all sugars generated from lignocellulose would be to produce hydrolytic enzymes, such as cellulases, from the pentose fraction remaining after consumption of hexoses by S. cerevisiae (Fig. 1). The cellulases produced can then be used on site in the next round of hydrolysis of the lignocellulosic feedstock and thereby reduce the dependence on externally produced enzymes.
FIG. 1.
Schematic representation of the experimental approach and on-site enzyme production in a cellulose-to-ethanol process.
Furthermore, it is desirable to recycle the process water in an ethanol production plant to minimize the production costs. However, lignocellulose hydrolysates are very complex and contain a wide range of different compounds. Some of these compounds, such as furan aldehydes, aliphatic acids, and phenolic compounds, inhibit the yeast S. cerevisiae, which results in lower ethanol yield and productivity. Recycling of the process water can lead to a buildup in the concentration of inhibitors, which is a phenomenon that has been pointed out as an obstacle to reusing the stillage stream (1, 35). There are several methods to avoid inhibitor-related problems, but they are often associated with additional process cost (40). However, A. niger is an organism that can utilize a broad range of compounds as nutrients, possibly including compounds that inhibit S. cerevisiae. It would be convenient if the A. niger cells could metabolize such compounds and thereby, due to the removal of inhibitors, make it more feasible to reuse the process water.
In this study, we explored the possibility of utilizing sugarcane bagasse and spruce wood for ethanol production and using the spent hydrolysates (stillage) for production of the Hypocrea jecorina cellulase Cel7B (formerly called endoglucanase I) by a recombinant strain of Aspergillus niger. Simultaneously, the Cel7B-producing recombinant A. niger strain also removed inhibitory lignocellulose-derived products, thus facilitating recycling of process water.
MATERIALS AND METHODS
Raw materials.
Sugarcane bagasse was air-dried to a dry-matter content of 96% and milled to pass a 2-mm screen. In addition, a previously prepared spruce hydrolysate was utilized. The spruce hydrolysate was produced by two-step dilute-acid hydrolysis as described by Alriksson et al. (2). The hydrolysate, which had an initial pH of about 2, was stored at 4°C prior to use.
Pretreatment of bagasse.
A bagasse prehydrolysate was prepared by using a previously described procedure (20). One hundred eighty grams of dried and milled raw material was mixed with 1,800 g of diluted sulfuric acid in each of three separate stainless steel cylinders, each with a total volume of 2.5 liters. The final concentration of sulfuric acid in the slurry was 2%. The cylinders were attached to a rotor in a polyethylene glycol heating bath controlled by a control unit (Jaako Pöyry AB, Karlstad, Sweden). The pretreatment was performed at 122°C for 60 min. Directly after the pretreatment had finished, the cylinders were rapidly cooled to room temperature in a water bath. The solids and the liquid of the pretreated slurry were separated by vacuum filtration. The solids from each cylinder were washed with 5 liters of distilled water (dH2O) and dried in a heating cabinet at 70°C for 72 h. The liquid fraction, hereafter referred to as bagasse prehydrolysate, was collected and stored at 4°C.
Enzymatic hydrolysis.
Pretreated solid material (80 g dry weight [DW]) was mixed with 800 g of bagasse prehydrolysate in a 2,000-ml Erlenmeyer glass flask closed with a cotton plug (experiment was done in quadruplicate). The pH of the slurries was adjusted to 4.8 with NaOH (12 M). Commercially available preparations of cellulase and cellobiase (Celluclast 1.5 L, with a manufacturer-stated activity of 700 endoglucanase units/g [Sigma-Aldrich, Steinheim, Germany], and Novozyme 188, with a stated activity of 250 cellobiase units/g [Sigma-Aldrich]) were added to the slurry at loadings of 319 endoglucanase units/g of solids (DW) and 23 cellobiase units/g of solids (DW), respectively. The enzyme dosages were based on the results of a set of small-scale optimization experiments. The slurries were incubated with shaking (incubator shaker model G25; New Brunswick Scientific, Edison, NJ) at 50°C and 150 rpm for 72 h. The pH of the slurries was measured and readjusted to 4.8 with NaOH every 10 hours. During the hydrolysis, the amount of released glucose in the slurries was monitored by measurements with a glucometer (glucometer Elite XL, Bayer AG, Leverkusen, Germany) every 10 hours. After the hydrolysis, the slurries were filtered. The pH of the liquid fraction, hereafter referred to as bagasse hydrolysate, was adjusted to pH 2.0 with HCl (12 M), and it was then stored at 4°C to prevent microbial growth during storage. The composition of sugarcane bagasse, the effect of the pretreatment, and the convertibility of the pretreated material by enzymatic hydrolysis have been reported by Martín et al. (20).
Fermentation with S. cerevisiae.
Prior to fermentation, the pH of the bagasse and the spruce hydrolysates was adjusted to 5.5 with NaOH. The hydrolysates were also supplemented with a nutrient solution, giving a final concentration of 0.5 g/liter (NH4)2HPO4, 0.025 g/liter MgSO4 · 7H2O, 1.38 g/liter NaH2PO4 · H2O, and 1 g/liter yeast extract. The fermentations of the bagasse and the spruce hydrolysates were carried out in 15 parallel 50-ml glass flasks equipped with magnets for stirring and sealed with rubber plugs pierced by cannulas for CO2 removal. The flasks were inoculated with baker's yeast (S. cerevisiae) (a baking strain obtained from Jästbolaget AB, Rotebro, Sweden). The flasks with bagasse hydrolysate were given an inoculum yielding a cell mass concentration of 1 g/liter (DW), whereas the flasks with the spruce hydrolysate were given a yeast inoculum of 9 g/liter (DW) due to the higher toxicity of the spruce hydrolysate. The flasks were incubated at 30°C for 5 h in a water bath with magnetic stirring (IKA-Werke, Staufen, Germany). The glucose levels during the course of fermentation were monitored by continuous measurements with a glucometer.
Distillation.
After the fermentation, the yeast cells were removed from the fermented hydrolysates by centrifugation (Sorvall RC26 Plus; Dupont, Newtown, CT) at 1,500 × g and 4°C for 6 min. The pH of the fermented hydrolysate liquids was adjusted to 7.0 with NaOH to prevent possible sugar degradation during the distillation. The fermented hydrolysate liquids were transferred to round-bottom flasks, and a few drops of antifoam were added to avoid excessive bubble formation during distillation. Standard distillation glassware was used for the distillation setup, and a polyethylene glycol heating bath was used as the heat source. The distillation was pursued until all ethanol was separated from the fermented hydrolysate liquids. The nonethanol fractions remaining in the round-bottom flasks, hereafter called spent bagasse hydrolysate (SBH) and spent spruce hydrolysate (SSH), were collected and stored at 4°C until further use.
A. niger recombinant strains.
In this study, a recombinant A. niger D15 transformant expressing the H. jecorina Cel7B gene was used (strain was designated A. niger D15[egI]). The expression of the Cel7B gene was under transcriptional control of the constitutive gpd promoter from A. niger and the glaA terminator from Aspergillus awamori (27). A transformant with the same promoter and terminator integrated in the chromosome but without the Cel7B gene was used as a negative control (the strain was designated A. niger D15[pGT]). The construction of the two recombinant strains is described in Rose and van Zyl (27).
Fermentation experiment A: A. niger cultivated on SBH.
For fermentation experiment A, four 100-ml Erlenmeyer flasks (named A to D) were filled with 48 ml of SBH, 1 ml of nutrient solution [25 g/liter (NH4)2HPO4, 1.25 g/liter MgSO4 · 7H2O, 69 g/liter NaH2PO4 · H2O, and 50 g/liter yeast extract], 0.05 ml of trace element solution (0.22 g/liter ZnSO4 · 7H2O, 0.11 g/liter H3BO3, 0.05 g/liter MnCl2 · 4H2O, 0.05 g/liter FeSO4 · 7H2O, 0.017 g/liter CoCl2 · 6H2O, 0.016 g/liter CuSO4 · 5H2O, 0.015 g/liter Na2MoO4 · 2H2O, and 0.5 g/liter EDTA) and closed with cotton plugs. The initial pH of this and other media used for cultivation of A. niger transformants was approximately 6.5. Flasks A and B were inoculated with 0.95 ml A. niger D15[egI] spores (final spore concentration was 1 × 106 spores/ml medium). Flasks C and D were inoculated with A. niger D15[pGT] spores at an equal concentration. For comparison, four 100-ml Erlenmeyer flasks (named E to H) closed with cotton plugs were filled with 49.05 ml of standard medium (5 g/liter yeast extract, 0.4 g/liter MgSO4 · 7H2O, 10 g/liter glucose, 2 g/liter Casamino Acids, 0.5 g/liter KCl, 1.5 g/liter KH2PO4, 6 g/liter NaNO3, and 1 ml/liter of the trace element solution). Flasks E and F were inoculated with 0.95 ml A. niger D15[egI] spores, and flasks G and H were inoculated with 0.95 ml A. niger D15[pGT] spores (final spore concentration was 1 × 106 spores/ml medium). The flasks were incubated for 11 days in an incubator with shaking (incubator shaker model G25; New Brunswick Scientific) at 30°C and 150 rpm. The endoglucanase activity was monitored during the fermentation experiment, and the biomass production was measured at the end of the experiment (see description below).
Fermentation experiment B: A. niger cultivated on SSH.
In fermentation experiment B, fermentation with A. niger D15[egI] was undertaken to investigate how the strain would perform in a different type of spent hydrolysate, viz., an SSH. For comparison, the standard medium and standard medium with 10 g/liter of xylose instead of glucose were used. The cultivation conditions and the inoculum sizes for A. niger D15[egI] were the same as described for fermentation experiment A. The experiment was ended after 10 days.
Fermentation experiment C: A. niger cultivated on standard medium with inhibitors.
In fermentation experiment C, an additional experiment was designed to determine whether A. niger D15[egI] was able to metabolize nonsugar compounds in lignocellulose hydrolysates. Eighteen 100-ml Erlenmeyer flasks were filled with 48 ml of standard medium, 1 ml of inhibitor solution, and 1 ml of spore inoculum (or 1 ml of sterile H2O for control flasks). The flasks were divided into six groups (three flasks per group). Each group had a different inhibitor solution added. Typical compounds found in lignocellulose hydrolysates, representing the three main inhibitor categories, aliphatic acids, furan aldehydes, and phenolic compounds, were chosen for the experiment. Group 1 (flasks 1A to 1C) had 5 g/liter of acetic acid added, group 2 (flasks 2A to 2C) had 1 g/liter of furfural added, group 3 (flasks 3A to 3C) had 2 g/liter of 5-hydroxymethylfurfural (HMF) added, group 4 (flasks 4A to 4C) had 0.5 g/liter of vanillin added, group 5 (flasks 5A to 5C) had 0.2 g/liter of coniferyl aldehyde added, and group 6 (flasks 6A to 6C) had no inhibitor but 1 ml sterile H2O as a volume equalizer added. The pH of all flasks was adjusted to 6.3 with NaOH. All the A and B flasks were inoculated with A. niger D15[egI] (1 × 106 spores/ml medium), while the C flasks (i.e., control flasks) were used to investigate if the concentrations of the inhibitors were affected in the absence of A. niger cells (for example, by degradation or evaporation). All 18 flasks were incubated for 9 days in an incubator with shaking at 150 rpm and 30°C. The endoglucanase activity was monitored during the experiment, and the biomass was measured at the end of the cultivation.
Biomass measurement.
To determine the DW of the A. niger biomass, pieces of Miracloth (Calbiochem, EMD Biosciences, La Jolla, CA) were dried in a microwave oven for 15 min and were thereafter placed in a desiccator. After 2 h, the Miracloth pieces were taken out of the desiccator and preweighed on an analytical scale. The A. niger culture suspension volumes from the fermentation experiments were measured, and the culture suspensions were then filtered with suction through dried Miracloth. Every piece of Miracloth with biomass was washed with 50 ml of dH2O, dried as previously described, and then weighed.
Enzyme activity assay.
The endoglucanase activity was monitored with a method based on the use of dinitrosalicylic acid as described by Bailey et al. (4). The buffer used for the assay was citrate (0.05 M, pH 5.5), the substrate was carboxymethyl cellulose (Fluka, ultralow viscosity; Sigma-Aldrich) (1% in citrate buffer [0.05 M, pH 5.5]), and the assay was carried out at 50°C. The enzyme activities are given in nkat/ml (nmol of reducing sugars produced/ml/s).
SDS-PAGE analysis.
Supernatants from the three fermentation experiments were diluted four times with dH2O and loaded onto 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels to determine the extracellular protein profile, as well as the size of the Cel7B protein. Subsequent to electrophoresis, the gel was stained with Sypro Red gel stain (Molecular Probes, Eugene, OR) according to the method of Steinberg et al. (33). The analysis of the gel was performed with a gel imager (ImageQuant 400; GE Healthcare, Uppsala, Sweden). The contrast of the gel images was enhanced by using the software ImageQuant 400 Capture (GE Healthcare). In addition, the Cel7B protein concentration was measured by running a concentration gradient of bovine serum albumin as a protein standard (Pierce, Rockford, IL) and the Cel7B supernatants on an SDS-PAGE gel and comparing the intensity of the Sypro Red-stained bands using the software ImageJ (http://rsb.info.nih.gov/ij/). The Cel7B protein concentration was calculated as the mean value of the results for two supernatants from separate cultivation flasks that had been analyzed as four replicates (i.e., on four separate gels). The Sypro Red dye exhibits low variability in the staining of different types of proteins due to its interaction with the dodecyl sulfate coat around the protein in the SDS-PAGE gel (34). The effect of the varying degree of glycosylation on the Sypro Red dye binding and, consequently, on the protein quantification was evaluated by comparing the intensities of Sypro Red-dyed deglycosylated and nondeglycosylated Cel7B protein samples containing the same amount of protein. Deglycosylation was performed as described in the section “Enzymatic deglycosylation” (see below). The specific activities reported are based on Cel7B protein measurements. The Cel7B protein measurements were made on samples taken at a fixed time for all samples in one fermentation experiment. The time of measurement was set at the time when the single highest endoglucanase activity was noted during each fermentation experiment.
Protein identification by MS.
The identities of the proteins in selected SDS-PAGE bands were verified by using mass spectrometry (MS). Two samples displaying different molecular masses based on the SDS-PAGE analysis were selected for MS analysis. These samples came from supernatants from the cultivations of the A. niger transformant with the gene coding for Cel7B and were obtained in experiments in which the fungus was grown on SBH medium (fermentation experiment A) and on standard medium with acetic acid (fermentation experiment C). The SDS-PAGE gels were stained with Coomassie brilliant blue R-250. After destaining, the putative Cel7B bands were excised from the gel and submitted to Alphalyse A/S (Odense, Denmark) for protein identification by peptide mass fingerprinting and by sequence analysis through tandem MS (MS-MS) sequencing. The protein samples were treated with iodoacetamide, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS for peptide mass fingerprinting. Peptide sequencing was performed by using MALDI-TOF-TOF MS. Mascot software (version 2.2.03; www.matrixscience.com) was used for database matching of the combined MS and MS-MS spectra.
Enzymatic deglycosylation.
To establish the type and degree of protein glycosylation, the Cel7B protein was treated with N-glucosidase F (Roche, Mannheim, Germany). The supernatant from the cultivation of A. niger D15[egI] in SBH was first concentrated with a centrifugal filter device (0.5 ml, 10-kDa cutoff) (Millipore, Bedford, MA) and dialyzed against dH2O (8-kDa cutoff). The sample was then treated in a solution of 20 mM sodium phosphate buffer (pH 8.6), 1% SDS, and 1% β-mercaptoethanol. The sample was then deglycosylated in a solution consisting of 25 mM sodium phosphate buffer (pH 7.2), 0.25% SDS, 1% β-mercaptoethanol, 1% Nonidet P-40, 0.5% EDTA, and 5 U N-glucosidase F. The reaction was carried out at 37°C for 7 h, and the treated sample was analyzed by SDS-PAGE according to the previous description. The enzymatic deglycosylation was exhaustive, since increase of the above-mentioned dosage of N-glycosidase F did not decrease the molecular mass of the recombinant Cel7B protein any further.
Chemical deglycosylation.
A chemical deglycosylation kit was used to achieve a complete removal of all types of carbohydrates attached to the protein core (GlycoProfile IV chemical deglycosylation kit; Sigma-Aldrich). The supernatant from the cultivation of A. niger D15[egI] in SBH was first dialyzed against dH2O (8-kDa cutoff) and concentrated with a centrifugal filter device (0.5 ml, 10-kDa cutoff). The concentrated culture supernatant was then lyophilized in a reaction vial to give a completely dry pellet containing approximately 1 mg of the Cel7B protein. An amount of 150 μl of precooled (2 to 8°C) trifluoromethanesulfonic acid was added to the reaction vial with the lyophilized pellet. The vial was sealed with a cap, and the pellet was dissolved with gentle shaking of the vial for 5 min. The vial was then incubated on ice for 25 min with occasional shaking. Four microliters of 0.2% bromophenol blue solution was added to the vial. The vial was then cooled to −20°C in a methanol-dry ice bath, and 150 μl of precooled (−20°C) 60% pyridine solution was added dropwise to the vial. To dissolve the precipitate formed, 20 μl of dH2O was added. An additional 150 μl of 60% pyridine solution was finally added. The deglycosylated protein was then desalted through dialysis (3.5-kDa cutoff) and analyzed by SDS-PAGE.
Chemical analyses.
The monosaccharide content in the bagasse prehydrolysate, the bagasse and spruce hydrolysates, the hydrolysates after fermentation with S. cerevisiae, the SBH and SSH, and the SBH and SSH after fermentation with A. niger was analyzed by anion exchange chromatography using a DX 500 system (Dionex, Sunnyvale, CA) equipped with a CarboPac PA-1 column (Dionex). The content of acetic acid was quantified by using a Dionex ICS-2000 chromatography system equipped with a conductivity detector. Separation was performed on an IonPac AS 15 (250 × 4 mm) column with an IonPac AG15 (50 × 4 mm) precolumn (Dionex). The total concentration of phenolic compounds was determined by using the Folin-Ciocalteu method (30) with vanillin as the standard. The concentrations of furfural and HMF were determined by high-pressure liquid chromatography. An XTerra MS C18 column (5 μm, 2.1 × 150 mm) (Waters, Milford, MA) was used in a Shimadzu VP series system (Shimadzu, Kyoto, Japan) with UV detection at 282 nm. Elution was conducted according to the description in Martin et al. (20). The vanillin and coniferyl aldehyde concentrations during fermentation experiment C were measured with high-pressure liquid chromatography. The XTerra MS C18 column was used in the Shimadzu VP series system with UV detection at 254 nm. Elution was performed at a flow rate of 0.4 ml/min with a gradient made of Milli-Q water and acetonitrile, both of which contained 3 mM formic acid. The gradient design consisted of four steps with a total time of 40 min: (i) 25% acetonitrile was applied for 6 min, (ii) the concentration of acetonitrile was increased linearly to 85% during 4 min, (iii) 85% acetonitrile was applied for 12 min, and (iv) the concentration of acetonitrile was instantly decreased to 25% and applied for 18 min.
RESULTS
The monosaccharide and inhibitor content in the bagasse prehydrolysate, the bagasse and spruce hydrolysates, the hydrolysates after fermentation with S. cerevisiae, the SBH and the SSH, and the SBH and SSH after fermentation with A. niger are shown in Table 1. The results show that S. cerevisiae, as expected, consumed glucose and mannose. Xylose, arabinose, and galactose were left in the medium. The contents of acetic acid and total phenolic compounds did not change much, while furan aldehydes were partially converted.
TABLE 1.
Concentrations of monosaccharides and inhibitors
| Hydrolysatea | Concnb (g/liter)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose | Xylose | Arabinose | Galactose | Mannose | Acetic acid | Phenolic compounds | Furfural | HMF | |
| BPH | 1.8 | 10.1 | 0.6 | 0.5 | 0.2 | 0.8 | 1.1 | 0.2 | 0.03 |
| BH | 15.9 | 11.1 | 0.6 | 0.6 | 0.4 | 0.8 | 0.9 | 0.1 | 0.02 |
| BHAFSc | ND | 10.0 | 0.5 | 0.5 | ND | 0.9 | 0.9 | <0.01 | 0.02 |
| SBHd | ND | 9.5 | 0.5 | 0.5 | ND | 0.9 | 1.0 | <0.01 | 0.02 |
| SBHAFAe | ND | ND | ND | ND | ND | ND | 0.7 | ND | ND |
| SH | 19.6 | 6.3 | 1.6 | 2.7 | 13.3 | 2.8 | 2.8 | 0.7 | 2.2 |
| SHAFSc | ND | 5.8 | 1.5 | 2.6 | ND | 3.0 | 2.8 | ND | 0.9 |
| SSHd | ND | 5.4 | 1.5 | 2.5 | ND | 3.0 | 2.9 | ND | 0.8 |
| SSHAFAe | ND | ND | ND | ND | ND | 0.1 | 2.0 | ND | 0.04 |
BPH, bagasse prehydrolysates; BH, bagasse hydrolysate; BHAFS, bagasse hydrolysate after fermentation with Saccharomyces cerevisiae; SBHAFA, SBH after 11 days of fermentation with Aspergillus niger; SH, spruce hydrolysate; SHAFS, spruce hydrolysate after fermentation with Saccharomyces cerevisiae; SSHAFA, SSH after 9 days of fermentation with Aspergillus niger.
Average values from two to three determinations are shown. Relative standard deviations of the results: monosaccharides and acetic acid, <10%; phenolic compounds, <12%; furfural and HMF, <5%. ND, not detected.
The hydrolysate was diluted about 5% compared to the concentration of BH or SH.
The hydrolysate was concentrated <2% compared to the concentration of BHAFS or SHAFS.
The hydrolysate was diluted about 4% compared to the concentration of SBH or SSH.
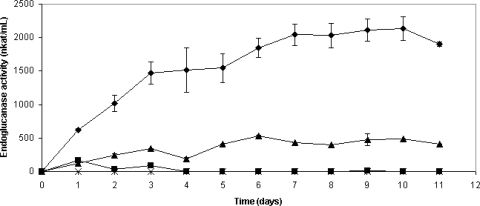
The SBH served as an excellent growth medium for the recombinant A. niger D15[egI] strain. No inhibition of the growth was noted, and all the acetic acid, furfural, and HMF, as well as about 30% of the phenolic compounds, were consumed or converted after 11 days of fermentation (Table 1). Fermentation with A. niger D15[egI] in SBH resulted in increasing endoglucanase activity for 10 days, and the highest activity monitored was 2,100 nkat/ml (Fig. 2 and Table 2). Fermentation with the same strain in standard medium resulted in an endoglucanase activity of 480 nkat/ml on day 10 (Fig. 2 and Table 2). The maximum activity, 530 nkat/ml, was observed on day 6 (Fig. 2). A. niger D15[pGT] did not give rise to endoglucanase activity whether grown on SBH or standard medium.
FIG. 2.
Endoglucanase activity during fermentation experiment A. ⧫, A. niger D15[egI] grown on SBH; ▴, A. niger D15[egI] grown on standard medium; ▪, A. niger D15[pGT] grown on SBH; ×, A. niger D15[pGT] grown on standard medium. The endoglucanase activities were calculated as the mean values of activity measurements of two separate cultures. Error bars indicate the standard deviations.
TABLE 2.
Results from fermentation series
| Fermentation expt | Strain and growth mediuma | Endoglucanase activity (nkat/ml)b | Cel7B protein concn (mg/ml)c | Endoglucanase activity/Cel7B protein (nkat/mg) | Biomass (DW g/liter)d |
|---|---|---|---|---|---|
| Ae | A. niger D15[egI] grown on SBH | 2,100 ± 130 | 0.41 ± 0.06 | 5,100 | 9.8 ± 1.6 |
| A. niger D15[pGT] grown on SBH | ND | ND | 6.8 ± 1.1 | ||
| A. niger D15[egI] grown on SM | 480 ± 22 | 0.11 ± 0.02 | 4,400 | 3.4 ± 0.3 | |
| A. niger D15[pGT] grown on SM | ND | ND | 4.1 ± 0.5 | ||
| Bf | A. niger D15[egI] grown on SSH | 820 ± 51 | 0.10 ± 0.01 | 7,500 | 4.5 ± 0.0 |
| A. niger D15[egI] grown on SM with xylose | 560 ± 16 | 0.06 ± 0.01 | 9,300 | 2.4 ± 0.2 | |
| A. niger D15[egI] grown on SM | 550 ± 52 | 0.11 ± 0.00 | 5,000 | 3.2 ± 0.1 | |
| Cg | A. niger D15[egI] grown on SM + acetic acid | 1,200 ± 43 | 0.10 ± 0.01 | 12,000 | 2.9 ± 0.4 |
| A. niger D15[egI] grown on SM + furfural | 340 ± 33 | 0.12 ± 0.00 | 2,800 | 3.5 ± 0.0 | |
| A. niger D15[egI] grown on SM + HMF | 500 ± 20 | 0.11 ± 0.00 | 4,500 | 5.2 ± 0.3 | |
| A. niger D15[egI] grown on SM + vanillin | 840 ± 79 | 0.15 ± 0.00 | 5,600 | 4.0 ± 0.1 | |
| A. niger D15[egI] grown on SM + coniferyl aldehyde | 670 ± 2 | 0.13 ± 0.00 | 5,200 | 4.1 ± 0.1 | |
| A. niger D15[egI] grown on SM | 590 ± 4 | 0.11 ± 0.01 | 5,400 | 4.4 ± 0.3 |
SM, standard medium.
The endoglucanase activity is calculated as the mean value ± standard deviation of the activity measurements of two separate cultures. ND, not detected.
The protein concentration is calculated as the mean value ± standard deviation for two supernatants from separate fermentation flasks repeatedly analyzed on four SDS gels. ND, not detected.
The biomass is calculated as the mean value ± standard deviation for two separate cultures made after 11, 10, and 9 days of cultivation for fermentation experiments A, B, and C, respectively.
Results from fermentation experiment A after 10 days of fermentation.
Results from fermentation experiment B after 9 days of fermentation.
Results from fermentation experiment C after 7 days of fermentation.
The volumetric activity, Cel7B protein concentration, specific Cel7B activity, and biomass production in fermentation experiments A, B, and C are presented in Table 2. A. niger D15[egI] grown on SBH reached a biomass concentration of 9.8 g/liter (DW), whereas A. niger D15[pGT] grown on SBH only reached a biomass of 6.8 g/liter (DW). When grown on standard medium, both strains reached a biomass of approximately 3 to 4 g/liter (DW). A. niger D15[egI] grown on SBH gave a Cel7B protein concentration of 0.41 mg/ml after 10 days of fermentation, which is almost four times higher than that of the same strain grown on standard medium.
The SSH also served as a good growth medium for A. niger D15[egI]. No inhibition was noted, and essentially all the acetic acid and HMF and about 30% of the phenolic compounds were consumed or converted after 9 days of fermentation (Table 1). The highest activity was noted after 9 days of cultivation and reached 820 nkat/ml (Table 2), while the same strain reached 550 nkat/ml (day 9) when grown on standard medium. The endoglucanase activity of A. niger D15[egI] grown on the standard medium with xylose instead of glucose followed that of the cultivation in ordinary standard medium, and the highest activity noted was 560 nkat/ml (day 9). Growth on the SSH, the standard medium, and the standard medium with xylose reached biomass concentrations of 4.5, 3.2, and 2.4 g/liter (DW), respectively. The Cel7B protein concentrations were 0.10 and 0.11 mg/ml for the fermentation in SSH and the fermentation in standard medium, respectively. The protein production of A. niger D15[egI] grown on standard medium with xylose was considerably lower, and it reached a Cel7B concentration of 0.06 mg/ml.
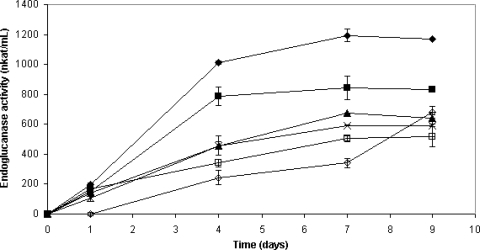
A. niger D15[egI] also grew well in all the inhibitor-containing standard media, except for the one that had furfural added, where a lag phase in the growth and enzyme activity was observed (Fig. 3). All the vanillin, acetic acid, coniferyl aldehyde, HMF, and furfural were consumed or converted within 9 days of fermentation. The fermentation in the medium with acetic acid added showed the highest endoglucanase activity and reached 1,200 nkat/ml after 7 days, whereas the activity of the reference cultivation in the medium without inhibitors reached 590 nkat/ml (day 7) (Table 2). The fermentations with vanillin and coniferyl aldehyde added also showed higher or slightly higher enzyme activities than the reference fermentation, while the HMF- and furfural-containing fermentations exhibited lower enzyme activities throughout most of the fermentation experiment. Fermentation in the presence of acetic acid showed the highest specific activity and reached 12,000 nkat/mg, which is about twice as high as that of the reference fermentation and about four times higher than that of the fermentation in the presence of furfural. The highest biomass concentration was observed for the cultivation where HMF was added (5.2 g/liter [DW]), while the cultivation with acetic acid added had the lowest biomass formation (2.9 g/liter [DW]). The reference fermentation had a biomass of 4.4 g/liter (DW). The Cel7B protein production was in the range of 0.10 to 0.15 mg/ml for all fermentations.
FIG. 3.
Endoglucanase activity during fermentation experiment C. ⧫, A. niger D15[egI] grown on standard medium containing 5 g/liter acetic acid; ▪, A. niger D15[egI] grown on standard medium containing 0.5 g/liter vanillin; ▴, A. niger D15[egI] grown on standard medium containing 0.2 g/liter coniferyl aldehyde; ×, A. niger D15[egI] grown on standard medium; □, A. niger D15[egI] grown on standard medium containing 2 g/liter HMF; ⋄, A. niger D15[egI] grown on standard medium containing 1 g/liter furfural. The endoglucanase activities were calculated as the mean values of the activity measurements of two separate cultures. Error bars indicate the standard deviations.
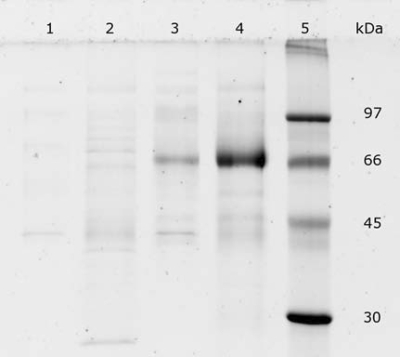
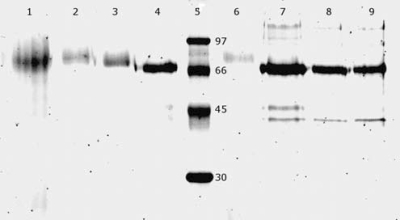
The SDS-PAGE analyses showed that the Cel7B protein was predominant in the supernatants of fermentation experiments A, B, and C. The recombinant Cel7B protein from A. niger D15[egI] grown on SBH and standard medium had a molecular mass of about 72 kDa (Fig. 4). However, the theoretical molecular mass of unglycosylated mature Cel7B is 46 kDa (ExPASy; http://www.expasy.org/uniprot/P07981) and native Cel7B from H. jecorina is only about 50 to 55 kDa (5, 24, 28, 38). Therefore, the recombinant Cel7B could be hyperglycosylated. A variation in molecular mass and heterogeneity of the Cel7B protein was seen during fermentation experiment A (Fig. 5). The Cel7B from the fermentation of SBH shifted gradually from large and heterogeneous in the beginning of the fermentation to a smaller and more homogeneous protein at the end of the fermentation. A shift in the molecular mass from larger to smaller was also observed for the Cel7B from the fermentation in standard medium (Fig. 5). However, the molecular mass was already stabilized after a few days of fermentation (Fig. 5). The molecular mass of Cel7B from fermentation experiment B was 72 kDa when the strain was grown on standard medium, 74 kDa when it was grown on SSH, and 76 kDa when it was grown on standard medium with xylose (Fig. 6). Fermentation experiment C gave Cel7B with a molecular mass of 72 kDa, with the exception of the cultures with added acetic acid, which rendered Cel7B with a molecular mass of 78 kDa (Fig. 6).
FIG. 4.
SDS-PAGE analysis of supernatants (diluted four times) from fermentation experiment A after 10 days of fermentation. Lanes: 1, supernatant from A. niger D15[pGT] grown on standard medium; 2, supernatant from A. niger D15[pGT] grow on SBH; 3, supernatant from A. niger D15[egI] grown on standard medium; 4, supernatant from A. niger D15[egI] grown on SBH; 5, molecular mass marker.
FIG. 5.
SDS-PAGE analysis showing the Cel7B protein profiles during fermentation in SBH and standard medium. Lanes: 1, supernatant (diluted two times) from A. niger D15[egI] grown on SBH for 1 day; 2, supernatant (diluted five times) from A. niger D15[egI] grown on SBH for 4 days; 3, supernatant (diluted eight times) from A. niger D15[egI] grown on SBH for 7 days; 4, supernatant (diluted eight times) from A. niger D15[egI] grown on SBH for 11 days; 5, molecular mass marker; 6, supernatant (undiluted) from A. niger D15[egI] grown on standard medium for 1 day; 7, supernatant (undiluted) from A. niger D15[egI] grown on standard medium for 4 days; 8, supernatant (diluted two times) from A. niger D15[egI] grown on standard medium for 7 days; 9, supernatant (diluted two times) from A. niger D15[egI] grown on standard medium for 11 days.
FIG. 6.
SDS-PAGE analysis showing variation in molecular mass of Cel7B depending on growth medium. Lanes: 1, supernatant from A. niger D15[egI] grown on standard medium with acetic acid for 7 days; 2, supernatant from A. niger D15[egI] grown on standard medium with xylose for 9 days; 3, supernatant from A. niger D15[egI] grown on SSH for 9 days; 4, supernatant from A. niger D15[egI] grown on standard medium for 7 days; 5, molecular mass marker.
The identity of the protein in two of the SDS-PAGE bands was verified by using MS. The two protein samples were from the cultivations on SBH and on standard medium with acetic acid. The molecular masses of these proteins, as judged by SDS-PAGE, were 72 and 78 kDa, respectively. As expected, the results of both MALDI-TOF peptide mass fingerprinting and MS-MS sequencing identified both protein samples as H. jecorina Cel7B. The analysis of the protein sample from the cultivation on SBH resulted in a sequence coverage of 15% and a Mascot score of 241. The analysis of the protein sample from the cultivation on acetic acid-enriched medium resulted in a sequence coverage of 17% and a Mascot score of 117.
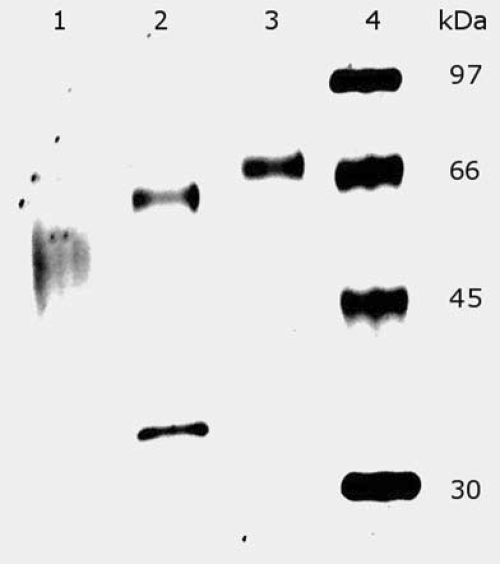
Treatment of the Cel7B protein with N-glucosidase F resulted in only moderate deglycosylation and gave a protein with a molecular mass of 65 kDa (Fig. 7, lane 2). A second protein band with a molecular mass of 35 kDa was also noted on the gel. This band is the N-glucosidase F protein used in the deglycosylation treatment. The chemical deglycosylation resulted in a heterogeneous protein band with a molecular mass ranging from 48 to 56 kDa (Fig. 7, lane 1). The degree of protein glycosylation had only a minor effect on the Sypro Red dye binding. The deglycosylated Cel7B from the cultivation on SBH displayed an apparent protein concentration that was 5% higher than that of a nondeglycosylated sample containing the same amount of protein. Since the molecular mass of the recombinant Cel7B differs relatively little (72 to 78 kDa), this result indicates that the values for specific activity are only slightly influenced by the effect of different degrees of glycosylation on the Sypro Red dye assay.
FIG. 7.
SDS-PAGE analysis of Cel7B protein from A. niger D15[egI] grown on SBH before and after different deglycosylation treatments. Lanes: 1, Cel7B protein treated with chemical deglycosylation; 2, Cel7B protein treated with N-glucosidase F; 3, untreated Cel7B protein; 4, molecular mass marker.
DISCUSSION
Cellulases can be produced by using prehydrolysates and hydrolysates of bagasse and wood as growth media for filamentous fungi, such as H. jecorina (e.g., see references 6, 12, 16, and 22). However, prehydrolysates and hydrolysates contain hexose sugars that instead could be utilized for ethanolic fermentation. Large amounts of hexose sugars, like glucose, mannose, and galactose, can be generated, especially if softwood is the raw material. For example, 16% (DW) of Norway spruce (Picea abies) consists of the polysaccharides mannan and galactan (31).
The suitability of the SBH as a growth medium for the Cel7B-expressing A. niger is clearly reflected by its high biomass production, heterologous protein production, and endoglucanase activity (Table 2). However, the SBH contained only about 10 g/liter of monosaccharides and xylose constituted nearly all of this amount (Table 1). The reference fermentation with standard medium with a glucose content of 10 g/liter resulted in considerably lower biomass production, protein production, and endoglucanase activity. Nevertheless, the activity/mg of Cel7B protein did not differ much between the reference and the SBH fermentation. The hypothesis that A. niger D15[egI] had a preference for xylose over glucose was not supported by the results from fermentation experiment B, where a standard medium with xylose and one with glucose were compared. The higher biomass production of A. niger D15[egI] than of A. niger D15[pGT] when grown on SBH suggests that the Cel7B produced facilitated additional liberation of nutrients from the SBH that was utilized by the strain.
A disadvantage of using H. jecorina as a cellulase producer is that its cellulase synthesis is regulated through induction by cellulose-derived compounds and repressed by glucose (14, 18). If the cellulase genes instead, as in our investigation, are under the control of constitutive promoters, there is no need for complex growth media with unique inducers.
It is possible that the expression of native cellulase genes in A. niger could be induced by various components in the SBH and SSH, which would result in higher values in the activity assay. However, A. niger D15[pGT] did not give rise to any endoglucanase activity when grown on SBH. Different cellulases work synergistically, and in order to be able to see the full effect of native exoglucanases of the strains, endoglucanases must be available to produce cellulose chain ends that the exoglucanases are able to attack. Nevertheless, it is likely that SBH contains substances other than monosaccharides that A. niger can utilize as nutrients, possibly oligosaccharides.
Fermentation in the presence of the phenolic compound vanillin had a positive effect on protein production which was reflected in the enzyme activity. Fermentation in the presence of acetic acid gave the highest values for both volumetric and specific enzyme activity in fermentation experiment C. However, the inclusion of acetic acid resulted in lower final biomass and Cel7B protein concentrations. A previous study of A. niger has shown a reduction in biomass formation upon the addition of acetic acid to the growth medium (37). Szengyel and Zacchi (36) investigated the effects of acetic acid and furfural and combinations of the two on the cellulase production of the filamentous fungus H. jecorina RUT C30. They concluded that furfural inhibits the enzyme production and that the addition of acetic acid increased the production of β-glucosidase but had no effect on cellulase production. They also stated that the average pH of the fermentation plays an important role in the enzyme production of H. jecorina RUT C30 (36).
One possible explanation for the high volumetric and specific activity of Cel7B in the fermentation with standard medium with acetic acid could be that the acetic acid might affect the protein glycosylation. The SDS-PAGE analysis of the culture supernatants showed that Cel7B from the cultivation with added acetic acid had a higher molecular mass (about 78 kDa) (Fig. 6) than Cel7B from the other cultivations (about 72 kDa) in fermentation experiment C. A similar explanation might also apply for the high specific Cel7B activity in the fermentations with SSH and standard medium with xylose, which both displayed Cel7B with higher molecular mass than that in the reference cultivation (Fig. 6). The SSH also contained quite high levels of acetic acid which might have affected the glycosylation of the Cel7B protein. The low volumetric and specific activity in the fermentation with added furfural can tentatively be explained by the fact that this fermentation displayed a lag phase in the beginning and did not reach its maximum activity until day 9. However, after 9 days of fermentation, when the activity had reached its maximum, the specific Cel7B activity was 5,700 nkat/mg, which is similar to that of the reference fermentation.
Glycosylation is an important posttranslational modification process of proteins that influences the stability, conformation, secretion, and biological activity of proteins (15, 39). The Cel7B protein consists of a cellulose-binding domain (C terminal), a linker region, and a catalytic domain (N terminal) (8). There are five potential N-glycosylation sites and one proposed O-glycosylation site on the catalytic domain (7). Usually O-glycosylation is a characteristic feature of the linker region, and the linker region of Cel7B has been described to be heavily O-glycosylated (7, 13). O-linked glycosylation has been shown to be important for enzyme stabilization (21, 41) and linker conformation (26). Eriksson and coworkers concluded that the degree of heterogeneity of the catalytic domain of H. jecorina Cel7B is dependent on N-glycosylation, O-glycosylation, and deamidation (7). In addition, the glycosylation machinery of filamentous fungi may involve several different trimming enzymes, such as endoglucosidases, mannosidase, and phosphatases, that together trim the glycosylation of the enzyme (32). These different kinds of trimming enzymes have also been detected in the extracellular growth medium of filamentous fungi (19, 32) and are believed to be able to alter the glycosylation pattern. The activity of these enzymes has been shown to be very dependent on the pH of the growth medium (7, 32) and the medium's composition (32).
The presence of acetic acid in the growth medium and the fact that A. niger consumes the acetic acid may affect the pH of the growth medium during the course of fermentation, since acetic acid can act as a buffer. This could affect the activity of the different trimming enzymes and lead to different glycosylation patterns (32). Furthermore, if filamentous fungi are grown in poor media, the fungi are likely to produce acid proteases, which will affect the trimming enzymes (32). This might be the case for the fermentation with standard medium supplemented with xylose. The shifting heterogeneity and molecular mass during fermentation experiment A support the idea that active extracellular trimming enzymes are important for the final glycosylation pattern of the protein. Medium composition has been shown to affect glycosylation of Cel7A in H. jecorina, with fully glycosylated enzyme found in minimal medium but extracellularly trimmed enzyme in nutrient-enriched medium (32).
It is not entirely surprising that Cel7B from H. jecorina is differently glycosylated when it is expressed in A. niger. It might be expected that the enzymes responsible for glycosylation and deglycosylation in A. niger would differ from those of H. jecorina. The expression of H. jecorina Cel7B in other organisms, such as S. cerevisiae (38) and Yarrowia lipolytica (23), has also resulted in hyperglycosylation. Nevertheless, the present study shows that the composition of the growth medium is an important factor for the final glycosylation of the recombinant Cel7B protein.
The enzyme N-glucosidase F is commonly used for deglycosylation of proteins, but it can only degrade N-linked carbohydrates. The fact that exhaustive N-glucosidase F treatment only led to a fractional deglycosylation of the recombinant Cel7B suggests that a considerable proportion of the glycosyl groups of the recombinant Cel7B consists of O-linked carbohydrates. The chemical deglycosylation method generated a heterogeneous protein product. Nevertheless, the lowest molecular mass of the deglycosylated protein was about 48 kDa, which is close to the calculated molecular mass of Cel7B (46 kDa) (ExPASy; http://www.expasy.org/uniprot/P07981).
Spent lignocellulose hydrolysates can probably be used as an inexpensive growth medium for several different microorganisms producing a wide array of industrially important enzymes, such as xylanases, amylases, ligninases, and different proteases. These enzymes could potentially be produced as high-value coproducts of a biorefinery. Nevertheless, hydrolytic enzymes, such as cellulases, could advantageously be produced at an ethanol production plant because they can also be utilized on-site in the hydrolysis of the lignocellulosic feedstock. Another potential coproduct is the A. niger biomass, which could be utilized as a protein source in cattle feed (29).
In conclusion, the recombinant A. niger strains were able to grow on spent hydrolysate from both sugarcane bagasse and spruce wood. Both served as good growth media, and the fermentations displayed higher endoglucanase activity than fermentations with standard medium with equal or higher monosaccharide content. The recombinant strains were also able to consume or convert essentially all furan aldehydes and acetic acid and about 30% of the phenolic compounds found in the two spent hydrolysates.
Model fermentations in standard medium with added lignocellulose-derived compounds that inhibit yeast confirmed that these types of compounds can be consumed or converted by A. niger. Some of these compounds (e.g., acetic acid) can presumably affect the protein glycosylation. Others can possibly also be utilized as nutrients by A. niger. The recombinant Cel7B protein was hyperglycosylated, with O-linked carbohydrates making up the major part of the attached carbohydrates. The study shows that spent hydrolysates can be used as growth media for the production of cellulases by A. niger, which simultaneously can reduce the content of inhibitory substances in the stillage stream.
Acknowledgments
This study was supported by Carl Fredrik von Horn's Foundation and the South African-Swedish Research Collaboration Programme of Sida/SAREC and NRF.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Alkasrawi, M., M. Galbe, and G. Zacchi. 2002. Recirculation of process streams in fuel ethanol production from softwood based on simultaneous saccharification and fermentation. Appl. Biochem. Biotechnol. 98:849-861. [DOI] [PubMed] [Google Scholar]
- 2.Alriksson, B., A. Sjöde, N.-O. Nilvebrant, and L. J. Jönsson. 2006. Optimal conditions for alkaline detoxification of dilute-acid lignocellulose hydrolysates. Appl. Biochem. Biotechnol. 130:599-611. [DOI] [PubMed] [Google Scholar]
- 3.Aristidou, A., and M. Penttilä. 2000. Metabolic engineering applications to renewable resource utilization. Curr. Opin. Biotechnol. 11:187-198. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, M. J., P. Biely, and K. Poutanen. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257-270. [Google Scholar]
- 5.Bhikhabai, R., G. Johansson, and G. Petterson. 1984. Isolation of cellulolytic enzymes from Trichoderma reesei QM9414. J. Appl. Biochem. 6:336-345. [PubMed] [Google Scholar]
- 6.Bigelow, M., and C. E. Wyman. 2002. Cellulase production on bagasse pretreated with hot water. Appl. Biochem. Biotechnol. 98:921-934. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, T., I. Stals, A. Collén, F. Tjerneld, M. Claeyssens, H. Stålbrand, and H. Brumer. 2004. Heterogeneity of homologously expressed Hypocrea jecorina (Trichoderma reesei) Cel7B catalytic module. Eur. J. Biochem. 271:1266-1276. [DOI] [PubMed] [Google Scholar]
- 8.García, R., J. A. Cremata, O. Quintero, R. Montesino, K. Benkestock, and J. Ståhlberg. 2001. Characterization of protein glycoforms with N-linked neutral and phosphorylated oligosaccharides: studies on the glycosylation of endoglucanase I (Cel7B) from Trichoderma reesei. Biotechnol. Appl. Biochem. 33:141-152. [DOI] [PubMed] [Google Scholar]
- 9.Hahn-Hägerdal, B., M. Galbe, G. Gorwa-Grauslund, G. Lidén, and G. Zacchi. 2006. Bio-ethanol: the fuel of tomorrow from the residues of today. Trends Biotechnol. 24:549-556. [DOI] [PubMed] [Google Scholar]
- 10.Hahn-Hägerdal, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jönsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 11.Hammerschlag, R. 2006. Ethanol's energy return on the investment: a survey of the literature 1990-present. Environ. Sci. Technol. 40:1744-1750. [DOI] [PubMed] [Google Scholar]
- 12.Hayward, T. K., J. Hamilton, D. Templeton, E. Jennings, M. Ruth, A. Tholudur, J. D. McMillan, M. Tucker, and A. Mohagheghi. 1999. Enzyme production, growth and adaptation of T. reesei strains QM9414, L-27, RL-P37, and rut C-30 to conditioned yellow poplar sawdust hydrolysate. Appl. Biochem. Biotechnol. 77:293-309. [Google Scholar]
- 13.Hui, J. P. M., T. C. White, and P. Thibault. 2002. Identification of glycan structure and glycosylation sites in cellobiohydrolase II and endoglucanase I and II from Trichoderma reesei. Glycobiology 12:837-849. [DOI] [PubMed] [Google Scholar]
- 14.Kubicek, C. P., R. Messner, F. Gruber, R. L. Mach, and E. M. Kubicek-Pranz. 1993. The Trichoderma cellulose regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microb. Technol. 15:90-99. [DOI] [PubMed] [Google Scholar]
- 15.Lis, H., and N. Sharon. 1993. Protein glycosylation: structural and functional aspects. Eur. J. Biochem. 218:1-27. [DOI] [PubMed] [Google Scholar]
- 16.Lo, C.-M., Q. Zhang, P. Lee, and L.-K. Ju. 2005. Cellulase production by Trichoderma reesei using sawdust hydrolysate. Appl. Biochem. Biotechnol. 122:561-573. [PubMed] [Google Scholar]
- 17.Lynd, L. R., M. S. Laser, D. Bransby, B. E. Dale, B. Davison, R. Hamilton, M. Himmel, M. Keller, J. D. McMillan, J. Sheehan, and C. E. Wyman. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169-172. [DOI] [PubMed] [Google Scholar]
- 18.Mandels, M., and E. T. Reese. 1960. Induction of cellulase in fungi by cellobiose. J. Bacteriol. 79:816-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maras, M., I. Van Die, R. Contreras, and C. A. M. J. J. van den Hondel. 1999. Filamentous fungi as production organism for glycoproteins of bio-medical interest. Glycoconjugate J. 16:99-107. [DOI] [PubMed] [Google Scholar]
- 20.Martín, C., B. Alriksson, A. Sjöde, N.-O. Nilvebrant, and L. J. Jönsson. 2007. Dilute-sulphuric acid prehydrolysis of agricultural and agro-industrial residues for ethanol production. Appl. Biochem. Biotechnol. 137-140:339-352. [DOI] [PubMed] [Google Scholar]
- 21.Neustroev, K. N., A. M. Golubev, L. M. Firsov, F. M. Ibatullin, I. Protasevich, and A. Makarov. 1993. Effect of modification of carbohydrate component on properties of glucoamylase. FEBS Lett. 2:157-160. [DOI] [PubMed] [Google Scholar]
- 22.Palmqvist, E., B. Hahn-Hägerdahl, Z. Szengyel, G. Zacchi, and K. Rèczey. 1997. Simultaneous detoxification and enzyme production of hemicellulose hydrolysates obtained after steam pretreatment. Enzyme Microb. Technol. 20:286-293. [Google Scholar]
- 23.Park, C. S., C. C. Chang, and D. D. Y. Ryu. 2000. Expression and high-level secretion of Trichoderma reesei endoglucanase I in Yarrowia lipolytica. Appl. Biochem. Biotechnol. 87:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Penttilä, M. E., L. André, M. Saloheimo, P. Lehtovaara, and J. K. Knowles. 1987. Expression of two Trichoderma reesei endoglucanases in the yeast Saccharomyces cerevisiae. Yeast 3:175-185. [DOI] [PubMed] [Google Scholar]
- 25.Percival Zhang, Y.-H., M. E. Himmel, and J. R. Mielenz. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24:452-481. [DOI] [PubMed] [Google Scholar]
- 26.Receveur, V., M. Czjzek, M. Schülein, P. Panine, and B. Henrissat. 2002. Dimension, shape, and conformational flexibility of a two domain fungal cellulase in solution probed by small angle X-ray scattering. J. Biol. Chem. 277:40887-40892. [DOI] [PubMed] [Google Scholar]
- 27.Rose, S. H., and W. H. van Zyl. 2002. Constitutive expression of the Trichoderma reesei β-1,4-xylanase gene (xyn2) and the β-1,4-endoglucanase gene (egI) in Aspergillus niger in molasses and defined glucose media. Appl. Microbiol. Biotechnol. 58:461-468. [DOI] [PubMed] [Google Scholar]
- 28.Shoemaker, S., K. Watt, G. Tsitovsky, and R. Cox. 1983. Characterization and properties of cellulases purified from Trichoderma reesei strain L27. Bio/Technology 1:687-690. [Google Scholar]
- 29.Singh, A., A. B. Abidi, A. K. Agrawai, and N. S. Darmwal. 1991. Single cell protein production by Aspergillus niger and its evaluation. Zentralbl. Mikrobiol. 146:181-184. [PubMed] [Google Scholar]
- 30.Singleton, V. L., R. Orhofer, and R. M. Lamuela-Raventos. 1999. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteau reagent. Methods Enzymol. 299:152-178. [Google Scholar]
- 31.Sjöström, E. 1993. Wood chemistry: fundamentals and applications. Academic Press, New York, NY.
- 32.Stals, I., K. Sandra, S. Geysens, R. Contreras, J. van Beeumen, and M. Claeyssens. 2004. Factors influencing glycosylation of Trichoderma reesei cellulases. I. Postsecretorial changes of the O- and N-glycosylation pattern of Cel7A. Glycobiology 14:713-724. [DOI] [PubMed] [Google Scholar]
- 33.Steinberg, T. H., R. P. Haugland, and V. L. Singer. 1996. Application of SYPRO orange and SYPRO red protein gel stains. Anal. Biochem. 239:238-245. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg, T. H., L. J. Jones, R. P. Haugland, and V. L. Singer. 1996. SYPRO orange and SYPRO red protein gel stains: one step fluorescent staining of denaturing gels for detection of nanogram levels of protein. Anal. Biochem. 239:223-237. [DOI] [PubMed] [Google Scholar]
- 35.Stenberg, K., C. Tengborg, M. Galbe, G. Zacchi, E. Palmqvist, and B. Hahn-Hägerdal. 1998. Recycling of process streams in ethanol production from softwoods based on enzymatic hydrolysis. Appl. Biochem. Biotechnol. 70-72:697-708. [DOI] [PubMed] [Google Scholar]
- 36.Szengyel, Z., and G. Zacchi. 2000. Effect of acetic acid and furfural on cellulose production of Trichoderma reesei RUT C30. Appl. Biochem. Biotechnol. 89:31-42. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi, K., K. Matsuda, H. Teramura, and K. Wake. 1977. Effect of acetic acid on growth of Aspergillus niger. J. Fac. Sci. Hokkaido Univ. Ser. V (Botany) 10:189-198. [Google Scholar]
- 38.van Arsdell, J. N., S. Kwok, V. L. Schweickart, M. B. Ladner, D. H. Gelfand, and M. A. Innis. 1987. Cloning, characterization, and expression in Saccharomyces cerevisiae of endoglucanase I from Trichoderma reesei. Bio/Technology 5:60-64. [Google Scholar]
- 39.Varki, A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3:97-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Sivers, M., G. Zacchi, L. Olsson, and B. Hahn-Hägerdahl. 1994. Cost analysis of ethanol production from willow using recombinant Escherichia coli. Biotechnol. Prog. 10:555-560. [DOI] [PubMed] [Google Scholar]
- 41.Williamson, G., N. J. Belshaw, and M. P. Williamson. 1992. O-glycosylation in Aspergillus glucoamylase. Conformation and role in binding. Biochem. J. 282:423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]