Abstract
The purpose of this study was the enrichment and phylogenetic identification of bacteria that dechlorinate 4,5,6,7-tetrachlorophthalide (commercially designated “fthalide”), an effective fungicide for rice blast disease. Sequential transfer culture of a paddy soil with lactate and fthalide produced a soil-free enrichment culture (designated the “KFL culture”) that dechlorinated fthalide by using hydrogen, which is produced from lactate. Phylogenetic analysis based on 16S rRNA genes revealed the dominance of two novel phylotypes of the genus Dehalobacter (FTH1 and FTH2) in the KFL culture. FTH1 and FTH2 disappeared during culture transfer in medium without fthalide and increased in abundance with the dechlorination of fthalide, indicating their growth dependence on the dechlorination of fthalide. Dehalobacter restrictus TEA is their closest relative, with 97.5% and 97.3% 16S rRNA gene similarities to FTH1 and FTH2, respectively.
4,5,6,7-Tetrachlorophthalide (commercially designated “fthalide”) is an effective fungicide for rice blast disease, which inhibits melanin biosynthesis and the formation of the mature appressorial cells of the rice blast pathogen on the host plant (5, 16). Fthalide has been reported to be reductively dechlorinated in soil (16) and compost (28), although its fates in paddy soil and the fthalide-dechlorinating bacteria are unknown. Besides fthalide, polychlorinated aromatic compounds are known to be reductively dechlorinated by the bacteria of several phyla. Six strains of Desulfitobacterium spp. of the phylum Firmicutes (2, 3, 6, 10, 23, 29) and Desulfomonile tiedjei DCB-1 of the phylum Proteobacteria (21) can dechlorinate polychlorinated phenols. Three strains of the phylum Chloroflexi can dechlorinate a variety of compounds, including polychlorinated phenols, benzenes, biphenyls, or dibenzo-p-dioxins: Dehalococcoides ethenogenes 195 (9, 19), Dehalococcoides sp. strain CBDB1 (1, 4), and strain DF-1 of Chloroflexi, collectively called the “o-17/DF-1 group” (18). Dehalococcoides spp. utilize hydrogen as an electron donor and acetate as a carbon source for growth coupled to the reductive dechlorination of chlorinated compounds (1, 12, 13, 19, 26). In contrast, Desulfitobacterium spp. can dechlorinate chlorinated compounds not only with hydrogen, but also organic acids, such as formate, pyruvate, lactate, or butyrate (3, 10, 23). Strain DF-1 can utilize hydrogen and formate for the dechlorination of polychlorinated biphenyls (PCBs) (18).
In this study, bacteria that dechlorinate fthalide were enriched from a paddy soil with sequentially transferred cultures using a soil-free medium supplemented with single organic acids. Acetate, formate, lactate, and butyrate were used in this study because they are frequently used in the enrichment of dechlorinators and release hydrogen at different concentrations (8, 11, 14). Fthalide-dechlorinating bacteria in the enriched culture were phylogenetically identified based on the 16S rRNA gene with PCR-denaturing gradient gel electrophoresis (DGGE), a 16S rRNA gene clone library, and quantitative real-time PCR (qPCR).
MATERIALS AND METHODS
Chemicals.
Fthalide was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan); 4,5,6-trichlorophthalide (4,5,6-TCPH), 4,5,7-trichlorophthalide (4,5,7-TCPH), 4,7-dichlorophthalide (4,7-DCPH), 4,6-dichlorophthalide (4,6-DCPH), and 4-chlorophthalide (4-MCPH) were gifts from Kureha Corp. (Tokyo, Japan).
Enrichment of fthalide-dechlorinating bacteria.
The paddy soil sample used as the inoculum was obtained from the ploughed layer of a paddy field located at Yatomi-cho in Aichi Prefecture, Japan, as described previously (36). Aliquots (200 μl) of 5 mM fthalide dissolved in acetone were put into dry sterilized glass vials (60 ml capacity) and crystallized by vacuum aspiration. Paddy soil samples (10 g wet weight) were placed in the glass vials, which were filled with 10 ml of distilled water containing 1 mg liter−1 resazurin. The samples were sparged with nitrogen gas for 15 min, neutralized to pH 7.0 with HCl, and sparged again with nitrogen gas for 15 min. The vials were closed with Teflon-coated butyl rubber caps and incubated at 30°C. Acetate, formate, lactate, or butyrate (each 20 mM) was added to the culture as the electron donor and carbon source. After incubation for 1 month, 500-μl aliquots of the incubated samples were transferred to 10 ml of a defined medium, designated “FTH medium,” which contained the following components (per liter): 1 g NaCl; 0.5 g KCl; 0.5 g NH4Cl; 0.1 g CaCl2 · 2H2O; 0.1 g MgCl2 · 6H2O; 0.2 g KH2PO4; 1 ml of 1 mg liter−1 resazurin solution; 1 ml of trace element solution SL10 (31); 10 ml of vitamin solution (15); 1 ml of Se/W solution (31); 10 ml of titanium(III) trinitriloacetic acid solution containing 25 mM titanium(III) and 100 mM trinitriloacetic acid (20); 10 ml of 0.2 mmol liter−1 FeS solution (7); 20 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.2); and 20 mM each acetate, formate, lactate, and butyrate. Yeast extract (0.01% to 0.0001%) was added to the transferred culture in decreasing concentrations until the 10th transfer. The transferred cultures were incubated for 10 to 15 days, and 5% of the culture was transferred when more than 90% of the fthalide was reduced. The enrichment culture supplemented with lactate, designated the “KFL culture,” was used in the following experiments. The 12th transferred KFL culture was also incubated in 100 ml in a large glass vial (600-ml capacity) to monitor the metabolites of lactate and fthalide and the biomass over time.
Effects of e− donors, e− acceptors, and inhibitory treatments on KFL culture.
The effects of different electron donors were examined in serially diluted cultures of the 11th transferred KFL culture. The KFL culture was cultured with 20 mM pyruvate, 20 mM acetate, 100% H2 (headspace) plus 10 mM acetate, or 80% H2 plus 20% CO2 (headspace) instead of lactate. Peptone (0.01%) was also added in some cases. SO42−, NO3−, or Fe(III) (each 10 mmol liter−1) was added to the medium as a 1% (vol/vol) concentration of 1 mol liter−1 stock solution. Fe(III) was added as Fe(OH)3, which was prepared as described previously (17). The effects of the following inhibitors on the dechlorination of fthalide were evaluated in the 11th transferred KFL culture: 50 mg liter−1 chloramphenicol, 50 mg liter−1 vancomycin, 5 mM bromoethanesulfonate (BES), or 10 mM sodium molybdate. The 11th transferred KFL cultures were occasionally steam sterilized at 121°C for 15 min or incubated at 80°C for 30 min before incubation.
Analytical procedures.
Fthalide and its metabolites were identified and quantified with a gas chromatography-mass spectrometry (GC-MS) system (Shimadzu, Kyoto, Japan) equipped with a DB-5MS column (J&W Scientific, Folsom, CA). In the sample preparation for GC-MS, 1-ml samples of the cultures were mixed with an equal volume of acetonitrile and then separated by mixing with 1 ml of ethyl acetate. The organic solvent phase was recovered and dehydrated by the addition of 1 g of anhydrous sodium sulfate. The extraction efficiencies for chlorinated phthalides using this procedure were in the range of 95% to 98%. An aliquot (2 μl) of the extract was injected manually into the GC-MS system with splitless injection. Helium was used as the carrier gas at a flow rate of 1.0 ml min−1. The injection and interface temperatures were maintained at 280°C and 320°C, respectively. The column temperature was initially set at 140°C for 0.25 min and then increased at a rate of 10°C min−1 to 320°C, where it was maintained for 3 min. The electron impact ionization mode was set at 70 eV. Chlorophthalides were identified in full-scan mode (m/z 50 to 400), and were quantified in selected ion mode (m/z 104, 139, 173, 207, and 243). Organic acids (i.e., lactate and its metabolites) were analyzed by injection of 20-μl aliquots of the filtered samples to a reversed-phase high-performance liquid chromatograph (Shimadzu) equipped with an ODS column (L-column; CERI, Tokyo, Japan), as described previously (36). The gaseous metabolites of lactate were analyzed with a GC-14B gas chromatograph (Shimadzu) equipped with a thermal conductivity detector and stainless-steel columns packed with Molecular Sieve 5A (GL-Science, Tokyo, Japan) or active carbon (GL-Science), as described previously (36).
Total cell counts.
The cells in culture were diluted with 0.20-μm filter-sterilized PBSE buffer and collected on a 0.22-μm polycarbonate black membrane filter (Millipore). Cells were stained with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) special packaging (Molecular Probes, Eugene, OR) and were observed under an epifluorescence microscope (Olympus, Tokyo, Japan) equipped with a DP7 digital camera (Olympus). About 1,000 DAPI-stained cells from 10 fields per sample were counted using the WinROOF program (Flovel Co., Tokyo, Japan).
DNA extraction.
DNA was extracted from 10 ml of the microcosm and the enriched culture by bead beating using the DNA extraction kit ISOIL for bead beating (Nippon Gene Co., Tokyo, Japan), according to the manufacturer's instructions.
PCR-DGGE.
PCR-DGGE and subsequent cloning of the major DGGE bands were performed as described previously (36). For PCR-DGGE, bacterial 16S rRNA gene fragments were amplified with primer 357f with a GC clamp at the 5′ terminus and primer 517r (22). PCR was performed by preheating the mixture to 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min. DGGE was performed with the Bio-Rad DCode system (Bio-Rad Laboratories, Hercules, CA) with polyacrylamide gel containing denaturant in the range from 40% to 60% at 100 V for 16 h in 0.5× Tris-acetate-EDTA buffer at 58°C. The major DGGE fragments were excised from the gel and cloned with the pT7Blue perfectly blunt cloning kit (Novagen, Madison, WI).
16S rRNA gene clone library and RFLP analysis.
Nearly full-length 16S rRNA gene fragments were PCR amplified with a pair of bacterial consensus primers, 27f and 1492r (30), from DNAs extracted from the 12th transferred KFL culture incubated with and without fthalide. PCR was performed under the same thermal cycling conditions used for PCR-DGGE, except with an annealing temperature of 55°C and 30 cycles. The PCR products were cloned as described above. The inserted regions were amplified from the plasmids by PCR using the T7 promoter forward primer and U19mer reverse primer. The PCR products (200 ng) were digested individually with 5 U each of endonucleases HhaI (Takara Bio, Otsu, Japan), HaeIII (Takara Bio), and SspI (Takara Bio) for 1 h at 37°C. SspI was used to distinguish the two phylotypes FTH1 and FTH2. The digested clones were separated electrophoretically on 3% agarose gel and visualized with ethidium bromide under UV illumination. The clones were categorized as phylotypes on the basis of their distinct restriction fragment length polymorphisms (RFLPs).
Sequencing and phylogenetic analysis.
The DGGE clones and representative clones with a unique RFLP pattern were sequenced with the BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster, CA) using an ABI 3100 DNA sequencer (Applied Biosystems). The DNA sequences were analyzed with the Genetyx program version 7 (Software Development Co., Tokyo, Japan) and aligned with 16S rRNA sequences obtained from the NCBI BLAST database using the BLAST search program (http://www.ncbi.nlm.nih.gov/BLAST/).
PCR detection of Dehalococcoides spp.
To check the presence of Dehalococcoides spp. in the KFL culture, the 16S rRNA gene fragments were PCR amplified using the primers DHC793f and DHC946r, which specifically amplify about 150 bp of the target region from the 16S rRNA genes of Dehalococcoides spp. and the “o-17/DF-1 group” (34, 35). These have been modified from the primers DHC 774 and DHC 946, respectively, designed previously for the detection of Dehalococcoides spp. (24). As the positive control, we used a cloned 1,434-bp amplicon of a 16S rRNA gene with 100% similarity to that of Dehalococcoides ethenogenes strain 195, which was obtained from an enrichment culture that dechlorinated 1,2-dichloroethane to ethene. The amplification of the positive control for the “o-17/DF-1 group” was not tested because no adequate control sample was available. PCR was performed using the thermal cycling program used for PCR-DGGE.
qPCR.
qPCR targeting FTH1 and FTH2, corresponding to fthalide-dechlorinating bacteria, was performed with DNAs extracted from the 12th transferred KFL culture and the following specific primers. Two forward primers, Dhb455IF (5′-CAAGCAATATTTTCACTTAAGC-3′) and Dhb456IIF (5′-CCAGTATGTAAATAATGTACTG-3′), were specific for FTH1 and FTH2, respectively, and were used in combination with the reverse primer Dhb645R (5′-CCTCTCCTGTCCTCAAGACT-3′), which is modified from the primer Dre645R specific for Dehalobacter sp., as reported previously (24). The bacterial universal primer set 357f and 517r was also used for the comparative amplification of the total bacterial 16S rRNA genes (22). qPCR was performed with a LightCycler FastStart DNA Master SYBR green I kit (Roche Molecular Biochemicals, Indianapolis, IN) and the LightCycler system (Roche Diagnostics, Mannheim, Germany), as described previously (35). Calibration curves (log DNA concentration versus an arbitrarily set cycle threshold value) for the estimation of the total and specific bacterial 16S rRNA gene copy numbers were constructed using serial dilutions of the amplicons of FTH1 and FTH2. The gene copy number of the amplicon was calculated by multiplying the molar concentration of amplicon by the Avogadro constant. The calibration curves were graphed and the copy numbers calculated with LightCycler software version 3.5 (Roche Diagnostics). The populations of bacteria corresponding to FTH1 and FTH2 were estimated using total cell counts, and the proportions of FTH1 and FTH2 in the total bacterial 16S rRNA genes were calculated by assuming that the copy number of the rRNA gene per genome was 1.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited under DDBJ/EMBL/GenBank (http://www.ddbj.nig.ac.jp/Welcome-j.html) accession no. AB294740 to AB294749.
RESULTS
Enrichment of bacteria that dechlorinate fthalide.
The paddy soils incubated individually with acetate, formate, lactate, or butyrate dechlorinated 50 mol liter−1 fthalide within 30 days, whereas the transferred cultures with acetate or butyrate suddenly lost the capacity to dechlorinate fthalide after the second transfer (data not shown). Although the enrichment culture with formate lost its dechlorination activity in the 10th transfer when not supplemented with yeast extract, the culture with lactate maintained its activity for more than 12 sequential transfers (data not shown). The enrichment culture with lactate, designated the “KFL culture,” was used in subsequent experiments.
Dechlorination of fthalide by KFL culture.
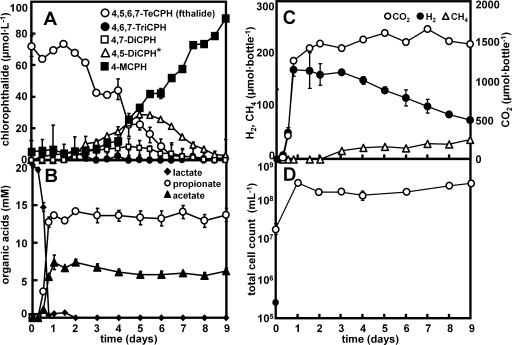
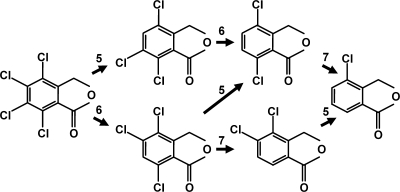
The KFL culture dechlorinated 100 μmol liter−1 fthalide to 4-MCPH within 10 days. The appearance of 4,6,7-TCPH, 4,7-DCPH, an unidentified dichlorophthalide, and 4-MCPH corresponded to the decrease in the concentration of fthalide in the KFL culture (Fig. 1A). The unidentified dichlorophthalide appeared at retention times different from those of the available standards for 4,6-DCPH and 4,7-DCPH and was considered to be 4,5-dichlorophthalide (4,5-DCPH), because it is the only dichlorophthalide with chloride at the 4 position that differed from 4,7-DCPH and 4,6-DCPH. 4,5-DCPH was tentatively quantified in this study using the calibration curve for 4,7-DCPH. 4-MCPH was not dechlorinated, even after incubation for 3 months. When the KFL culture was incubated with fthalide at a much higher concentration, 600 μmol liter−1, 4,5,7-TCPH appeared (data not shown), indicating two different pathways of fthalide dechlorination to 4,6,7-TCPH or 4,5,7-TCPH. Propionate, acetate, H2, CO2, and CH4 were detected as the metabolites of lactate (Fig. 1B and C). The total cell number in the KFL culture, as measured by DAPI staining, increased within 1 day from (1.7 ± 0.5) × 107 to (2.5 ± 0.9) × 108 cells ml−1 during the transformation of lactate and then was relatively stable at an order of magnitude of 108 cells ml−1, with only minor changes during the dechlorination of fthalide (Fig. 1D). The possible dechlorination pathways of fthalide in the KFL culture were determined, as shown in Fig. 2.
FIG. 1.
Dechlorination of fthalide by the KFL culture supplemented with 20 mM lactate. (A) Changes in the concentrations of fthalide and its dechlorinated metabolites. (B) Lactate and the transformed organic acids. (C) Gaseous metabolites. (D) Total cell counts. The data are averages and standard deviations of determinations from three different culture bottles, independently prepared in parallel. See Materials and Methods for the abbreviations of the chemicals. *, 4,5-DCPH was tentatively quantified using the calibration curve for 4,7-DCPH, because no chemical standard is available.
FIG. 2.
Possible pathways by which fthalide is dechlorinated in the KFL culture.
Effects of electron donors.
The serially diluted KFL cultures dechlorinated fthalide with 20 mM lactate and H2 at dilutions of 10−5, whereas the activity was lost with 5 mM lactate, 20 mM acetate, 20 mM pyruvate, or 20 mM propionate at dilutions of 10−1 or 10−2. The KFL culture dechlorinated fthalide with H2 only in the presence of 0.01% peptone diluted 10−6, whereas it did not dechlorinate fthalide with only 0.01% peptone. Based on these results and the changes in the concentrations of lactate and its metabolites (Fig. 1), the reactions that transformed lactate, fthalide, and their metabolites are expected to be as follows (27): (i) 3 lactate− → 2 propionate− + acetate− + HCO3− + H+, ΔG°′ = −165 kJ reaction−1 (pH 7.0); (ii) lactate− + 2H2O → acetate− + HCO3− + H+ + 2H2, ΔG°′ = −4.2 kJ reaction−1 (pH 7.0); (iii) fthalide + 3H2 → 4-CPH + 3H+ + 3Cl−, with ΔG°′ of chlorophthalides not available; (iv) 4H2 + HCO3− + H+ → CH4 + 3H2O, ΔG°′ = −136 kJ reaction−1 (pH 7.0); and (v) acetate− + H2O → HCO3− + CH4, ΔG°′ = −31.0 kJ reaction−1 (pH 7.0).
Effects of inhibitory treatments.
The dechlorination of fthalide was completely inhibited in the KFL cultures spiked with chloramphenicol, vancomycin, BES, molybdate, or heating at 80°C for 30 min in the KFL cultures (Table 1). The 4-MCPH detected in all samples was carried over from the dechlorination products present in the 5% (vol/vol) transferred inocula. SO42− or NO3− made the fthalide dechlorination slower, and Fe(OH)3 completely inhibited the fthalide dechlorination in the KFL culture.
TABLE 1.
Effects of inhibitory treatments and electron acceptors on fthalide dechlorination in KFL cultures
| Treatment | Concn (mol liter−1) of:
|
||||
|---|---|---|---|---|---|
| Fthalide | 4,6,7-TCPH | 4,7-DCPH | 4,5-DCPH | 4-MCPH | |
| None | 0.6 ± 0.9 | 0.0 ± 0.1 | 3.2 ± 0.1 | 0.3 ± 0.4 | 98 ± 3.3 |
| Autoclave | 71 ± 22 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.5 ± 0.8 | 8.0 ± 2.2 |
| Heating at 80°C (30 min) | 50 ± 16 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 2.5 ± 2.9 |
| Chloramphenicol | 44 ± 4.6 | 0.7 ± 1.0 | 0.7 ± 1.1 | 0.0 ± 0.0 | 4.6 ± 0.1 |
| Vancomycin | 45 ± 1.7 | 0.25 ± 0.36 | 0.44 ± 0.63 | 0.27 ± 0.39 | 4.4 ± 0.66 |
| Molybdate | 44 ± 11 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 4.7 ± 7.0 |
| BES | 55 ± 26 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 5.5 ± 1.0 |
| Ampicillin | 39 ± 10 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 12 ± 1.0 |
| NO3 | 29 ± 6.7 | 12 ± 1.8 | 19 ± 4.8 | 4.8 ± 5.3 | 7.3 ± 1.7 |
| SO4 | 20 ± 1.0 | 13 ± 4.8 | 30 ± 5.8 | 7.6 ± 5.3 | 11 ± 1.7 |
| Fe(III) | 39 ± 9.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 6.0 ± 1.9 |
Microbial community analysis based on 16S rRNA genes.
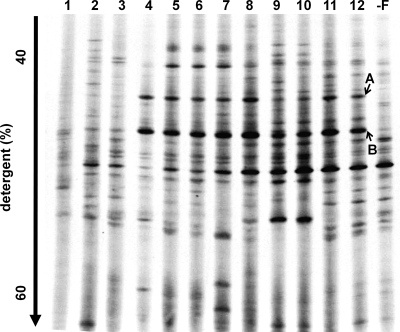
The succession of microbial communities in the KFL cultures during transfer was determined by PCR-DGGE targeted to the domain Bacteria (Fig. 3). The microbial community structure changed dramatically during the first four transfers. The earlier transferred cultures were too diverse to identify the dominant bacteria because of the carryover from the soil. However, after the fifth transfer, the microbial community became relatively stable, with minor changes in the subsequent incubations. Two major fragments, A and B, always appeared in the KFL culture and disappeared in the nonfthalide culture, which had been transferred twice without fthalide (“−F” culture shown in Fig. 3). These two major bands were regarded as the major bacteria dependent for growth on the dechlorination of fthalide. The two bands A and B had 94.9% and 95.9% similarity to the 16S rRNA sequence of Dehalobacter restrictus strains PER-K23 (15) and TEA (32), respectively, in the phylum Firmicutes, their closest relatives, which have previously been isolated as bacteria that dechlorinate tetrachloroethene.
FIG. 3.
DGGE fragment patterns of the 16S rRNA genes, targeted to the domain Bacteria, amplified from the total DNAs of the KFL cultures during the sequential transfer of the cultures. The number given on each DGGE pattern is the number of transfers. “−F” indicates a culture transferred twice without fthalide, produced using the 10th transferred KFL culture as the inoculum. Bands A and B were cloned and sequenced.
To obtain the nearly complete sequences of the 16S rRNA genes of the dominant bacteria in the KFL culture, two clone libraries were constructed using 16S rRNA gene fragments amplified from the KFL cultures incubated with and without fthalide (corresponding to lanes 12 and “−F,” respectively, in Fig. 3). Fifty clones from each culture (100 clones in total) were isolated and were classified into eight phylotypes based on their RFLP patterns: two phylotypes (phylotype 1 and phylotype 2) of the genus Dehalobacter, five phylotypes of Clostridium sp., and one phylotype of Sedimentibacter sp. (Table 2). Phylotype 1 and phylotype 2 only appeared in the KFL culture supplemented with fthalide and had 100% similarities to the sequences of DGGE bands B and A, respectively. According to these results, bacteria corresponding to phylotype 1 and phylotype 2 grew on the dechlorination of fthalide in the KFL culture. Phylotype 1 and phylotype 2 were designated FTH1 and FTH2, respectively, to easily distinguish the bacteria that dechlorinate fthalide in this study. The closest relative of FTH1 and FTH2 is Dehalobacter restrictus TEA (32), with which they shared 16S rRNA gene similarities of 97.5% and 97.3%, respectively. FTH1 and FTH2 are themselves 99.0% similar. No fragment was observed in the PCR amplification targeting Dehalococcoides sp. using DNAs extracted from the KFL culture, although the positive control yielded amplicons of the expected size (data not shown).
TABLE 2.
Phylogenetic identification of phylotypes detected in 16S rRNA gene clone libraries constructed from the 12th KFL cultures incubated with and without fthalide
| Phylotype | Frequency (%)a
|
Closest relative | Similarity (%) | Accession no. | |
|---|---|---|---|---|---|
| +F | −F | ||||
| 1 | 44 | 0.0 | Dehalobacter restrictus TEA | 97.5 | U84497 |
| 2 | 12 | 0.0 | Dehalobacter restrictus TEA | 97.3 | U84497 |
| 3 | 12 | 10 | Sedimentibacter sp. strain C7 | 97.4 | AY766466 |
| Sedimentibacter saalensis ZF2T | 96.5 | AJ404680 | |||
| 4 | 6.0 | 42 | Uncultured bacterium TANB101 | 99.5 | AY667266 |
| Clostridium hemolyticum DSM 5565T | 96.1 | Y18173 | |||
| 5 | 24 | 6.0 | Clostridium pascui DSM 10365T | 93.7 | X96736 |
| 6 | 0.0 | 30 | Uncultured bacterium IB-18 | 99.0 | AJ488078 |
| Clostridium methylpentosum DSM 5476T | 90.9 | Y18181 | |||
| 7 | 2.0 | 8.0 | Clostridium propionicum DSM 1682T | 96.9 | X77841 |
| 8 | 0.0 | 4.0 | Uncultured bacterium Ac051 | 94.9 | AY336985 |
| Clostridium aldrichii DSM 6159T | 89.5 | X71846 | |||
+F, fthalide-containing culture; −F, culture not containing fthalide.
Population dynamics of phylotypes FTH1 and FTH2 in the KFL culture.
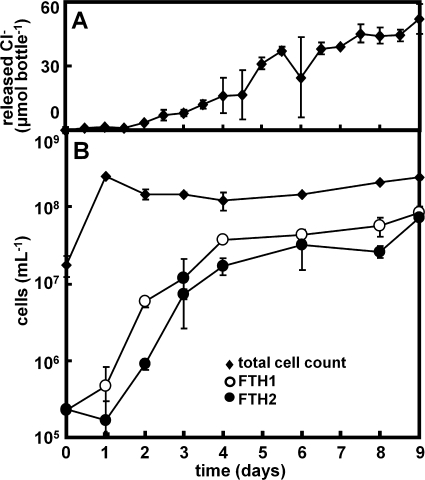
The relative proportions of FTH1 and FTH2 in the total bacterial 16S rRNA of the KFL culture showed similar dynamics and were about 1% at days 0 and 1 and increased to about 30% at days 4 to 9, corresponding to 2.3 × 105 cells ml−1 at day 0 to 8.3 × 107 cells ml−1 at day 9 for FTH1 and 2.3 × 105 cells ml−1 at day 0 to 7.1 × 107 cells ml−1 at day 9 for FTH2 (Fig. 4). The population dynamics of FTH1 and FTH2 correlate well with the dechlorination activity determined from the calculated amount of chloride released from fthalide (Fig. 4). The r2 linear correlation coefficients between the calculated amount of chloride released from fthalide and the populations of FTH1 and FTH2 were 0.79 and 0.74, respectively, although the correlation with the total counts of DAPI-stained cells was 0.19 (data not shown). These results clearly indicate that the growth of FTH1 and FTH2 depends on the dechlorination of fthalide.
FIG. 4.
Population dynamics of Dehalobacter sp. phylotypes FTH1 and FTH2 in the KFL culture. (A) The amount of chloride released from fthalide was calculated from the concentration of the dechlorinated products of fthalide. (B) The total cell count was determined by directly counting the DAPI-stained cells, and the populations of FTH1 and FTH2 were estimated using total cell counts and the proportions of FTH1 and FTH2 in the total bacterial 16S rRNA genes, assuming that the copy number of the rRNA gene per genome was 1.
DISCUSSION
In this study, fthalide dechlorination and the involvement of novel phylotypes FTH1 and FTH2 of the genus Dehalobacter were demonstrated in a sequentially transferred culture of paddy soil. The pathway of fthalide dechlorination in the KFL culture was the same as that in the paddy soil or in the transferred cultures containing formate, acetate, or butyrate and was almost identical to that previously reported in compost, in which fthalide was dechlorinated to 4,7-DCPH and a minor 4-MCPH component via the 4,6,7-TCPH products (28). However, it differs from the pathway observed in soil, which dechlorinates fthalide to 4,6-DCPH via 4,6,7-TCPH (16). No oxidation or thiomethylation of fthalide was observed in the original paddy soil or the KFL culture, although they have been observed in compost at different stages of maturation (28). The disappearance of the fthalide dechlorination from the transferred culture containing butyrate, acetate, or formate was probably caused by a lack of hydrogen production or trace nutrients. The deficiency of hydrogen probably also occurred in the KFL culture with 5 mM lactate, because most of the lactate was converted to propionate without the production of hydrogen in the KFL culture.
Bacteria corresponding to the phylotypes FTH1 and FTH2 of Dehalobacter spp. grew in the KFL culture using fthalide. Dehalobacter spp. are known obligate halorespiring bacteria, which only grow with the reductive dechlorination of chlorinated compounds, as do Dehalococcoides spp. In the genus Dehalobacter, three strains have been isolated: D. restrictus strain PER-K23 (15) and D. restrictus strain TEA (32), which grow on the dechlorination of tetrachloroethene and trichloroethene; and Dehalobacter sp. strain TCA1 (25), which grows on the dechlorination of 1,1,2-trichloroethane and 1,1-dichloroethane. There have been no reports of the capacities of isolates of Dehalobacter spp. to dechlorinate chlorinated aromatic compounds. Recently, Dehalocobacter spp. were detected in a sediment culture containing Dehalococcoides sp. as the dominant bacteria that dechlorinated 2,3,4,5-tetrachlorobiphenyl (2,3,4,5-TeCB) (33). The KFL culture also dechlorinated 2,3,4,5-TeCB (data not shown) but did not contain Dehalococcoides spp. and the “o-17/DF-1 group,” known dechlorinators of PCBs. These results suggested the involvement of Dehalobacter spp. in the dechlorination of PCBs, although other dechlorinators that could grow independently on fthalide dechlorination or existed as minor populations were possibly missed in this study. Fthalide may be a good alternative with which to enrich bacteria that dechlorinate PCBs because of its aromatic structure and higher solubility in water (10 μM).
Microbial community analysis of the KFL culture revealed a predominance of Clostridium sp. and Sedimentibacter sp., together with FTH1 and FTH2. The KFL culture required peptone for the fthalide dechlorination when the cultures were supplemented with hydrogen but not with lactate. Dehalobacter restrictus strain PER-K23 is reported to require arginine, histidine, and threonine for growth (15). These results suggest that Clostridium sp. and Sedimentibacter sp. present with Dehalobacter sp. in the KFL culture probably supply hydrogen and the trace nutrients essential for the growth of Dehalobacter sp.
In conclusion, we obtained a soil-free enrichment culture of bacteria that dechlorinate fthalide from a paddy soil and for the first time demonstrated that Dehalobacter sp. grows on a chlorinated aromatic compound, fthalide.
Acknowledgments
We thank the Aichi-ken Agricultural Research Center for collecting the paddy soils and Kureha Corp. for providing the chlorophthalides.
This research was partly supported by the Environmental Technology Development Fund of the Ministry of the Environment; a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (B2:17310045); the New Energy and Industrial Technology Development Organization; and the Kurita Water and Environment Foundation.
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Gorisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard, B., R. Beaudet, R. Villemur, G. McSween, F. Lépine, and J.-G. Bisaillon. 1996. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int. J. Syst. Bacteriol. 46:1010-1015. [DOI] [PubMed] [Google Scholar]
- 3.Breitenstein, A., A. Saano, M. Salkinoja-Salonen, J. R. Andreesen, and U. Lechner. 2001. Analysis of a 2,4,6-trichlorophenol-dehalogenating enrichment culture and isolation of the dehalogenating member Desulfitobacterium frappieri TCP-A. Arch. Microbiol. 175:133-142. [DOI] [PubMed] [Google Scholar]
- 4.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Gorisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 5.Chida, T., T. Uekita, K. Stake, K. Hirano, K. Aoki, and T. Noguchi. 1982. Influence of fthalide in penetration process of Pyricularia oryzae. Nippon Shokubutsu Byori Gakkaiho 48:58-63. (In Japanese.) [Google Scholar]
- 6.Christiansen, N., and B. K. Ahring. 1996. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46:442-448. [Google Scholar]
- 7.Ehrenreich, A., and F. Widdel. 1994. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl. Environ. Microbiol. 60:4517-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fennell, D. E., J. M. Gossett, and S. H. Zinder. 1997. Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ. Sci. Technol. 31:918-926. [Google Scholar]
- 9.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 10.Gerritse, J., V. Renard, T. M. Pedro-Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 11.Gibson, S. A., and G. W. Sewell. 1992. Stimulation of reductive dechlorination of tetrachloroethene in anaerobic aquifer microcosms by addition of short-chain organic acids and alcohols. Appl. Environ. Microbiol. 58:1392-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, J., K. Ritalahti, K. Yang, S. Koenigsberg, and F. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 13.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. Loffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 14.Heimann, A. C., D. J. Batstone, and R. Jakobsen. 2006. Methanosarcina spp. drive vinyl chloride dechlorination via interspecies hydrogen transfer. Appl. Environ. Microbiol. 72:2942-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and J. A. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 16.Ishida, M., and K. Nambu. 1975. Fthalide (Rabcide). Nouyakukagaku 3:10-26. (In Japanese.) [Google Scholar]
- 17.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May, H. D., G. S. Miller, B. V. Kjellerup, and K. R. Sowers. 2008. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl. Environ. Microbiol. 74:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maymó-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 20.Moench, T. T., and J. G. Zeikus. 1983. An improved preparation method for a titanium (III) media reductant. J. Microbiol. Methods 1:199-202. [Google Scholar]
- 21.Mohn, W. W., and K. J. Kennedy. 1992. Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1. Appl. Environ. Microbiol. 58:1367-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanford, R. A., J. R. Cole, F. E. Löffler, and J. M. Tiedje. 1996. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling the oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl. Environ. Microbiol. 62:3800-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smits, T. H., C. Devenoges, K. Szynalski, J. Maillard, and C. Holliger. 2004. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57:369-378. [DOI] [PubMed] [Google Scholar]
- 25.Sun, B., B. N. Griffin, H. L. Ayala-del-Rio, S. A. Hashsham, and J. M. Tiedje. 2002. Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298:1023-1025. [DOI] [PubMed] [Google Scholar]
- 26.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. E. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuda, T., M. Nishiki, H. Hoshi, K. Shinoda, M. Ishida, and T. Misato. 1976. Metabolic fate of fthalide (4,5,6,7-tetra-chlorophthalide) in compost. J. Pestic. Sci. 1:283-294 (In Japanese.) [Google Scholar]
- 29.Utkin, I., C. Woese, and J. Wiegel. 1994. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int. J. Syst. Bacteriol. 44:612-619. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widdel, F. G., W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-293. [Google Scholar]
- 32.Wild, A., R. Hermann, and T. Leisinger. 1996. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511. [DOI] [PubMed] [Google Scholar]
- 33.Yan, T., T. M. LaPara, and P. J. Novak. 2006. The effect of varying levels of sodium bicarbonate on polychlorinated biphenyl dechlorination in Hudson River sediment cultures. Environ. Microbiol. 8:1288-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, N., K. Asahi, Y. Sakakibara, K. Miyake, and A. Katayama. 2007. Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes. J. Biosci. Bioeng. 104:91-97. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida, N., N. Takahashi, and A. Hiraishi. 2005. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 71:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, N., Y. Yoshida, Y. Handa, H.-K. Kim, S. Ichihara, and A. Katayama. 2007. Polyphasic characterization of a PCP-to-phenol dechlorinating microbial community enriched from paddy soil. Sci. Total Environ. 381:233-242. [DOI] [PubMed] [Google Scholar]