Abstract
Fibronectin (FN) forms the primitive fibrillar matrix in both embryos and healing wounds. To study the matrix in living cell cultures, we have constructed a cell line that secretes FN molecules chimeric with green fluorescent protein. These FN–green fluorescent protein molecules were assembled into a typical matrix that was easily visualized by fluorescence over periods of several hours. FN fibrils remained mostly straight, and they were seen to extend and contract to accommodate movements of the cells, indicating that they are elastic. When fibrils were broken or detached from cells, they contracted to less than one-fourth of their extended length, demonstrating that they are highly stretched in the living culture. Previous work from other laboratories has suggested that cryptic sites for FN assembly may be exposed by tension on FN. Our results show directly that FN matrix fibrils are not only under tension but are also highly stretched. This stretched state of FN is an obvious candidate for exposing the cryptic assembly sites.
Assembly of the fibronectin (FN) matrix has been studied most extensively in cell cultures, in which a network of extended fibrils is demonstrated by antibody staining. The matrix consists of interconnected fibrils up to 1 μm or more in diameter. Electron microscopy shows that these fibrils are bundles of thinner filaments, ≈5 nm in diameter, and that the fibrils can vary from ≈10 nm in diameter (and contain only a few filaments) to 100–1,000 nm in diameter (and contain many parallel filaments) (1, 2). The 5-nm diameter of the thin filaments is close to the ≈3-nm diameter of individual FN molecules (3), but the exact molecular arrangement of molecules within filaments and fibrils is not known.
Visualizing the FN matrix by immunofluorescence requires fixation of the cultures and does not reveal dynamics of a living culture. Green fluorescent protein (gfp) has been used as a tag to localize many intracellular proteins in living cells. Visualization of the cytoskeleton has been particularly dramatic, and localization of proteins to the nucleus or specialized membranous compartments has had many applications. Surprisingly, we were unable to find any references using gfp to localize extracellular matrix proteins. It seemed a useful approach and feasible, and, indeed, a recent study reported localization of the protein SPARC-gfp in Caenorhabditis elegans (4). This study and our localization of FN–gfp reported below suggest that gfp should be generally useful to localize extracellular matrix molecules.
To visualize the matrix in living cultures, we have made chimeras of FN and gfp. An eventual goal is to follow the assembly of the matrix, starting with freshly plated cells. In preliminary observations of more established matrices, we observed surprising movements of the FN–matrix fibrils that suggest an elasticity never before demonstrated. We report here the design of the successful FN–gfp chimera and the observations of matrix fibril elasticity.
MATERIALS AND METHODS
Construction of Expression Vector.
The vector for transfecting cells to secrete FN (pAIPFN) was kindly provided by Kiyotoshi Sekiguchi, Osaka Medical Center (5). Site-directed mutagenesis was performed to create a NotI restriction enzyme site between FN-III domains 3 and 4 of the FN. Cycle 3 mutant gfp DNA (ref. 6; Affimax, Palo Alto, CA) was amplified by PCR, adding a NotI restriction site at each end. The gfp was then ligated into the NotI site of the FN vector, and clones were selected with the correct orientation. The sequence at the insertion site was (FN-III 3) . . . TTGT-GGRMASK . . . (gfp) . . . ELYKGGR-PRSD . . . (FN-III 4) [the gfp sequence is underlined; the NotI restriction site added three extra amino acids (GGR) at each end of the gfp]. The pAIPFN-gfp vector and the pEE 14 vector for selection (7) were cotransfected into Chinese hamster ovary (CHO) cells and selected for resistance to l-methionine sulfoximine. High expressing clones were first identified by screening media for secreted FN and then by checking for assembly of an FN matrix. Not all high secreting clones assembled a matrix, but one clone was identified that assembled a substantial FN matrix and was used for these experiments. In addition to FN matrix assembly, this clone secreted substantial quantities of FN into the medium. FN–gfp (6–8 mg/liter) was obtained by gelatin affinity purification from 5-day conditioned medium of confluent cultures.
Microscopy.
Cell suspensions (100 μl; 5 × 105 cells per ml) were dropped onto 25-mm circular coverslips, incubated for 30–60 min at 37°C to allow the cells to settle, and then covered with 2 ml of medium [phenol red-free OPTI-MEM (GIBCO/BRL], including 1% FN–free fetal calf serum). After the cells were cultured for 15–36 h to allow assembly of an FN matrix, coverslips were mounted in a windowed chamber with 1 ml of medium to allow long-term observation. The living culture was observed with a Zeiss LSM410 microscope at 37°C (see Figs. 1–3 and 5) or a Bio-Rad MRC600 confocal scanning microscope at room temperature (see Fig. 4). Laser beam radiation was performed with a Laser Science (Cambridge, MA) LSI (VSL-337ND).
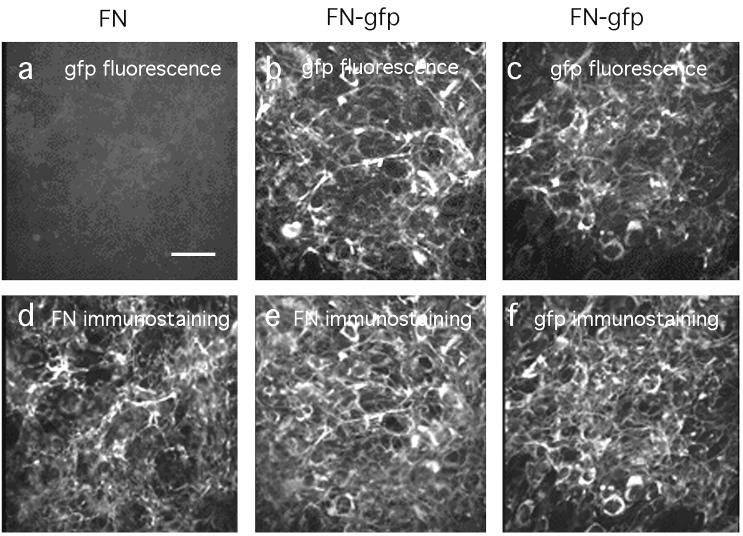
Figure 1.
Identical FN matrices are assembled by CHO cells transfected with native plasma FN (a and d) or with FN–gfp (b, c, e, and f). Cells were cultured 36 h to allow matrix assembly and were fixed with 3.7% formaldehyde in PBS, treated with 0.2% Triton X-100, and then immunostained using polyclonal antibodies for FN (d and e) or gfp (f). (a–c) gfp fluorescence is visualized; (d–f) the rhodamine-labeled second antibody is visualized. (Bar = 50 μm.)
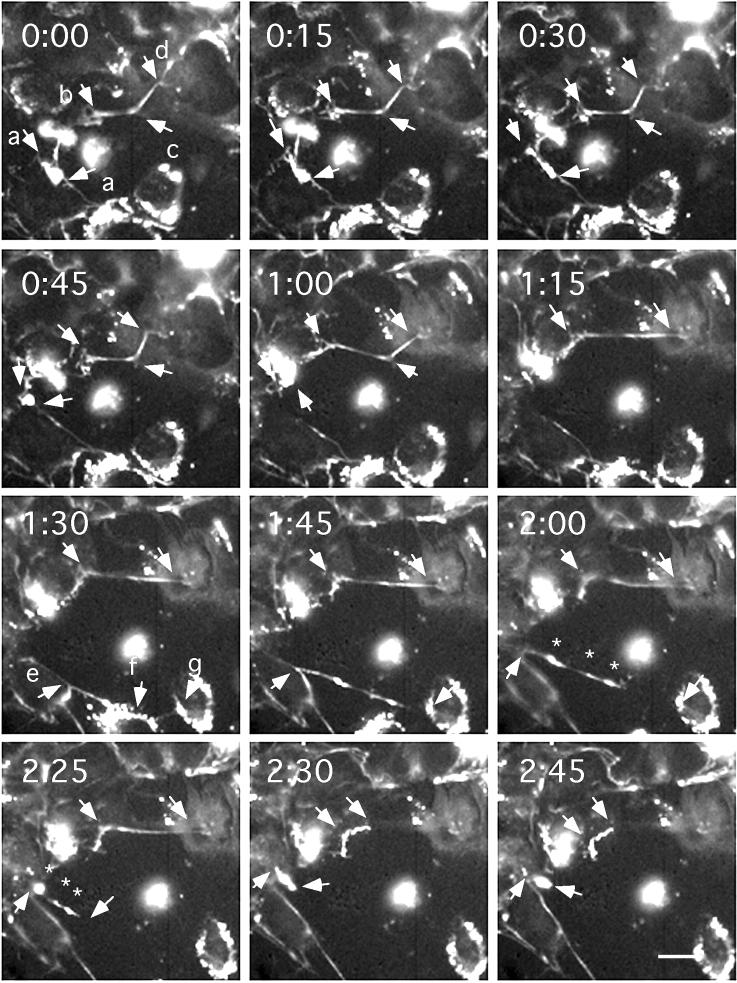
Figure 3.
The elasticity of FN matrix fibrils demonstrated by exceptionally large movements and breakages. Specific movements are discussed in the text. Images were originally taken at 5-min intervals after 15 h of culture. Arrows indicate fibril ends or predicted cell adhesion sites. (Bar = 20 μm.) A digital movie of this sequence is published as supplemental material on the PNAS web site (www.pnas.org).
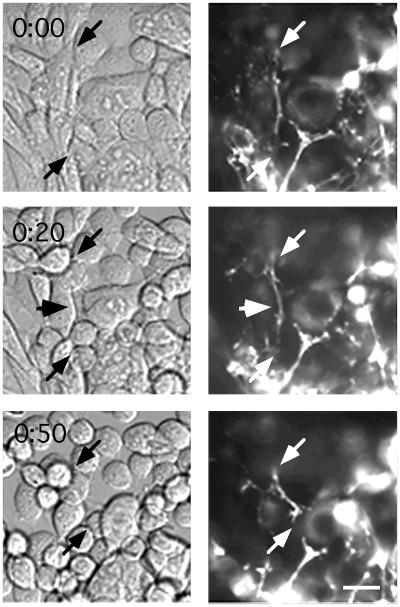
Figure 5.
Movement of cells and FN fibrils after EDTA treatment. A cell culture is imaged by Nomarski optics (Left) and gfp fluorescence (Right) before and 20 and 50 min after adding 5 mM EDTA. At 0 min, the fibril indicated by arrows runs near, but not precisely along, some cell edges. At 20 min, the fibril runs closely along the cell edge at the arrowhead, but its other attachments are not clear. At 50 min that cell has rounded; the fibril still contacts the cell at one point, but it maintains a straight path rather than following the rounded edge of the cell. The bottom right end of the fibril has moved about 20 μm from 20 to 50 min, but what it is attached to is not clear. (Bar = 20 μm.)
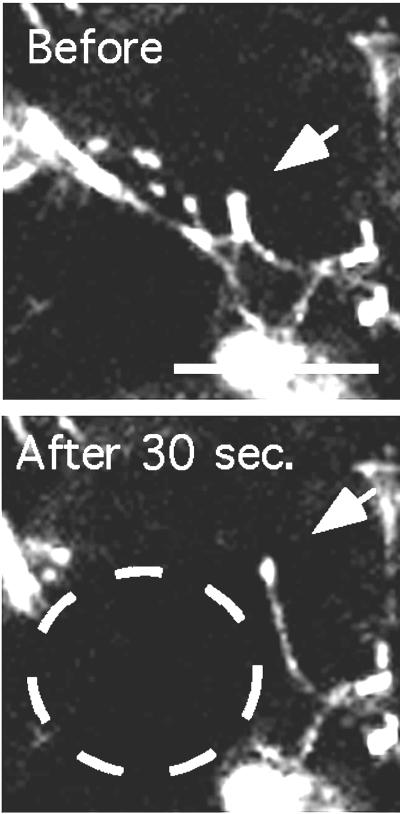
Figure 4.
The mobility of an FN matrix fibril after laser beam radiation. The spot burned by the laser is indicated by the white circle. (Bar = 20 μm.)
RESULTS AND DISCUSSION
We made and tested chimeras with gfp located at three different places on the FN molecule. When the gfp was placed at the N terminus or the C terminus, we obtained very low levels or no secreted protein from transfected cells. In the third construct, we placed the gfp between FN-III domains 3 and 4, a region of the FN molecule that seems to be unimportant for FN matrix assembly (8, 9). The particular FN molecule coded by this vector is missing the alternatively spliced A and B domains and contains the V (III-CS) domain (5).
For transfection we chose a CHO cell line, which makes very little FN. This untransfected CHO cell line can assemble an FN matrix when grown in medium containing FN (10), but in the absence of exogenous FN, it assembles no detectable matrix (unpublished observations). We produced a CHO cell line transfected with pAIPFN that secreted native FN into the matrix and assembled an FN matrix (Fig. 1). We also produced a CHO cell line transfected with the FN–gfp construct, which secreted as much FN–gfp as cells transfected with pAIPFN, and also assembled an FN matrix. The matrix could be visualized by gfp fluorescence or by antibodies against FN or gfp (Fig. 1). The gfp fluorescence colocalized with the antibody-stained fibrils, but there were some differences in intensity. Some fibrils were relatively more intense in gfp, and others were more intense in immunofluorescence, perhaps because of variable thickness of fibrils and limited penetration of antibodies. The matrix assembled from FN–gfp was indistinguishable from the normal FN matrix.
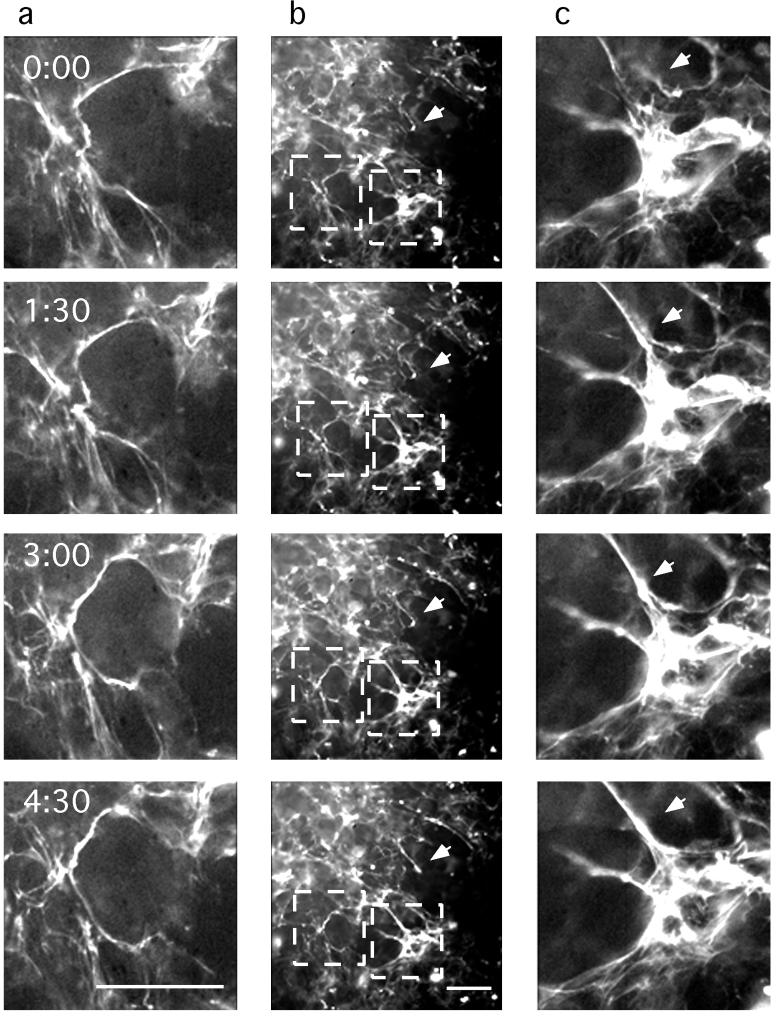
Matrix fibrils were seen predominantly on the apical cell surface, but the cell layer was only about 4 μm thick; therefore, most fibrils could have had attachments to the substrate. When observed over time, some FN–gfp matrix fibrils did not move at all, but some showed active movements both parallel and perpendicular to the substrate. We believe the stationary fibrils are attached primarily to the substrate, whereas the moving fibrils are attached at some points to cells, which are moving. Fig. 2 shows motions of a typical cell culture over 4.5 h. The brightest fibril in Fig. 2a was most extended at time 0, and then it rotated, shortened, and assumed two or three bends after 4.5 h. In Fig. 2b, the arrow indicates a short fibril extending upward and to the left from a globular patch. At the upper end, this fibril is close to but not quite contacting another fibril at 0:00 and 1:30. By 3:00 these fibrils had fused, and at 4:00 they formed a uniform, straight fibril. Brightness increased substantially over the 4.5 h. In Fig. 2c, the arrow indicates a short fibril that appears to move left and merge with the larger, tautly extended fibril.
Figure 2.
The mobility of the FN matrix network. Selected images show active fibril movement (arrows). Images were originally taken at 5-min intervals after 36 h of culture. White squares in b are magnified in a and c. (Bar = 50 μm.)
Fig. 3 shows the most dramatic movements that we observed for matrix fibrils. Arrows a–a indicate a patch that extended into a straight fibril from 0–30 min and then collapsed back to a small spot between 30 and 45 min. The fibril indicated by b–c–d is particularly interesting. It appears to be attached at each of these points and to be stretched taut between b–c and c–d. The b–c segment was stretched about 50% longer between 0:45 and 1:00, and then the attachment at c appeared to break, leaving the fibril tautly stretched between b and d at 1:15. This fibril was fairly static until 2:25, when it appeared to detach from the right end and rapidly contracted to about one-fourth of its stretched length. The contracted fibril appeared thicker and brighter, consistent with a contraction. The arrow at e indicates a prominent branched fibril, one branch going to f and then to g. From 1:30 to 1:45, the point e moved substantially left and upward; at 1:45 the fibril appeared to detach from f and run straight from e to g. Between 2:00 and 2:25, the fibril appeared to detach from g and contracted toward e. Note that the three bright globular domains of this contracted fibril can be related to the more extended bright spots before contraction (asterisks in 2:00 and 2:25). Thus, the detached fibril appeared to contract uniformly along its length.
Images were taken at 5-min intervals, but longer intervals are presented to show the most interesting movements and best focus. When a fibril detached and contracted, as in Fig. 3 b–d at 2:25, the contraction usually appeared complete in the 5-min interval and may have happened much faster.
Fig. 4 shows a fibril movement induced by laser damage. The bright patch of FN indicated by the arrow is attached at its bottom to several fibrils. The laser appears to destroy the fibrils on the left and the patch moves sharply upward. It remains attached to the fibril on the right, which it pulls almost vertical. Some of the FN in this extended fibril may have been extruded from the patch, which is much dimmer after the movement.
We attempted to visualize the attachment of FN fibrils to cells by Nomarski optics. However, in contrast to some cell lines that initiate matrix fibrils on individual cells in sparse culture (11, 12), our CHO cell line developed a matrix only after reaching confluence. In Nomarski images of these confluent cultures, the shapes of some cells were clear, but some cell edges were not visible (Fig. 5, 0 min). Sometimes FN fibrils were seen along cell borders, but the most prominent FN fibrils were often not associated with any obvious feature in the Nomarski images. Thus, for most fibrils we could not identify attachment points or correlate fibril movements with cellular structures.
Treatment of the cell culture with 5 mM EDTA caused the cells to round up and move dramatically (Fig. 5). Because calcium is essential for integrin–FN interaction, the EDTA should also disrupt all cellular attachments to FN fibrils. Surprisingly, most of the FN matrix showed only small movements after EDTA treatment. We assume that these static fibrils were attached primarily to the substrate, because they were not altered as the cells rounded and moved. However, some fibrils showed larger movements, and these moving fibrils could sometimes be localized along cell edges (Fig. 5, 20 min, arrowhead). It seems that most movements of FN fibrils must be caused by movement of attached cells or by detachment of fibrils from the cells, but we could only rarely visualize the site of attachment.
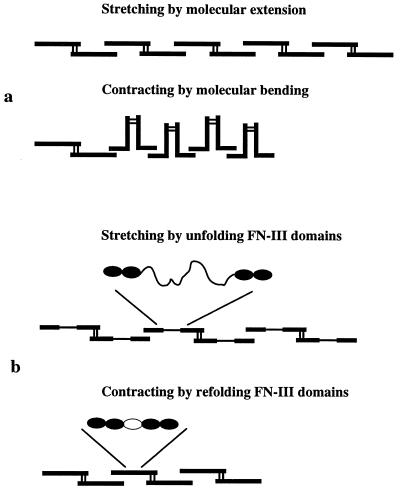
Fig. 6 shows two possible models for extension and contraction of FN matrix fibrils. The first is based on the observation that single dimers of FN can fold into a compact conformation or can be quite extended (16). If the FN dimers in the fibril were attached to each other at their ends, but each dimer tended to fold upon itself, this might generate a contracted fibril (Fig. 6a). An alternative elastic mechanism is based on the possibility of unfolding individual FN-III domains by stretching, giving an extended polypeptide about 8 times the length of the folded domain (Fig. 6b). This was originally suggested as a theoretical possibility for titin and FN (13, 17) and has now been dramatically demonstrated by stretching titin or tenascin with laser tweezers or the atomic force microscope (14, 15, 18, 19).
Figure 6.
Schematic diagram of possible mechanisms for stretching and contracting an FN matrix fibril. These diagrams illustrate single FN molecules; it is important to remember that the fluorescent FN fibrils are ten to hundreds of molecules thick, and hundreds of molecules in legnth. (a) In the contracted fibril the molecules are in a compact conformation, with bends between FN-III domains and stabilized by intramolecular bonds. Stretching the fibril breaks these bonds and extends the FN molecules. (b) The fibril could be stretched by unfolding FN-III domains and contracted by refolding the domains. The unfolded domain is 28.5 nm compared with the 3.5-nm length of the folded domain (13–15).
In either model two mechanisms could contribute to the restoring force. First would be the entropic spring effect, in which the molecule or extended polypeptide contracts to maximize its possible configurations (14, 15, 18, 19). The physiological elasticity of titin appears to be largely because of an entropic spring effect, operating on the bends between Ig domains and on the unfolded peptide of the PEVK domain (20, 21). A second source of restoring force would be an enthalpic contribution from the bonds stabilizing the folded conformation of the molecules or from the energy of refolding a denatured module. The enthalpic contribution was very small in the atomic force microscope measurements of titin and tenascin (14, 15) but may be sufficient to contribute to restoring force in the FN fibrils. The matrix fibrils we are visualizing are probably on the order of 100 molecules thick (1, 2), providing a complex array of elastic units in parallel and in series.
Halliday and Tomasek (22) showed that cells cannot assemble an FN matrix unless the matrix is anchored and tension can develop. The first FN-III module of FN has been implicated in matrix assembly, but its assembly site appears to be cryptic, exposed only in denatured or partially assembled modules (23–25). Remarkably, Zhong et al. (12) recently showed that this cryptic site can be exposed by cell contractility. They proposed that the tension could stretch FN and expose a cryptic site by separating intramolecular contacts of modules (Fig. 6a); alternatively, a cryptic site could be exposed by unraveling a module (Fig. 6b). Our results now show that some, if not most, FN fibrils in a cell culture are indeed highly stretched, up to 4 times their relaxed length. This stretched FN is an ideal candidate for exposing the cryptic assembly sites.
Supplementary Material
Acknowledgments
This work was supported by research Grant N00014-97-1-0911 from the U.S. Office of Naval Research and Grant CA07456 from the National Cancer Institute.
ABBREVIATIONS
- FN
fibronectin
- gfp
green fluorescent protein
- CHO
Chinese hamster ovary
References
- 1.Chen L B, Murray A, Segal R A, Bushnell A, Walsh M L. Cell. 1978;14:377–391. doi: 10.1016/0092-8674(78)90123-x. [DOI] [PubMed] [Google Scholar]
- 2.Singer I I. Cell. 1979;16:675–685. doi: 10.1016/0092-8674(79)90040-0. [DOI] [PubMed] [Google Scholar]
- 3.Leahy D J, Aukhil I, Erickson H P. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald M C, Schwarzbauer J E. Curr Biol. 1998;8:1285–1288. doi: 10.1016/s0960-9822(07)00540-4. [DOI] [PubMed] [Google Scholar]
- 5.Akamatsu H, Ichihara-Tanaka K, Ozone K, Kamiike W, Matsuda H, Sekiguchi K. Cancer Res. 1996;56:4541–4546. [PubMed] [Google Scholar]
- 6.Crameri A, Whitehorn E A, Tate E, Stemmer W P. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 7.Bebbington C R, Hentschel C C G. In: DNA Cloning. Glover D M, editor. III. Oxford: IRL; 1987. pp. 163–188. [Google Scholar]
- 8.Ichihara-Tanaka K, Maeda T, Titani K, Sekiguchi K. FEBS Lett. 1992;299:155–158. doi: 10.1016/0014-5793(92)80236-a. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzbauer J E. J Cell Biol. 1991;113:1463–1473. doi: 10.1083/jcb.113.6.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung C Y, Erickson H P. J Cell Sci. 1997;110:1413–1419. doi: 10.1242/jcs.110.12.1413. [DOI] [PubMed] [Google Scholar]
- 11.Dzamba B J, Wu H, Jaenisch R, Peters D M. J Cell Biol. 1993;121:1165–1172. doi: 10.1083/jcb.121.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong C L, Chrzanowskawodnicka M, Brown J, Shaub A, Belkin A M, Burridge K. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson H P. Proc Natl Acad Sci USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 15.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 16.Erickson H P, Carrell N A. J Biol Chem. 1983;258:14539–14544. [PubMed] [Google Scholar]
- 17.Soteriou A, Clarke A, Martin S, Trinick J. Proc R Soc London Ser B. 1993;254:83–86. doi: 10.1098/rspb.1993.0130. [DOI] [PubMed] [Google Scholar]
- 18.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 19.Kellermayer M S Z, Smith S B, Granzier H L, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 20.Trombitás K, Greaser M, Labeit S, Jin J P, Kellermayer M, Helmes M, Granzier H. J Cell Biol. 1998;140:853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linke W A, Stockmeier M R, Ivemeyer M, Hosser H, Mundel P. J Cell Sci. 1998;111:1567–1574. doi: 10.1242/jcs.111.11.1567. [DOI] [PubMed] [Google Scholar]
- 22.Halliday N L, Tomasek J J. Exp Cell Res. 1995;217:109–117. doi: 10.1006/excr.1995.1069. [DOI] [PubMed] [Google Scholar]
- 23.Morla A, Zhang Z, Ruoslahti E. Nature (London) 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- 24.Hocking D C, Sottile J, McKeown-Longo P J. J Biol Chem. 1994;269:19183–19191. [PubMed] [Google Scholar]
- 25.Ingham K C, Brew S A, Huff S, Litvinovich S V. J Biol Chem. 1997;272:1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.