Abstract
The xylem-limited, insect-transmitted bacterium Xylella fastidiosa causes Pierce's disease in grapes through cell aggregation and vascular clogging. GacA controls various physiological processes and pathogenicity factors in many gram-negative bacteria, including biofilm formation in Pseudomonas syringae pv. tomato DC3000. Cloned gacA of X. fastidiosa was found to restore the hypersensitive response and pathogenicity in gacA mutants of P. syringae pv. tomato DC3000 and Erwinia amylovora. A gacA mutant of X. fastidiosa (DAC1984) had significantly reduced abilities to adhere to a glass surface, form biofilm, and incite disease symptoms on grapevines, compared with the parent (A05). cDNA microarray analysis identified 7 genes that were positively regulated by GacA, including xadA and hsf, predicted to encode outer membrane adhesion proteins, and 20 negatively regulated genes, including gumC and an antibacterial polypeptide toxin gene, cvaC. These results suggest that GacA of X. fastidiosa regulates many factors, which contribute to attachment and biofilm formation, as well as some physiological processes that may enhance the adaptation and tolerance of X. fastidiosa to environmental stresses and the competition within the host xylem.
Xylella fastidiosa is a fastidious, xylem-limited, nonflagellated, insect-transmitted, gram-negative bacterium that causes many plant diseases, including Pierce's disease (PD) (7), a disease which is threatening the grape industry in California in particular. The disease process of PD is related to specific features of X. fastidiosa. It has the ability to adhere to the host (plant and insect) cell surfaces and to form biofilms that enable it to be specifically transmitted by insect vectors and to survive in and colonize the xylem tissue of plants (40). Cells of X. fastidiosa aggregate, form biofilms, and probably clog the host's vascular system, resulting in disease symptoms (32). To understand disease progression and to develop an effective disease control strategy, a better understanding of the complex interactions among the pathogen, plant, and insect vector is critical (21). However, very little is known about the basis of these complex interactions.
Pathogenic bacteria use gene regulatory mechanisms to rapidly respond to and survive in changing environments (47). Inside the xylem of plants, X. fastidiosa is exposed to a range of variable stress factors, such as changes in osmolarity, availability of nutrients, and agents generating reactive oxygen intermediates (1). To ensure survival, X. fastidiosa may respond to these stress situations via specific regulatory mechanisms. We are investigating regulatory pathways that contribute to the success of X. fastidiosa as a pathogen through mutagenesis of “global” regulatory genes that are known to coordinate expression of virulence-related factors in other pathogenic species. In a previous study, we constructed a mutant of X. fastidiosa defective in algU, encoding an alternate sigma factor that is highly conserved in gram-negative bacteria. The algU mutant had reduced cell-cell aggregation, attachment, and biofilm formation and lower virulence in grapevines (43). Microarray analysis showed that 42 genes had significantly lower expression in the algU mutant than in the wild type. This work identified several genes that could contribute to aggregation and biofilm formation as well as other physiological processes, such as virulence, competition, and survival.
An additional regulatory system identified in pathogenic and environmental bacteria is the two-component system of GacS and GacA, involved in sensing environmental signals (19). GacS is a putative sensor kinase that perceives environmental signals, and GacA is a response regulator, which functions as the transcriptional activator of one or more genes. Genes regulated by GacA include regulators of pathogenicity factors, and genes involved in quorum sensing, toxin production, motility, biofilm formation, and extracellular polysaccharide production in a wide range of pathogenic bacterial species, including Pseudomonas syringae, Erwinia carotovora, and Pseudomonas aeruginosa (4, 8, 38). The similarity between gacA of X. fastidiosa (designated gacAXf) and gacA of P. syringae (designated gacADC3000) suggests that, like gacADC3000, gacAXf may regulate the pathogenicity of X. fastidiosa by acting as a global regulator during infection and the process of disease development. While a gacA homolog was identified in X. fastidiosa, a gacS homolog was not found, which suggests that there may be a specific regulatory role for gacA in X. fastidiosa (44). In this study, we cloned and characterized gacAXf and analyzed the phenotypic effects of a gacA deletion in X. fastidiosa (DAC1984). We also performed whole-genome microarray analysis of gene expression in the mutant in comparison with that in the parent strain and identified genes whose expression in vitro is controlled by GacA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this work are listed in Table 1. For growth rate measurements, aggregation, adhesion, colony morphology determination, and biofilm formation, strains of X. fastidiosa were cultured on PD3 Gelrite medium (10, 43). After 7 days at 28°C, cells were harvested using a scraper (Fisher Scientific, CA), washed and resuspended in 1 ml of PD3 broth, and adjusted to an optical density at 600 nm (OD600) of 0.10. Cells used for pathogenicity tests were cultured for 5 days at 28°C on PW Gelrite medium (25, 43), then harvested, and adjusted to the same OD as mentioned above with sterile water. Pseudomonas syringae pv. tomato DC3000 and P. syringae strains AC811, AC812, and AC813 were maintained on Kings medium B (KmB) agar (27) at 28°C. E. amylovora strains EC19, EC191, EC192, and EC193 were maintained on LB agar at 28°C. When required, antibiotics were added as follows: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 10 μg/ml; gentamicin (Gm), 10 μg/ml; spectinomycin, 50 μg/ml; and tetracycline (Tc), 10 μg/ml. All bacteria were stored in 15% glycerol at −80°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | DH1 F−Φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | N. T. Keen |
| X. fastidiosa | 7 | |
| A05 | Wild type | |
| DAC1984 | Gentamicin cassette replacing most of gacA ORF (ΔgacA::Gm) of X. fastidiosa A05 | This work |
| P. syringae pv. tomato | A. Chatterjee | |
| DC3000 | Wild type | |
| AC811 | Kmr; gacADC3000 derivative of DC3000 | A. Chatterjee |
| AC812 | Tcr Kmr; AC811 carrying pCPPgacAXf-Exp | This work |
| AC813 | Tcr Kmr; AC811 carrying plasmid pCPP47 | This work |
| E. amylovora | A. Chatterjee | |
| EC19 | Wild type | |
| EC191 | Gmr; gacA derivative of EC19 | A. Chatterjee |
| EC192 | Tcr Gmr; EC191 carrying pCPPgacAXf-Exp | This work |
| EC193 | Tcr Gmr; EC191 carrying plasmid pCPP47 | This work |
| Plasmids | ||
| pUC129 | Apr; cloning vector | New England Biolabs |
| pUCgacAXf-Exp | Apr; a fragment including the gacA promoter and ORF of X. fastidiosa (gacAXf-Exp) cloned into pUC129 | This work |
| pCPP47 | Tcr; broad-range plasmid | A. Chatterjee |
| pCPPgacAXf-Exp | Tcr; gacAXf-Exp cloned into pCPP47 constructed as gacAXf-Exp; expression plasmid | This work |
| pUC19841 | Apr; mutagenized PCR fragment of the flanking regions of gacA ORF of X. fastidiosa cloned into pUC129 | This work |
| pUC19842 | Apr Gmr; Gm cassette from pGEM-T-GM cloned into the AscI site of pUC19841 | This work |
| pGEM-T Easy | Apr; cloning vector | Promega |
| pBBR1MCS-5 | Gmr; broad-range plasmid | S. Lindow |
| pGEM-T-GM | Apr Gmr; Gm cassette from pBBR1MCS-5 cloned into pGEM-T | This work |
Cloning of gacA of X. fastidiosa (gacAXf-Exp).
A 1,025-bp region of X. fastidiosa A05 genomic DNA containing the gacA promoter and an open reading frame (ORF) (PD1984) was amplified by PCR with Vent polymerase (New England Biolabs, MA) and primers GacAExpFor and GacAExpRev (see Table S1 in the supplemental material). The PCR-amplified fragment was cloned into the SmaI site of pUC129 to make pUCgacAXf-Exp. The PCR fragment insert was released from pUCgacAXf-Exp and cloned into the NotI and SpeI sites of pCPP47 to construct pCPPgacAXf-Exp (Table 1). The presence of the cloned PCR fragment (gacA promoter and ORF) in pUCgacAXf-Exp and pCPPgacAXf-Exp was confirmed by sequence comparison with the genomic DNA of X. fastidiosa.
Electroporation of bacteria.
Electrocompetent cells of P. syringae pv. tomato AC811 and E. amylovora EC191 were prepared as described previously (4, 8). One microgram of the plasmid pCPPgacAXf-Exp DNA in a volume of 5 μl was electroporated into 50 μl of P. syringae pv. tomato AC811 or E. amylovora EC191 electrocompetent cells in a 0.1-cm-gap cuvette at 1.8 kV, 200 Ω, and a capacitance of 25 μF in a GenePulser (Bio-Rad, CA) with time constants of about 4 ms. Pseudomonas cells were plated on KmB agar (4, 27) supplemented with Km and Tc. One Km- and Tc-resistant clone was selected as P. syringae pv. tomato AC812. Erwinia cells were plated on LB agar (8) supplemented with Gm and Tc. One Gm- and Tc-resistant clone was selected as E. amylovora EC192. The pCPP47 DNA was electroporated into P. syringae pv. tomato AC811 or E. amylovora EC191 electrocompetent cells as negative controls to yield P. syringae pv. tomato AC813 or E. amylovora EC193 (Table 1). Cloned inserts in P. syringae pv. tomato AC812 and E. amylovora EC192 carrying pCPPgacAXf-Exp were confirmed by PCR with primers GacAExpFor/Rev (see Table S1 in the supplemental material).
HR and pathogenicity tests.
Strains of P. syringae pv. tomato were grown on KmB agar overnight at 28°C, and E. amylovora was grown in LB broth overnight at 28°C. Bacterial cells at an approximate OD600 of 0.1 were pelleted and resuspended in water for hypersensitive reaction (HR) and pathogenicity tests. The procedure for HR in tobacco leaves was as previously described (4). Leaves of tobacco (Nicotiana tabacum L. cv. Samsun) were infiltrated with P. syringae cell suspensions at 5 × 106 to 1 × 107 CFU/ml. For pathogenicity tests, leaves of 5-week-old African violet plants were infiltrated with E. amylovora cell suspensions (1 × 106 CFU/ml) (49). Five plants with a total of 10 leaves were inoculated for each assay. Tobacco and African violet plants were kept on the benches in a greenhouse with 75% humidity and a photoperiod of 16 h at 28°C.
Construction of a gacA deletion mutant of X. fastidiosa (DAC1984).
A crossover PCR strategy (49) was used to construct a ΔgacA::Gm mutant of X. fastidiosa. Two different asymmetric PCRs were performed to generate fragments to the left side (primers GacAA and GacAB) and right side (primers GacAC and GacAD) of the gacA ORF (PD1984) (see Table S1 in the supplemental material). The left and right PCR fragments were mixed, denatured at 95°C for 5 min, and annealed at overlapping barcode regions (indicated with italics in Table S1 in the supplemental material), including an AscI recognition site in primers GacAB and GacAC, at 25°C for 10 min. The mixture was further amplified by PCR with primers GacAA and GacAD to generate the final, mutagenized 1.1-kb fragment, which was cloned into pUC129 to make pUC19841 (Table 1). The DNA sequence of the PCR fragment in pUC19841 was confirmed by comparison with the genomic sequences of X. fastidiosa. A Gm cassette from pGEM-T-GM (Table 1) was excised and cloned into the AscI site of a 1.1-kb PCR fragment in pUC19841, resulting in the mutant construct pUC19842 (Table 1).
Electrocompetent cells of X. fastidiosa strain A05 (7) were prepared according to published procedures (15). One to two micrograms of pUC19842 DNA in a volume of 5 μl was electroporated into the cells under the conditions described earlier. The electrocompetent cells alone and PD3 broth with no bacterial cells served as negative controls. Electroporated cells were grown for 24 h in PD3 broth with shaking and plated on PD3 Gelrite medium supplemented with 10 μg/ml Gm to select for replacement of the wild-type gacA ORF with Gm by homologous recombination. A Gm-resistant clone was selected as a potential ΔgacA::Gm mutant strain and named X. fastidiosa DAC1984.
X. fastidiosa A05 or DAC1984 was cultured in 50 ml PD3 broth at 28°C for 7 to 10 days with or without antibiotics. The genomic DNAs were extracted with a MasterPure DNA purification kit (Epicentre Technologies, WI). The insertion of Gm in the genome of DAC1984 was confirmed by PCR using primers M13For/M13Rev and GacAORF P1/P2, respectively (see Table S1 in the supplemental material). A fragment of 0.831 kb from A05 and a 2.22-kb fragment from DAC1984 were cut from gels, cloned into pGEM-T Easy (Promega, WI), and sequenced. The location of Gm in DAC1984 genomic DNA was determined by comparing the sequences of the cloned PCR fragment from DAC1984 with the sequences from A05, using Vector NTI (Invitrogen, CA).
Phenotypic analyses.
The colony morphologies of X. fastidiosa A05 and DAC1984 were observed after 10 to 14 days of growth at 28°C on PD3 Gelrite plates. For cell attachment analysis, A05 and DAC1984 were grown in 50 ml of PD3 broth in 125-ml glass flasks on a shaker at 28°C for 6 to 10 days. In vitro growth curves in 3 ml of PD3 broth were determined after 3 to 21 days of growth at 28°C. Due to the aggregation of the cells in broth, immediately after inoculation and 3, 6, 9, 12, 15, 18, and 21 days later, the cells were dispersed by repeated pipetting or vortexing. Cell concentration was determined by measuring turbidity at OD600. Cell aggregation, biofilm formation, and lipopolysaccharide (LPS) gel analyses were done as described previously (3, 16, 29), with modifications as described previously for a comparison of the X. fastidiosa A05 wild-type strain and an algU mutant (43).
Tolerance of DAC1984 to desiccation stress in vitro.
The sensitivities of X. fastidiosa A05 and DAC1984 to desiccation on filters were assessed using a modification of a previous procedure (37). Seven- to ten-day-old cultures were collected and adjusted to an OD600 of 0.10 with sterile distilled water and serially diluted to 1 ×10 4 CFU/ml. One milliliter of each dilution was vacuum filtered onto Millipore filters (no. HAWP04700; pore size, 0.25 μm; diameter, 3.5 cm). The filters were placed in petri dishes at 25°C for slow drying. At 0, 2, 4, 6, 8, 10, 12, and 14 days, filters were placed onto PD3 Gelrite plates and incubated at 28°C for 3 weeks. The filters containing water only and incubated for the same period of time served as controls. The number of colonies on each filter was recorded. Each treatment consisted of five filters and was repeated three times.
Susceptibility to oxidative stress in vitro.
Sensitivity to hydrogen peroxide (H2O2) or sodium hypochlorite (NaOCl) was examined as previously described (33). Millipore filter disks (diameter, 6 mm) were soaked with 10 μl of H2O2 (3 or 12%, vol/vol) or NaOCl (3 or 6%, vol/vol) and placed on PD3 plates on which 100 μl of 7-day-old cultures of X. fastidiosa A05 or DAC1984 was spread with a glass rod. The diameters of the inhibition zones surrounding the impregnated disks were measured after 14 to 21 days of incubation at 28°C. Three disks were used in each treatment, each treatment was repeated three times, and the results were averaged.
Pathogenicity assays with grapes.
X. fastidiosa A05 and DAC1984 were grown on PW Gelrite medium for 5 days at 28°C, suspended in sterile deionized water, and adjusted to an OD600 of 0.10. Five to ten 20-μl drops of each suspension were used to inoculate five to ten canes on seedlings of Vitis vinifera L. cv. Pinot Noir by using a needle inoculation procedure as previously described (22). A water inoculation served as a negative control. The inoculated grapevines were kept on benches in a greenhouse and were observed for symptom development approximately every 2 weeks for 5 months after inoculation. The symptoms were rated on a visual scale from 0 to 5 as described previously (16), with 0 representing healthy grapevines without scorched leaves (water control) and 5 representing plants with heavy scorching or numerous matchstick symptoms, where the petiole remains attached to the cane after scorched leaf blades abscise and fall. The final disease index was an average for 10 independent replications for each X. fastidiosa strain.
Recovery and determination of populations of X. fastidiosa from inoculated grapes.
To recover and confirm the bacteria in inoculated grapes, 12 weeks after inoculation, petiole tissues (2 to 3 cm) from each vine inoculated with either X. fastidiosa A05 or DAC1984 cells were harvested at the inoculated points as well as 25 cm and 50 cm above the inoculation points. Tissues were washed once with deionized water containing Tween 20; surface sterilized for 1 min in 20% commercial bleach, 1 min in 2% sodium hypochlorite, and 1 min in 70% ethanol; and rinsed three times in sterile deionized water. The samples were ground in 100 μl of sterile deionized water and cultured on PD3 and PW Gelrite media with or without Gm. After incubation for 21 days at 28°C, the identity of X. fastidiosa cells on PD3 Gelrite plates was confirmed by PCR using specific primers for X. fastidiosa A05, GacAORFP1/P2, and tapBPD1993P1/P2 (see Table S1 in the supplemental material).
To determine the bacterial populations, 16 weeks after inoculation, 2- to 3-cm petiole tissues of each vine inoculated with X. fastidiosa A05 and DAC1984 were harvested and treated as mentioned above. Tissues were tested using an enzyme-linked immunosorbent assay (ELISA) with a PathoScreenXF kit according to the manufacturer's instructions (Agdia, Inc., IN). The antibodies used in the Agdia ELISA system are a mixture of polyclonal antibodies raised to whole cells of three serologically distinct isolates of X. fastidiosa cells (Agdia, Inc., IN). The PD3-cultured X. fastidiosa A05 and DAC1984 cells were resuspended in phosphate-buffered saline (PBS) buffer (Agdia, Inc., IN) and used to confirm that ELISA worked equally well for quantifying the populations of the wild type and the mutant. Developed plates were measured at 650 nm using a SpectraMax microplate reader via SoftMaxPro (version 3.1.2; Molecular Devices Corp., CA). Bacterial populations were calculated by comparing the OD650 to that of the positive control (purified X. fastidiosa cells in PBS suspension).
RNA isolation, quantification, and RT-PCR.
A modified hot-phenol RNA preparation procedure was used to extract total RNA from X. fastidiosa A05 and DAC1984 (28). Bacterial cultures were incubated in 50 ml of PD3 broth at 28°C for 5 days under constant agitation. After the hot-phenol extraction, RNA was dissolved in RNase-free distilled H2O and DNase treated using Turbo DNA-free DNase (2 U/μl) (Ambion, TX). To ensure that the RNA preparation was DNA free, a 1-μl aliquot of RNA (500 ng/μl) was used to amplify the gacA ORF by using GacAORFP1/P2 primers (see Table S1 in the supplemental material). The quality of isolated RNAs was determined by denaturing RNA formaldehyde gel electrophoresis (5). The expression of gacA was analyzed by reverse transcription-PCR (RT-PCR) with primers GacAORFmRNAFor/Rev (see Table S1 in the supplemental material), using the AccessQuick RT-PCR System per the manufacturer's instructions (Promega, WI).
Microarray hybridizations and microarray data analysis.
The gene expression profiles of X. fastidiosa A05 and DAC1984 were analyzed by NimbleGen prokaryotic gene expression arrays (NimbleGen System, Inc., WI). DNA microarray chips were designed with a 24-mer oligonucleotide according to the available X. fastidiosa genomic sequences. The expression levels of RNA were averaged from three technical replications in a single hybridization experiment. The raw data were analyzed using the ArrayStar FirstLight software program. The expression levels of 2,188 genes under treatment (DAC1984) and control (A05) were analyzed (17). The hybridization signal intensity obtained from A05 or DAC1984 RNA was normalized according to the total signal strength. The normalized hybridization signals were log plot analyzed for signal reliability (17) and were statistically analyzed by Student's t test (P < 0.001) for differential expression. The normalized signal intensity of DAC1984 was divided by that of A05 to calculate the mutant/wild-type (M/W) ratio. M/W ratios obtained from individual hybridization experiments were averaged to give the final M/W number. Genes having final M/W ratios of ≥1.5 or ≤0.66 were selected as positively or negatively regulated mutated genes, respectively.
Validation of microarray data.
To validate the differential expression data obtained in microarray analysis, RT-PCR experiments were performed with specific primers designed to amplify internal regions of the ORFs of the target genes (see Table S1 in the supplemental material). Several positively regulated or negatively regulated genes were chosen, and primers were designed for their ORFs according to the X. fastidiosa Temecula1 genome sequences. cDNA was amplified from stored RNA by using the AccessQuick RT-PCR system (Promega, WI) according to the manufacturer's instructions. The amplification conditions used were as follows: 45 min at 45°C for reverse transcription; 35 cycles of 2 min at 55°C for initial denaturation, 1 min at 55°C for annealing, and 2 min at 72°C for extension; and a final extension of 10 min at 72°C.
RESULTS
Complementation of the HR and pathogenicity in heterologous pathogens with gacAXf.
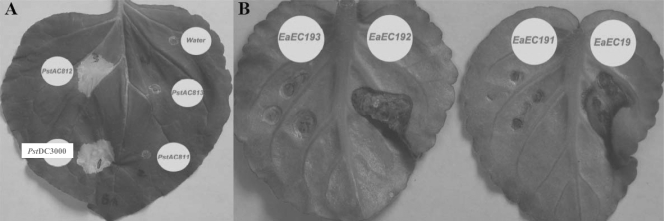
An alignment of the predicted amino acid sequences of GacA from X. fastidiosa and P. syringae pv. tomato DC3000 showed that the sequences are 211 amino acids in length and are 43% identical and 69% similar overall (data not shown). In the HR and pathogenicity test experiments, P. syringae pv. tomato DC3000 (P. syringae wild type) and P. syringae pv. tomato AC812 (P. syringae gacA mutant complemented with cloned gacA from X. fastidiosa) elicited typical HR in tobacco (Nicotiana tabacum L. cv. Samsun), whereas the water control, P. syringae pv. tomato AC811 (P. syringae gacA mutant) and P. syringae pv. tomato AC813 (P. syringae gacA mutant carrying only the plasmid vector used to clone X. fastidiosa gacA) did not (Fig. 1), confirming that gacAXf restored the elicitation of HR in tobacco. E. amylovora EC19 (the E. amylovora wild type) and E. amylovora EC192 (an E. amylovora gacA mutant complemented with cloned gacA from X. fastidiosa) produced disease symptoms in African violet leaves. In contrast, E. amylovora EC191 (an E. amylovora gacA mutant) and E. amylovora EC193 (an E. amylovora gacA mutant carrying only the plasmid vector used to clone X. fastidiosa gacA) failed to produce disease symptoms (Fig. 1), suggesting that gacAXf restored the ability of E. amylovora EC192 to cause disease in African violet leaves. This demonstrated that gacAXf can complement gacA deficiencies of P. syringae and E. amylovora in regulating the HR and pathogenicity.
FIG. 1.
Complementation of GacA function by gacAXf in gacA mutants of P. syringae and E. amylovora. (A) Effect of gacAXf in gacA mutant P. syringae pv. tomato AC811 (PstAC811) on the elicitation of the HR in tobacco leaf (Nicotiana tabacum L. cv. Samsun). Leaf panels were infiltrated with bacterial cell suspensions at 5 × 106 to 1 × 107 CFU/ml. Site 1, P. syringae pv. tomato DC3000 (wild type); site 2, P. syringae pv. tomato AC811 (gacA mutant); site 3, P. syringae pv. tomato AC812 (gacA mutant complemented with cloned gacAXf); site 4, P. syringae pv. tomato AC813 (gacA mutant with cloning vector alone); and site 5, water. (B) Effect of gacAXf in gacA mutant E. amylovora EC191 (EaEC191) in causing of disease symptoms in Africa violet. Leaves were infiltrated with bacterial cell suspensions at 1 × 106 CFU/ml. Site 1, E. amylovora EC19 (wild type); site 2, E. amylovora EC191 (gacA mutant); site 3, E. amylovora EC192 (gacA mutant complemented with cloned gacAXf); site 4, E. amylovora EC193 (gacA mutant with the cloning vector alone).
Physiological properties of DAC1984 in vitro.
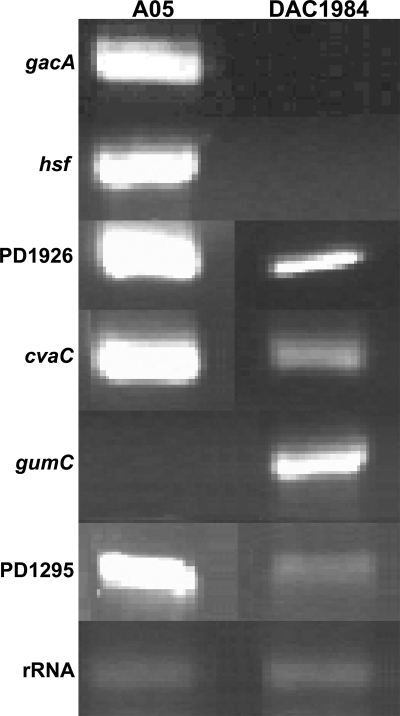
The replacement of the gacA ORF with a Gm cassette in the genome of DAC1984 was confirmed by electrophoresis after PCR amplification of the modified gacA gene (data not shown). Sequence analysis indicated that Gm physically replaced most of the gacA ORF, from only 25 bp downstream from the ATG start codon to 35 bp upstream from the TGA terminal codon of the gacA ORF. RT-PCR analysis using GacAORFmRNAFor/Rev (see Table S1 in the supplemental material) showed that there was no gacA-sized RT-PCR fragment within DAC1984 cells (insertion of the Gm cassette made the fragment too large to be detectable in this assay), but strong expression was detected in A05 cells (Fig. 2). After being streaked five to eight times on PD3 Gelrite medium with 10 μg/ml Gm, DAC1984 still grew well and was indistinguishable from the parent, indicating that the mutant was stable.
FIG. 2.
RT-PCR of genes differentially expressed between wild-type X. fastidiosa A05 and gacA mutant DAC1984.
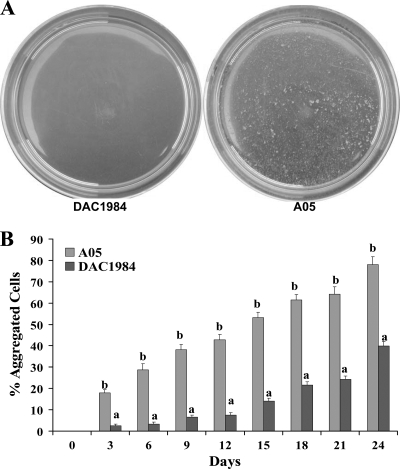
In vitro growth curves of X. fastidiosa A05 and DAC1984 over 21 days were similar, with cell concentrations of both the wild type and the gacA mutant increasing from an OD600 of approximately 0.15 at 3 days to an OD600 of 0.55 at 21 days (data not shown). There was no obvious difference in colony morphology between A05 and DAC1984, but DAC1984 had less sticky colonies on PD3 Gelrite medium when touched with a bacteriological loop (data not shown). In PD3 broth, visual observation showed that colonies of A05 formed large aggregates, whereas DAC1984 grew in less aggregated clumps (Fig. 3). An OD assay was used to quantify the effect of the gacA deletion on cell-to-cell aggregation and showed that the percentage of aggregated cells of DAC1984 was significantly lower than that of A05 (Fig. 3).
FIG. 3.
Cell-to-cell aggregation of wild-type X. fastidiosa A05 and gacA mutant DAC1984. (A) Cell-to-cell aggregation of X. fastidiosa DAC1984 (left) and A05 (right) in PD3 broth in petri dishes. (B) Quantitative assessment of cell-to-cell aggregation of X. fastidiosa A05 or DAC1984 by an OD assay as described previously (3, 29). Three replicates were used in each experiment. For each assay time, different letters indicate significant differences (Student's t test; P < 0.05) between the wild type and the mutant.
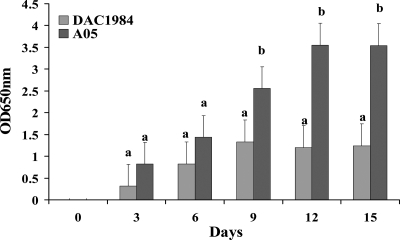
As is typical of X. fastidiosa, cells of A05 attached to the surfaces of flasks and formed wide rings, but DAC1984 cells attached to the surfaces formed rather light rings (data not shown). This suggested that DAC1984 has a reduced surface attachment ability, resulting in reduced biofilm formation. Biofilm formation by DAC1984 was investigated further with a crystal violet staining method. X. fastidiosa A05 formed more biofilm in PD3 broth than did DAC1984 (Fig. 4). Deoxycholate-polyacrylamide gel electrophoresis analysis showed that there was no significant difference between the purified-LPS profile in DAC1984 grown in vitro and LPS from A05 (data not shown).
FIG. 4.
Analysis of biofilm formation of wild-type X. fastidiosa A05 and gacA mutant DAC1984 by a crystal violet staining method (29). Biofilm cells were stained with crystal violet, the amount of stain was dissolved with ethanol, and the resulting absorbance was measured at OD650. Three replicates were used in each experiment. For each assay time, different letters indicate significant differences (Student's t test; P < 0.05) between the wild type and the mutant.
Tolerance to oxidative and desiccation stresses.
No significant differences were observed in tolerance to oxidative stress (sensitivity to hydrogen peroxide or sodium hypochlorite) between mutant and wild-type strains (data not shown). When X. fastidiosa A05 and DAC1984 were exposed to desiccation stresses in vitro in petri dishes, the survival rate of DAC1984 was significantly lower than that of A05 (Table 2).
TABLE 2.
Tolerance of wild-type X. fastidiosa and DAC1984 to desiccation stress in vitro
| Genotype | Survival rate (%) of X. fastidiosa cells after drying on filtersa for:
|
||||
|---|---|---|---|---|---|
| 0 Days | 2 Days | 4 Days | 6 Days | 8 Days | |
| Wild type | 100 | 51.4§ | 7.9§ | 0.7¶ | 0¶ |
| DAC1984 | 100 | 20.6¶ | 0.7¶ | 0¶ | 0¶ |
Data are averages for three independent replications for each treatment. Five filters per dilution were used in each treatment. For each assay date, the presence of different symbols indicates a significant difference (Student's t test; P < 0.05) between the wild-type and mutant survival rates. The data for days 10 to 14 were all 0 and are not shown.
Pathogenicity tests and recovery from infected plants.
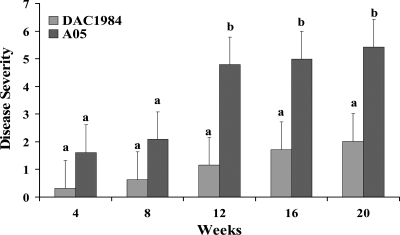
Compared with grapevines inoculated with wild-type X. fastidiosa A05, grapevines inoculated with DAC1984 developed significantly less severe disease symptoms 12 to 20 weeks after inoculation (Fig. 5). Water-inoculated control grapevines did not show any PD symptoms. All diseased grapevines were positive, and the asymptomatic water control vines were negative, for the presence of X. fastidiosa when the plants were tested by ELISA. Bacteria were reisolated from macerated, inoculated grapevine petioles on PD3 and PW Gelrite media. The bacterial genotypes were confirmed as X. fastidiosa A05 or DAC1984 by PCR amplification with primers for GacAORFP1/P2 and primers tapBPD1993P1/P2 (see Table S1 in the supplemental material) (data not shown).
FIG. 5.
PD progression in grapevines inoculated with wild-type X. fastidiosa A05 and gacA mutant DAC1984. Disease severity was based on a visual disease scale of 0 to 5 and was assessed 4, 8, 12, 16, and 20 weeks after inoculation (16). The data are averages for 10 independent replications. For each time point, different letters indicate significant differences (Student's t test; P < 0.05) between the wild type and the mutant.
To gain further understanding of the reduced virulence of the gacA mutant, bacterial populations and bacterial movement in infected grapevines were estimated from ELISAs. The ELISA showed no cross-reaction with any healthy tissue tested. Preliminary experiments showed that the ELISA used to quantify the X. fastidiosa population worked equally well for A05 and DAC1984 cultures. Bacterial populations at inoculation points as well as at 25 cm and 50 cm above inoculation points were estimated from ELISAs by comparing the OD650 with that of the X. fastidiosa positive control with known concentrations (Table 3). One OD650 of the X. fastidiosa positive control (purified X. fastidiosa cells in PBS suspension) represented approximately 1 × 104 CFU/ml. The average bacterial populations were calculated by comparing the OD650 to that of the positive control and divided by the average weight of 2- to 3-cm sampled petioles. There were no X. fastidiosa cells detected in the asymptomatic water-inoculated control grapevines. The cell populations of DAC1984 were less than those of A05 at 25 cm and 50 cm above inoculation points (Table 3). The actual populations could have been higher than we reported, since those were calculated based on X. fastidiosa cultures in PBS buffer rather than plant sap. Plant sap could lower ELISA detection. However, it is the relative difference between the wild type and the mutant that is significant (Table 3). Those data suggest that the mutated gacA gene may affect the growth and possibly the movement of X. fastidiosa inside the xylem, resulting in reduced pathogenicity.
TABLE 3.
Bacterial populations in grapevines 16 weeks after inoculation
| Genotype | Population (106 CFU/g of tissue)a
|
||
|---|---|---|---|
| At inoculation point | Above inoculation point
|
||
| 25 cm | 50 cm | ||
| Wild type | 9.087 ± 2.6§ | 0.609 ± 0.18§ | 0.646 ± 0.28§ |
| DAC1984 | 1.004 ± 3.1¶ | 0.028 ± 0.012¶ | 0.002 ± 0.0023¶ |
Data are averages for 10 independent replications for each treatment. Each treatment had five samples. For each assay point, the presence of different symbols indicates a significant difference (Student's t test; P < 0.05) between the wild-type and mutant populations. The data for the asymptomatic water control were all 0 and are not shown.
In vitro gene expression profiling of DAC1984.
The expression levels of 2,188 genes in X. fastidiosa A05 and DAC1984 were monitored using a cDNA microarray. The expression levels of RNA were averaged from three replications in a single hybridization experiment. The normalized hybridization signals formed a linear pattern after the log plot analysis, indicating that the hybridization signals were stable, repeatable, and reliable (data not shown). Twenty-seven genes were differentially expressed in DAC1984, compared with what was found for A05 (Table 4). Several putative pathogenicity-related genes, such as xadA (PD0731), encoding an outer membrane protein, hsf (PD0744), encoding a surface protein, and gumC (PD1395), involved in the biosynthesis of fastidian gum, were regulated by gacA in X. fastidiosa. oprO (PD0264), actP (PD1294), cvaC (PD0216), fimbrial protein gene PD1926, and hypothetical protein gene PD1295 were positively regulated by gacA in X. fastidiosa. Several genes were negatively regulated by gacA in X. fastidiosa, including genes encoding phage-related proteins, hypothetical conserved proteins, biotin synthesis (bioI), lysophospholipase-secretory lipase (PD1702 and PD1703), membrane fusion proteins, acriflavin resistance proteins, membrane-bound glucose dehydrogenase, and polyvinylacohol dehydrogenase, according to the functional groups (Table 4). RT-PCR was used to validate that there were lower expression levels of hsf, cvaC (PD0216), fimbrial protein gene PD1926, and PD1295 in the gacA mutant and higher expression levels of gumC (Fig. 2). Detection of rRNA at similar levels in X. fastidiosa A05 and DAC1984 (Fig. 2) indicated that the RT-PCR condition was reliable.
TABLE 4.
Genes differentially expressed in X. fastidiosa DAC1984 in vitro, organized by functional groups
| Functional group and gened | ORF | Description | M/W ratioa,b | Expression level in mutantc |
|---|---|---|---|---|
| Cell structure | ||||
| Membrane components/outer membrane; oprO | PD0264 | Porin O precursor | 0.24 | Lower |
| Surface structures | PD1926 | Fimbrial protein-pilus assembly protein | 0.12 | Lower |
| Cellular processes; transport/other; actP | PD1294 | Acetate permease | 0.337 | Lower |
| Intermediary metabolism; energy metabolism and carbon/electron support | PD2039 | Oxidoreductase | 2.75 | Higher |
| Biosynthesis of small molecules; | PD1688 | Cytochrome P450-like enzyme | 2.45 | Higher |
| cofactors, prosthetic groups, and carrier biosynthesis/biotin; bioI | PD1703 | Hypothetical protein-conserved domains: lysophospholipase | 6.31 | Higher |
| PD1702 | Hypothetical protein-conserved domains: secretory lipase | 5.29 | Higher | |
| Pathogenicity, virulence, and adaptation | ||||
| Toxin production and detoxification | ||||
| cvaC | PD0215 | Colicin V precursor: antibacterial polypeptide toxin | 3.63 | Higher |
| cvaCe | PD0216 | Colicin V precursor: antibacterial polypeptide toxin | 0.51 | Lower |
| Outer membrane protein | ||||
| xadA | PD0731 | Outer membrane protein: autotransporter adhesion | 0.11 | Lower |
| hsf | PD0744 | Surface protein: autotransporter adhesion | 0.31 | Lower |
| Exopolysaccharides; gumC | PD1395 | Exopolysaccharide biosynthesis | 3.17 | Higher |
| Hypothetical conserved/hypothetical proteins | PD0243 | Conserved domains: membrane fusion protein | 3.01 | Higher |
| PD0244 | Conserved domains: acriflavin resistance protein | 2.7 | Higher | |
| PD0956 | V8-like Glu-specific endopeptidase | 4.77 | Higher | |
| PD1299 | Conserved domains: polyvinyl alcohol dehydrogenase | 6.18 | Higher | |
| PD1295 | Putative membrane protein: unknown function | 0.20 | Lower | |
| PD0521 | Unknown | 3.24 | Higher | |
| PD0657 | Unknown | 3.28 | Higher | |
| PD0743 | Unknown | 6.62 | Higher | |
| PD0955 | Unknown | 2.67 | Higher | |
| Mobile genetic elements/phage-related | PD0911 | Phage-related proteins: unknown function | 6.04 | Higher |
| functions and prophages | PD0912 | Phage-related proteins: unknown function | 8.41 | Higher |
| PD0917 | Phage-related proteins: unknown function | 3.94 | Higher | |
| PD0924 | Phage-related proteins: unknown function | 6.39 | Higher | |
| PD0925 | Phage-related proteins: unknown function | 9.12 | Higher | |
| PD0930 | Phage-related proteins: unknown function | 3.96 | Higher |
The hybridization signal intensity (mean for three technical replicates) obtained with the mutant was divided by that obtained with the wild type to obtain the M/W ratio.
The normalized hybridization signals for those genes between the wild type and the mutant are all statistically significantly different, as analyzed by Student's t test (P < 0.001).
Genes having final M/W ratios of >1.5 or <0.66 were designated as having higher or lower expression levels in the mutant, respectively.
Genes were detected on the basis of X. fastidiosa Temecula1 genomic sequences at the NCBI website.
Currently annotated as a colicin V precursor.
DISCUSSION
A gacA mutant of X. fastidiosa (DAC1984) did not differ in growth rate in vitro from A05, but it had reduced abilities to aggregate, attach to surfaces, form biofilms, and incite disease symptoms in grapevine. DAC1984 had decreased movement inside plants, associated with decreased disease development (23). The populations of DAC1984 at 25 cm and 50 cm above the inoculation points were reduced compared with those of A05 but were also significantly lower at the inoculation points. Thus, we cannot determine from these data specifically whether DAC1984 was defective for vessel-to-vessel movement versus growth rate inside the xylem.
It was reported that cell surface structures such as LPSs and extropolysaccharides play an important role in the attachment process and biofilm formation (14). However, the purified LPS profile of DAC1984 was not significantly altered compared with LPS from A05 by the assay used.
The extracellular polysaccharide, fastidian gum, of X. fastidiosa is thought to be synthesized by nine enzymes encoded by the gumBCDEFHJKM operon (44). In Xanthomonas campestris pv. campestris, a related operon is responsible for production of the extracellular polysaccharide xanthan gum, and xanthan-deficient mutants have reduced virulence (26). The finding of a related operon in X. fastidiosa suggests that fastidian gum may also be an important virulence factor, as in X. campestris pv. campestris (9). Fastidian gum in X. fastidiosa may be involved in biofilm formation that is thought to benefit X. fastidiosa survival inside xylem of plant and insect vectors (9). GumC may be responsible for regulating gum polymerization or secretion of fastidian gum (12). There was an increased expression level of gumC in DAC1984, indicating that gumC is repressed by GacA in vitro. The expression of gumC was significantly suppressed under high cell densities of X. fastidiosa, while other gum genes were induced (42), suggesting that gumC may be involved in the early stage of regulating or secreting fastidian gum, which would benefit the early steps of biofilm formation in planta.
Genes involved in surface structures and attachment components, such as hsf, xadA, and fimbrial protein gene PD1926, were positively regulated by GacA in X. fastidiosa. Several Hsf-like surface proteins are predicted to occur in the X. fastidiosa genome (44). hsf (PD0744) has a high level of similarity to the hsf adhesin gene of the human pathogen Haemophilus influenzae (46). hsf of H. influenzae encodes surface fibrils with an adhesion mechanism different from that of type I fimbriae and is responsible for the attachment to human epithelial cells (46), thus allowing a tight contact between the pathogen and the host cell. Previous studies showed that fimbriae, pili, and Hsf surface fibrils were coexpressed in X. fastidiosa (45). Higher expression levels of hsf were detected under the pathogenic condition but not under the nonpathogenic condition (13), suggesting that Hsf may play a role in the virulence of X. fastidiosa through initial adhesion to xylem cell walls and may be involved in the initial process of biofilm formation.
xadA homologs are present in the genomes of the plant pathogens X. fastidiosa, Xanthomonas oryzae pv. oryzae, and Xanthomonas campestris pv. vesicatoria (36, 41, 44), encoding a predicted fimbrial outer membrane protein. XadA is predicted to be an autotransporter, secreted by the type V secretion system (20). A xadA mutant of the rice pathogen X. oryzae pv. oryzae was deficient for virulence and had changes in colony morphology (41). A xadA mutant of X. fastidiosa had a 100-fold reduction in ability to attach to surfaces, compared with the wild type (31). The expression of xadA was decreased in DAC1984, possibly resulting in a reduced ability to adhere to xylem cell walls.
A gene encoding a predicted fimbrial protein (PD1926) was shown to be positively regulated by GacA. PD1926 is located in the gene cluster comprising PD1922 to PD1928 (48), which includes homologs of PilD (PD1922), PilC (PD1923), PilA (PD1924), PilB (PD1927), PilR (PD1928), and PilS (PD1929), thought to function in the biogenesis of type IV pili and twitching motility in P. aeruginosa (24, 34). Mutations in pilA, pilB, and pilR of X. fastidiosa, which still possessed type I pili only, resulted in a twitching-negative phenotype and could not colonize upstream vascular regions in planta (11, 30, 35). However, the mutants of pilB had enhanced biofilm formation (35). It is predicted that PD1926 is a gene involved in the formation or function of type IV pili of X. fastidiosa, but its specific contribution is not known. If PD1926 contributes to the formation of type IV pili, then our finding that the gacA mutant had reduced cell-cell aggregation and biofilm formation is not consistent with the recent finding by Li et al. (30), indicating that mutants of X. fastidiosa lacking type IV pili had enhanced cell-cell aggregation and biofilm formation. The long type IV pili are thought to partially mask the adhesion functions of shorter type I pili (30). Our previous work with AlgU, which negatively regulates PD1926, was more consistent with the findings of Li et al., since an algU mutant had reduced cell-cell aggregation and biofilm formation (43). However, in addition to PD1926, expression levels of hsf and xadA were reduced in the gacA mutant, which would be expected to reduce the initial surface attachment and the process of biofilm formation. Therefore, the reduced-adherence phenotype of the gacA mutant is consistent with the reduced expression of several genes encoding surface structures and attachment.
One gene predicted to encode a colicin V precursor, cvaC (PD0215), was negatively regulated by GacA, but another cvaC gene (PD0216) was positively regulated by GacA in X. fastidiosa in this study. Colicin V is an antibacterial polypeptide toxin that acts against closely related sensitive bacteria (18). X. fastidiosa has three colicin-like precursor protein genes, PD0215, PD0216, and PD0217, and genes that should encode a complete colicin V secretory machinery (44, 48). There is also a cvi homolog, PD0214, in X. fastidiosa, predicted to encode a colicin V immunity protein (44, 50). In other studies of cvaC gene expression in X. fastidiosa, cvaC (PD0215) and cvaC (PD0217) were induced by glucose (39), and expression levels of cvaC (PD0216) were higher in later stages of colonization of citrus (13). In addition, cvaC (PD0216) was positively regulated by AlgU in our previous study (43). Since there are diverse endophytic bacterial populations inside the xylems of grape and citrus plants that may influence colonization by X. fastidiosa (2, 6), cvaC may play a role in competing with indigenous microbes in colonization of the xylem.
GacA in X. fastidiosa was shown to regulate genes contributing to attachment and biofilm formation as well as various physiological processes. Our previous work showed that AlgU also regulates genes involved in similar phenotypes, but there was little overlap in the genes regulated by these two proteins (43). GacA may contribute to the attachment process by positively regulating production of the nonfimbrial adhesion proteins Hsf and XadA and by regulating gumC, which may be involved in early stages of regulating or secreting fastidian gum. AlgU positively regulates different surface proteins, such as MopB and OmpW, and negatively regulates the predicted fimbrial protein PD1926, which is positively regulated by GacA.
GacA and AlgU also regulated genes predicted to contribute to various metabolic processes and antimicrobial competition. Most of the specific genes predicted to encode these functions were different for the GacA-versus-AlgU regulatory pathways, except for the colicin V precursor gene cvaC (PD0216), a gene encoding a putative membrane protein of unknown function (PD1295), and an additional ORF of unknown function (PD0521). Specific roles of genes regulated by GacA and AlgU are being investigated through mutagenesis and functional assays.
Supplementary Material
Acknowledgments
Thanks to Steven Lindow (Department of Plant and Microbial Biology, University of California, Berkeley, CA) for providing plasmid pBBR1MCS-5 and Arun K. Chatterjee (Department of Plant Microbiology & Pathology, University of Missouri, Columbia, MO) for providing gacA mutants of P. syringae and E. amylovora and related plasmids. Thanks to Zhenyu Jia (Department of Pathology & Laboratory Medicine, University of California, Irvine, CA) for helping with analysis of the microarray data.
This project was supported by grants from the California Department of Food and Agriculture and the University of California Agricultural Experiment Station.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alves, G., T. Ameglio, A. Guilliot, P. Fleurat-Lessard, A. Lacointe, S. Sakr, G. Petel, and J. L. Julien. 2004. Winter variation in xylem sap pH of walnut trees: involvement of plasma membrane H+-ATPase of vessel-associated cells. Tree Physiol. 24:99-105. [DOI] [PubMed] [Google Scholar]
- 2.Araújo, W. L., J. Marcon, W. Maccheroni, Jr., J. D. van Elsas, J. W. L. van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdman, S., E. Jurkevitch, M. E. Soria-Diaz, A. M. G. Serrano, and Y. Okon. 2000. Extracellular polysaccharide composition of Azospirillum brasilense and its relation with cell aggregation. FEMS Microbiol. Lett. 189:259-264. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee, A., Y. Cui, H. Yang, A. Collmer, J. R. Alfano, and A. K. Chatterjee. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16:1106-1117. [DOI] [PubMed] [Google Scholar]
- 5.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooksey, D. A., and J. Borneman. 2005. Culture-independent analysis of endophytic microbial communities in grapevine in relation to Pierce's disease, p. 155-157. In Proceedings of the Pierce's Disease Research Symposium. California Department of Food and Agriculture, San Diego, CA.
- 7.Costa, H. S., E. Raetz, T. R. Pinckard, C. Gispert, R. Hernandez-Martinez, C. K. Dumenyo, and D. A. Cooksey. 2004. Plant hosts of Xylella fastidiosa in and near southern California vineyards. Plant Dis. 88:1255-1261. [DOI] [PubMed] [Google Scholar]
- 8.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol. Plant-Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 9.da Silva, F. R., A. L. Vettore, E. L. Kemper, A. Leite, and P. Arruda. 2001. Fastidian gum: the Xylella fastidiosa exopolysaccharide possibly involved in bacterial pathogenicity. FEMS Microbiol. Lett. 203:165-171. [DOI] [PubMed] [Google Scholar]
- 10.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony peach disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 11.De La Fuente, L., E. Montanes, Y. Meng, Y. Li, T. J. Burr, H. C. Hoch, and M. Wu. 2007. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl. Environ. Microbiol. 73:2690-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Pieri, C., L. M. Beltramini, H. S. Selistre-de-Araujo, A. L. Vettore, F. R. da Silva, P. Arruda, G. Oliva, and D. H. F. de Souza. 2004. Overexpression, purification, and biochemical characterization of GumC, an enzyme involved in the biosynthesis of exopolysaccharide by Xylella fastidiosa. Protein Expr. Purif. 34:223-228. [DOI] [PubMed] [Google Scholar]
- 13.de Souza, A. A., M. A. Takita, E. O. Pereira, H. D. Coletta-Filho, and M. A. Machado. 2005. Expression of pathogenicity-related genes of Xylella fastidiosa in vitro and in planta. Curr. Microbiol. 50:223-228. [DOI] [PubMed] [Google Scholar]
- 14.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilhabert, M. R., L. M. Hoffman, D. A. Mills, and B. C. Kirkpatrick. 2001. Transposon mutagenesis of Xylella fastidiosa by electroporation of Tn5 synaptic complexes. Mol. Plant-Microbe Interact. 14:701-706. [DOI] [PubMed] [Google Scholar]
- 16.Guilhabert, M. R., and B. C. Kirkpatrick. 2005. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to Xylella fastidiosa biofilm maturation and colonization and attenuate virulence. Mol. Plant-Microbe Interact. 18:856-868. [DOI] [PubMed] [Google Scholar]
- 17.Gusnanto, A., A. Ploner, and Y. Pawitan. 2005. Fold-change estimation of differentially expressed genes using mixture mixed-model. Stat. Appl. Genet. Mol. Biol. 4:Article 26. [DOI] [PubMed] [Google Scholar]
- 18.Havarstein, L. S., H. Holo, and I. F. Nes. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology 140:2383-2389. [DOI] [PubMed] [Google Scholar]
- 19.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendson, M., A. H. Purcell, D. Chen, C. Smart, M. Guilhabert, and B. Kirkpatrick. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Martinez, R., H. S. Costa, C. K. Dumenyo, and D. A. Cooksey. 2006. Differentiation of strains of Xylella fastidiosa infecting grape, almonds, and oleander using a multiprimer PCR assay. Plant Dis. 90:1382-1388. [DOI] [PubMed] [Google Scholar]
- 23.Hill, B. L., and A. H. Purcell. 1995. Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology 85:1368-1372. [Google Scholar]
- 24.Hobbs, M., E. S. R. Collie, P. D. Free, S. P. Livingston, and J. S. Mattick. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7:669-682. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Q., and J. L. Sherald. 2004. Isolation and phylogenetic analysis of Xylella fastidiosa from its invasive alternative host, porcelain berry. Curr. Microbiol. 48:73-76. [DOI] [PubMed] [Google Scholar]
- 26.Katzen, F., D. U. Ferreiro, C. G. Oddo, M. V. Ielmini, A. Becker, A. Puehler, and L. Ielpi. 1998. Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J. Bacteriol. 180:1607-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 28.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leite, B., P. C. Andersen, and M. L. Ishida. 2004. Colony aggregation and biofilm formation in xylem chemistry-based media for Xylella fastidiosa. FEMS Microbiol. Lett. 230:283-290. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., G. Hao, C. D. Galvani, Y. Meng, L. De la Fuente, H. C. Hoch, and T. J. Burr. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology 153:719-726. [DOI] [PubMed] [Google Scholar]
- 31.Lindow, S. E., and A. H. Purcell. 2004. Role of attachment of Xylella fastidiosa to grape and insects in its virulence and transmissibility, p. 210-213. In Proceedings of the Pierce's Disease Research Symposium. California Department of Food and Agriculture, San Diego, CA.
- 32.Marques, L. L. R., G. P. Manfio, D. M. Reid, H. Ceri, and M. E. Olson. 2001. Xylella fastidiosa are biofilm-forming phytopathogenic bacteria. Phytopathology 91:S58. [Google Scholar]
- 33.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to σE and stress response. J. Bacteriol. 176:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 35.Meng, Y., Y. Li, C. D. Galvani, G. Hao, J. N. Turner, T. J. Burr, and H. C. Hoch. 2005. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noeel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41:1271-1281. [DOI] [PubMed] [Google Scholar]
- 37.Ophir, T., and D. L. Gutnick. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215-1226. [DOI] [PubMed] [Google Scholar]
- 39.Pashalidis, S., L. M. Moreira, P. A. Zaini, J. C. Campanharo, L. M. C. Alves, L. P. Ciapina, R. Z. N. Vencio, E. G. M. Lemos, A. M. Da Silva, and A. C. R. da Silva. 2005. Whole-genome expression profiling of Xylella fastidiosa in response to growth on glucose. OMICS 9:77-90. [DOI] [PubMed] [Google Scholar]
- 40.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 41.Ray, S. K., R. Rajeshwari, Y. Sharma, and R. V. Sonti. 2002. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 46:637-647. [DOI] [PubMed] [Google Scholar]
- 42.Scarpari, L. M., M. R. Lambais, D. S. Silva, D. M. Carraro, and H. Carrer. 2003. Expression of putative pathogenicity-related genes in Xylella fastidiosa grown at low and high cell density conditions in vitro. FEMS Microbiol. Lett. 222:83-92. [DOI] [PubMed] [Google Scholar]
- 43.Shi, X. Y., C. K. Dumenyo, R. Hernandez-Martinez, H. Azad, and D. A. Cooksey. 2007. Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by algU. Appl. Environ. Microbiol. 73:6748-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, A. J. G., F. C. Reinach, P. Arruda, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 45.Smolka, M. B., D. Martins, F. V. Winck, C. E. Santoro, R. R. Castellari, F. Ferrari, I. Brum, E. Galembeck, H. D. C. Filho, M. A. Machado, S. Marangoni, and J. C. Novello. 2003. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics 3:224-237. [DOI] [PubMed] [Google Scholar]
- 46.St. Geme, J. W., III, and D. Cutter. 1995. Evidence that surface fibrils expressed by Haemophilus influenzae type b promote attachment to human epithelial cells. Mol. Microbiol. 15:77-85. [DOI] [PubMed] [Google Scholar]
- 47.Storz, G., and R. Hengge-Aronis (ed.). 2000. Bacterial stress responses. ASM Press, Washington, DC.
- 48.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, et al. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, C. H., M. Gavilanes-Ruiz, Y. Okinaka, R. Vedel, I. Berthuy, M. Boccara, J. W.-T. Chen, N. T. Perna, and N. T. Keen. 2002. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant-Microbe Interact. 15:472-480. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, L. H., M. J. Fath, H. K. Mahanty, P. C. Tai, and R. Kolter. 1995. Genetic analysis of the colicin V secretion pathway. Genetics 141:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.