Abstract
The proper design of DNA microarray experiments requires knowledge of biological and technical variation of the studied biological model. For the filamentous fungus Aspergillus niger, a fast, quantitative real-time PCR (qPCR)-based hierarchical experimental design was used to determine this variation. Analysis of variance components determined the contribution of each processing step to total variation: 68% is due to differences in day-to-day handling and processing, while the fermentor vessel, cDNA synthesis, and qPCR measurement each contributed equally to the remainder of variation. The global transcriptional response to d-xylose was analyzed using Affymetrix microarrays. Twenty-four statistically differentially expressed genes were identified. These encode enzymes required to degrade and metabolize d-xylose-containing polysaccharides, as well as complementary enzymes required to metabolize complex polymers likely present in the vicinity of d-xylose-containing substrates. These results confirm previous findings that the d-xylose signal is interpreted by the fungus as the availability of a multitude of complex polysaccharides. Measurement of a limited number of transcripts in a defined experimental setup followed by analysis of variance components is a fast and reliable method to determine biological and technical variation present in qPCR and microarray studies. This approach provides important parameters for the experimental design of batch-grown filamentous cultures and facilitates the evaluation and interpretation of microarray data.
Culturing filamentous organisms such as Aspergillus niger is difficult to reproduce compared to culturing unicellular organisms. Filamentous growth is characterized by the elongation and branching of hyphae, cylindrical cells that increase in length by growth at one end. De novo biosynthesis and active enzyme production occur mainly at the hyphal tips. In regions of distance from the tip, the hyphae age and become biologically less active (38). This hyphal growth is the result of adaption to the habitat of the organism, which enables it to spread over and penetrate surfaces and cross over nutrient-depleted gaps (6). However, under laboratory conditions, attachment of fungal mycelium to fermentor baffles and other extremities introduces heterogeneous growth that can be suppressed only to a certain extent (for instance, by cooling the fermentor headplate). The growing mycelium increases the culture broth viscosity, which reduces the mass transport of nutrients, oxygen, and heat, and affects the mixing characteristics in a fermentor or shake flask over time. Physical agitation and shear stress can cause uncontrolled breakage and fragmentation of the mycelia (37).
Recent technologies such as global transcriptome analysis by DNA microarrays or quantitative real-time PCR (qPCR) require the use of replicate biological samples for high-quality data. Given the difficulties in culturing A. niger, obtaining transcript data without wasting resources requires proper experimental design. The key to designing such experiments is to determine how much replication is needed—the sample size (36). A larger amount of replicates leads to increased statistical accuracy of measurement, whereas insufficient replication impedes data analysis. The required number of independent samples depends on a variety of factors as follows: the organism under study and its biological variation, the magnitude of the expected gene response, the statistical power to detect the genuine gene response to a condition, and the false discovery rate (13, 36).
Consensus is emerging on what comprises a “proper microarray experiment” (13). According to this consensus, defining the experimental objectives and requirements is a necessity before actually starting an experiment (39). During the experiment, standardized protocols and methods reduce the variability that is introduced in each process step (27). A selected experimental design makes data analysis and interpretation as simple and as powerful as possible. Finally, for publication of the microarray results and data, the minimum information about a microarray experiment (MIAME) guidelines (4) are adopted. Notwithstanding this consensus, Jafari and Azuaje have published an extensive review on papers describing microarray methods and applications of microarray technology (15) and concluded that recently published gene expression data analysis studies often lack key information that is required to interpret and evaluate published data.
The goal of the present study was to minimize the variation in A. niger batch fermentations by optimization of protocols and procedures. The variation between fermentations was determined with an analysis of variance components of data obtained by qPCR. This relatively inexpensive technology is used to measure transcript levels for few genes in many samples simultaneously. Furthermore, qPCR is routinely used to validate microarray results (11, 21).
The effects of the optimization and quality control measures for our experimental setup were assessed by examination of the global transcriptional response toward induction with d-xylose. The xylanolytic system of A. niger is under the control of the transcriptional activator XlnR, and the genes under its control are well documented (34). Recently, the transcriptional response toward d-xylose was examined by microarray analysis for three Aspergillus species (2). The availability of these data on the transcriptional response toward d-xylose allows for validation of the biological response observed during our studies.
MATERIALS AND METHODS
Strain and spore preparations.
A. niger 872.11 (ΔargB pyrA6 prtF28 goxC17 cspA1) is derived from CBS 120.49. All media were based on minimal medium (25), contained 100 mM sorbitol as the carbon source, and were supplemented with uridine and arginine. To obtain spores, 20 spores per mm2 were plated onto complete medium plates (25), incubated for 5 days at 30°C, and allowed to mature at 4°C for 24 h. The spore suspension was washed and stored at 4°C until use.
Fermentation.
Four 2.5-liter glass fermentors (Applikon) with 2.2 liters of minimal medium were kept at a constant temperature of 30 ± 0.5°C, while fermentor headplates were kept at 8°C. A total of 1.0 × 106 spores per ml were added to the fermentor. During germination, each fermentor was aerated through the headspace (50 liters/h) and stirred at 300 rpm. When dissolved oxygen levels dropped below 60% for over 5 min, the stirrer speed was set to 750 rpm, and aeration was switched to sparger inlet. This switching time point was defined as T equals 0 h. Each fermentor was induced with either 0.1 mM sorbitol or d-xylose at T equals 14 h. Samples were snap-frozen in liquid nitrogen directly after filtration, with less than 20 s between sampling and snap-freezing.
RNA isolation.
Frozen mycelium was ground for 40 s using a dismembrator (Braun Melsungen). A Trizol-chloroform extraction preceded total RNA extraction with RNeasy minicolumns (Qiagen), according to the manufacturer's protocol for yeast. RNA integrity was assessed with an Experion system (Bio-Rad) by visual inspection of the electropherograms. Graphs depicting RNA integrity categories were used as visual aids (28). Electropherograms approximating an RNA integrity number of 8 or lower or with a 28S/18S ratio below 1.8 were discarded.
qPCR measurements.
Variation in transcript levels was determined for seven A. niger genes and a synthetic control RNA transcript (Table 1). This synthetic control RNA transcript, a bacterial kanamycin synthetase-encoding gene fused to a eukaryotic poly(A) tail (Promega), is spiked to total RNA prior to cDNA synthesis and can correct for various efficiencies of reverse transcription or PCR itself (12). The first four genes of Table 1 were used as endogenous reference genes. These A. niger reference genes showed little variation in transcript levels on more than 100 microarrays that were run in our laboratory prior to this study (D. van der Veen, J. M. Oliveira, E. S. Martens-Uzunova, and L. H. de Graaff, unpublished data) and were selected using the method suggested by Lee and coworkers (18). No elevated expression levels are expected for these genes (10, 16, 19, 22). Expression levels for malate synthase, whose expression is not influenced by addition of d-xylose, were also measured. Finally, the transcriptional response upon the addition of d-xylose was measured by determining the transcript levels of two genes, xlnB and xlnD. Primers were designed using the Primer3 web interface (26) and are given in Table 1. Primer design criteria were as follows: length, 20 to 22 bp; melting temperature, 60 ± 1°C; and GC content, 50% ± 5%. Amplicons ranged from 125 to 150 bp and had a melting temperature of 80 ± 5°C.
TABLE 1.
Primers used in this study
| Probe set IDa | Gene or transcript | Description | Primers (5′ to 3′)c |
|---|---|---|---|
| An00g08669_at | An08g06940b | Histone, strong similarity to A. nidulans histone H4.1 (10) | Fw-ATCTTGCGTGACAACATCCA |
| Rev-CACCCTCAAGGAAGGTCTTG | |||
| An00g10935_at | An08g05910b | Strong similarity to Aspergillus nidulans SagA, which is involved in endocytosis and DNA repair (16) | Fw-CCAGGATGAAGAGTGGGAGA |
| Rev-GCAGCTGGAGTGCTTCTTTC | |||
| An00g13122_at | An02g04120b | Similarity to S. cerevisiae Atx2, which is a Golgi transporter involved in manganese homeostasis (19) | Fw-TTTTCAGTCTGGCTGCTCCT |
| Rev-CTGTTTTCCTGCATCGTGTG | |||
| An00g07894_at | An14g05050b | Strong similarity to Schizosaccharomyces pombe Dma1, which is a component of the spindle assembly point involved in mitotic division (22) | Fw-ACTCCAGAGGACAAGCAGGA |
| Rev-GCAGACGCATGCTCTCAATA | |||
| An00g09644_at | An15g01860 | Malate synthase | Fw-TGATTAAGACGTGTCACCGC |
| Rev-GGAGTGGGCATGTAGGTGTT | |||
| An00g14004_at | An01g00780 | Endoxylanase B (xlnB) | Fw-CAACTTTGTCGGTGGAAAGG |
| Rev-GGGTAGCCGTGTAGATATCG | |||
| An00g14006_at | An01g09960 | β-Xylosidase (xlnD) | Fw-TAATCTACGCCGGTGGTATC |
| Rev-TTCTTGAGCGAAGAGGAATC | |||
| Kanamycin | Kanamycin synthetase-encoding poly(A)-tailed synthetic gene | Fw-AGCATTACGCTGACTTGACG | |
| Rev-AGGTGGACCAGTTGGTGATT |
The probe set ID refers to the DNA microarray probe set identifier corresponding to that gene.
Endogenous reference genes.
Fw-, forward primer; Rev-, reverse primer.
Total isolated RNA was diluted in two steps to a concentration of 200 ng/μl. A total of 1.00 μg of total RNA was spiked with 0.1 ng of the synthetic RNA transcript, followed by DNase I treatment. cDNA was synthesized using the Omniscript reverse transcriptase kit (Qiagen). Four units of reverse transcriptase enzyme was added to the DNase-treated total RNA at a final volume of 20 μl. The cDNA synthesis reaction was kept at 37°C, and after 1 hour, the synthesized cDNA was diluted 20-fold and stored at −20°C until use. For qPCR measurements, PCR primers (final concentration, 1.4 μM) and 4 μl of diluted cDNA were pipetted to 2× SYBR PCR mastermix (ABgene) using a CAS-1200 pipetting robot (Corbett Life Science).
A Rotor-Gene 3000 qPCR machine (Corbett Life Science) was used for thermocycling under the following conditions: 15 min at 95°C, followed by 40 amplification cycles (denaturation, 15 s at 95°C; annealing, 15 s at 58°C; elongation, 20 s at 72°C). For each single run, nontemplate control samples for every primer pair used in that run were included. After the amplification cycles, a melting step was performed. Quality control was done by inspection of the melting curve, and samples with significant primer-dimer formation were not considered for analysis. The cycle threshold value and amplification efficiency were determined with the Rotor-Gene software using the comparative quantification method (the cycle threshold value corresponds to the Rotor-Gene software “take-off” value). The relative expression ratio of gene expression was calculated by following Pfaffl (24) as follows: ratio = (Egene)ΔCt (pre − post)/(Ekana)ΔCt (control − sample). In this formula, Ekana denotes the amplification efficiency of the synthetic kanamycin transcript, and Egene denotes that of the gene for which the ratio is determined. ΔCt denotes the cycle threshold difference between pre- and postinduced fermentor samples for the gene and kanamycin transcripts.
Microarray processing.
cDNA and cRNA synthesis and labeling and array hybridization were performed by following the Affymetrix users' manual (1) using the one-cycle target labeling and control reagent kit and starting with 5 μg of total RNA as template material. Fifteen micrograms of fragmented and labeled cRNA was hybridized to custom-made Aspergillus niger arrays at 45°C for 16 h. Washing and staining were done using the hybridization, wash, and stain kit (Affymetrix), using a GeneChip FS-450 fluidics station and an Agilent G2500A gene array scanner. Scanned images were converted into .CEL files using Microarray Suite version 5 software (Affymetrix).
Data analysis.
For the 1,920 qPCR measurements obtained from the 5-week fermentor experiment, variance components were calculated by restricted maximum likelihood (REML) variance components analysis (REML sparse algorithm with average information optimization) (23) using GenStat 9.2 software (VSN International). Per gene, three REML analyses were run using each gene's cycle threshold, amplification efficiency, and expression ratio values as response variates. A random model, yw.f.b.r.d.q.s = μ + ɛweek + ɛw.fermentor + ɛw.f.biomass + ɛw.f.b.RNA + ɛw.f.b.r.cDNA + ɛw.f.b.r.c.qPCR, was applied, using ɛw.f.b.r.c.qPCR as the residual term (subscripts are abbreviated after first usage; e.g., ɛw.f.biomass is ɛweek.fermentor.biomass).
For microarray data analysis, .CEL files of the individual array images were imported into GeneSpring 7.3 (Agilent Technologies) using its robust multichip average (RMA) preprocessor to obtain RMA-normalized signal values for all arrays (14). Probe sets with an RMA-normalized signal below 37.7—three times the lowest value detected—on all arrays were discarded, leaving 9,320 probe sets (64%). In comparison, when using the Affymetrix MAS 5.1 software-derived flag calls, an average of 5,948 probe sets (41%) are called “present” per array. For the six microarrays used in this study, statistically significant differentially expressed genes were determined using the limma package (29). A Student's two-sample t test between the sorbitol and d-xylose arrays was executed, using empirical Bayesian statistics to moderate within-gene standard errors, Benjamini and Hochberg's “false discovery rate” to correct for multiple testing (3), and an adjusted P value of <0.05 to discriminate between differentially expressed genes. To check for the influence of unequal sample size bias, testing was recalculated with the inclusion of four additional sorbitol-grown samples derived from cultures grown identically (our unpublished data), giving similar results.
Microarray data accession number.
Raw and RMA-normalized array data were deposited in NCBI's GEO database (9) under series entry GSE11405.
RESULTS
Selection of conditions for the response system.
The xylanolytic system of A. niger consists of enzymes that degrade cellulose and hemicellulose, is under the control of the transcriptional regulator XlnR (34), and can be induced by the monosaccharide d-xylose. XlnR-controlled genes are subject to carbon catabolite repression by d-xylose (8). To reduce this repressing effect, the d-xylose concentration that induced the xylanolytic system but had the least repressing effect on XlnR-controlled genes was determined prior to the analysis of variance components experiment. Three fermentor cultures were induced with either 0.1, 1, or 50 mM d-xylose and sampled for up to 4 h. Expression levels for two XlnR-controlled genes, xlnB and xlnD, were followed by qPCR. These genes encode endoxylanase B (17) and β-xylosidase (35), respectively.
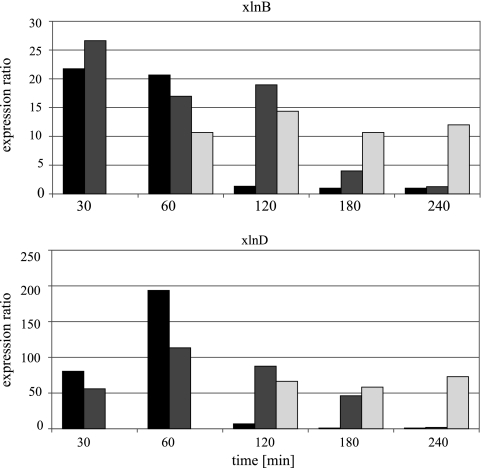
The observed transcript levels for xlnB and xlnD are given in Fig. 1. A d-xylose concentration of 0.1 mM is able to induce both genes, with an expression ratio for xlnD of 190-fold at 60 min after induction. This lowest concentration of 0.1 mM d-xylose also discriminates best between noninduced and induced states: for this concentration, transcript levels increase for 60 min and return to preinduced levels in the next hour. For both the 1-mM and 50-mM concentrations, elevated transcript levels for xlnB and xlnD could be detected up to 3 hours after induction. We decided to induce fermentor cultures with 0.1 mM d-xylose and to sample them 1 h after induction.
FIG. 1.
Expression ratios of xlnB and xlnD. The expression ratios of the xlnB and xlnD genes measured after induction with either 0.1 (black), 1 (dark gray), or 50 (light gray) mM of d-xylose. The sampling time in minutes is presented on the horizontal axis. No transcript levels were determined for the 50-mM fermentations for both genes at 30 min. At 60 min, a bad qPCR run prevented xlnD ratio calculation.
Experimental design.
The aim of this experiment was both to minimize all variation introduced in each of the steps preceding a qPCR measurement or microarray scan and to determine their magnitude. After optimization of the processing protocols, we investigated the variation in our experimental setup quantitatively by qPCR. For this, a hierarchical factorial experiment was designed (Fig. 2). We performed one fermentation cycle per week for 5 weeks, using three identical fermentations per week. These cultures were induced with 0.1 mM of d-xylose. A fourth culture was randomly selected as the noninduced control. Each processing step following induction was executed in duplicate, starting with the independent harvesting of mycelial samples. This led to 128 individual qPCR measurements for a single fermentation or a total of 1,920 individual measurements. Another 768 measurements originated both from noninduced cultures and from culture samples taken before induction. In total, 2,688 qPCR measurements were made (see the supplemental material).
FIG. 2.

Experimental design. Schematic representation of hierarchical experimental design used. In a 5-week period (gray boxes), four fermentors were run. Three fermentors were induced to 0.1 mM d-xylose, and the fourth fermentor was induced to 0.1 mM sorbitol. Each fermentation run was sampled twice, and from each mycelial sample, total RNA was extracted twice independently. The total RNA obtained was split in two, and independent cDNA synthesis reactions were performed. Each cDNA sample that was made was used to measure eight genes in duplicate using qPCR. The gray arrow indicates an identical processing step.
In the elongation phase of a qPCR, SYBR green dye molecules intercalate with double-stranded DNA that is formed during the reaction. The dye-DNA complex absorbs blue light and emits green light (32), which is measured and plotted against the cycle run number. The resulting sigmoid-shaped curve is used to extract the cycle threshold and amplification efficiency values. The cycle threshold value represents the amount of transcripts for a specific gene in the cDNA template upon the start of the PCR, with lower values representing more copies of a transcript present in the cDNA pool. The amplification efficiency is a primer pair-specific value that represents the efficiency of the PCR and hence is independent from the experimental conditions apart from the qPCR run itself (31). Insight into the variation that is introduced in all steps except the day-to-day and fermentor-to-fermentor variations is deduced from the expression ratio. This ratio expresses the relative change of a gene's transcription level compared to that of a noninduced sample. A ratio greater than 1 indicates elevated transcription of a gene, while a ratio of 1 indicates no change. Table 2 gives descriptive statistics of the data set. All results of the qPCR measurements are given in the supplemental material. Variance components were estimated for each of the three variables and were subdivided by the eight genes followed in this study (Table 3).
TABLE 2.
Descriptive statistics
| Variance component measuring indicated gene or transcript | No. of measured samplesa | Avg value ± SD |
|---|---|---|
| Cycle threshold | ||
| An08g05910 | 230 | 24.08 ± 1.22 |
| An02g04120 | 232 | 21.4 ± 0.98 |
| An14g05050 | 236 | 25.69 ± 1.47 |
| An08g06940 | 236 | 16.49 ± 0.57 |
| An15g01860 | 233 | 24.83 ± 1.43 |
| Kanamycin | 234 | 16.08 ± 0.49 |
| xlnB | 236 | 22.62 ± 1.38 |
| xlnD | 235 | 16.08 ± 0.82 |
| Amplification efficiency | ||
| An08g05910 | 230 | 1.70 ± 0.026 |
| An02g04120 | 232 | 1.72 ± 0.044 |
| An14g05050 | 236 | 1.68 ± 0.032 |
| An08g06940 | 236 | 1.70 ± 0.024 |
| An15g01860 | 233 | 1.71 ± 0.015 |
| Kanamycin | 234 | 1.74 ± 0.028 |
| xlnB | 236 | 1.73 ± 0.018 |
| xlnD | 235 | 1.73 ± 0.027 |
| Expression ratio | ||
| An08g05910 | 226 | 1.06 ± 0.43 |
| An02g04120 | 228 | 1.05 ± 0.43 |
| An14g05050 | 232 | 1.02 ± 0.42 |
| An08g06940 | 232 | 0.88 ± 0.32 |
| An15g01860 | 229 | 2.45 ± 1.74 |
| Kanamycin | 234 | 1.00 |
| xlnB | 232 | 16.42 ± 9.35 |
| xlnD | 231 | 137.73 ± 94.90 |
qPCR runs that did not meet quality control standards account for differences between the theoretical number of samples possible (240) and the actual number of measured samples.
TABLE 3.
Relative variance components estimatesa
| Variance component measuring indicated gene or transcript | Estimates by:
|
|||||
|---|---|---|---|---|---|---|
| Wk | Fermentor | Biomass sample | RNA sample | cDNA sample | qPCR measurement | |
| Cycle threshold | ||||||
| An08g05910 | 78.8 | 5.9 | 1.3 | 2.5 | 4.2 | 7.3 |
| An02g04120 | 66.2 | 1.9 | 0.0 | 0.3 | 11.5 | 20.1 |
| An14g05050 | 81.4 | 3.0 | 2.7 | 3.9 | 3.2 | 5.9 |
| An08g06940 | 68.1 | 5.5 | 1.0 | 14.1 | 8.3 | 2.9 |
| An15g01860 | 29.6 | 60.4 | 0.6 | 2.8 | 3.1 | 3.6 |
| Kanamycin | 67.5 | 0.0 | 0.0 | 18.1 | 10.1 | 4.3 |
| xlnB | 69.1 | 10.7 | 0.0 | 4.7 | 9.3 | 6.2 |
| xlnD | 4.8 | 19.4 | 2.9 | 47.7 | 8.2 | 17.0 |
| Amplification efficiency | ||||||
| An08g05910 | 68.9 | 2.1 | 2.8 | 0.0 | 1.1 | 25.1 |
| An02g04120 | 75.9 | 3.1 | 0.0 | 0.0 | 2.2 | 18.7 |
| An14g05050 | 74.7 | 0.1 | 0.0 | 0.0 | 0.3 | 24.9 |
| An08g06940 | 77.8 | 0.1 | 0.0 | 0.0 | 4.6 | 17.5 |
| An15g01860 | 30.1 | 5.3 | 0.0 | 1.5 | 8.2 | 54.8 |
| Kanamycin | 71.5 | 0.8 | 0.0 | 7.5 | 2.1 | 18.1 |
| xlnB | 31.2 | 6.2 | 7.1 | 0.0 | 11.3 | 44.3 |
| xlnD | 32.6 | 25.2 | 1.9 | 9.1 | 6.6 | 24.7 |
| Expression ratio | ||||||
| An08g05910 | 0.0 | 36.4 | 10.7 | 0.0 | 24.6 | 28.3 |
| An02g04120 | 2.1 | 28.9 | 0.0 | 4.3 | 21.5 | 43.2 |
| An14g05050 | 0.0 | 30.6 | 9.8 | 2.8 | 24.5 | 32.2 |
| An08g06940 | 0.3 | 58.5 | 2.3 | 12.7 | 19.8 | 6.3 |
| An15g01860 | 6.0 | 75.9 | 0.5 | 6.8 | 1.9 | 9.0 |
| Kanamycin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| xlnB | 11.7 | 62.0 | 0.0 | 6.0 | 9.4 | 10.9 |
| xlnD | 0.0 | 80.7 | 1.6 | 3.6 | 11.5 | 2.6 |
Columns indicate the terms of the random model used to describe variance and correspond to the experimental procedure outlined in Fig. 2.
DNA microarray analysis results.
To investigate the differences in global transcript levels between fermentations done in this study, six fermentor samples were selected for hybridization onto DNA microarrays. These samples were selected based on differences in their distribution over the weeks and fermentor vessels used, their biomass density, and the xlnD expression levels observed in these samples by qPCR (Table 4).
TABLE 4.
RNA sample properties
| Column in supplemental materiala | Carbon source | Wk | Fermentor | Biomass | RNA sample | xlnD ratiob | Dry wt (g/liter) |
|---|---|---|---|---|---|---|---|
| A | d-Xylose | 2 | 3 | 2 | 1 | 20 | 1.84 |
| B | 3 | 2 | 2 | 1 | 300 | 1.41 | |
| C | 5 | 3 | 1 | 1 | 50 | 2.20 | |
| D | 5 | 3 | 1 | 1 | 50 | 2.20 | |
| E | 4 | 5 | 2 | 1 | 120 | 1.93 | |
| F | Sorbitol | 5 | 5 | 1 | 1 | 1 | 1.64 |
| G | 3 | 4 | 1 | 1 | 1 | 1.92 |
Coding corresponds to columns of the qPCR-measured data in the supplemental material.
The average of four expression ratios calculated from the four replicate qPCR measurements.
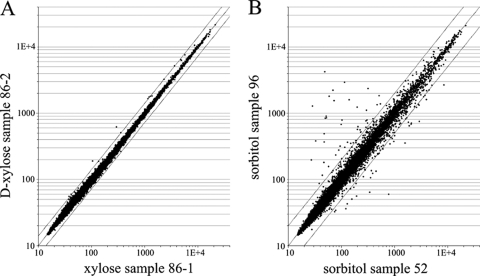
To analyze technical variation that is introduced within microarray processing steps, the total RNA of one mycelial sample was split (Table 4, C and D), processed independently, and hybridized onto two microarrays. RMA signal values were calculated and plotted (Fig. 3). Only the RMA signal values for 5 of 14,554 probe sets differ more than 1.5-fold but not more than 2.1-fold between the technical replicates. No mRNA sequences hybridize to these probe sets: 28S rRNA hybridizes to four of five probe sets, while the fifth probe set hybridizes to one of the control transcripts used in the Affymetrix array platform, the 5′ region of the Escherichia coli bioD transcript. This result indicates that the specific microarray handling and processing steps contribute little to overall variation.
FIG. 3.
Technical variation between samples. Scatter plot of microarray data. (A) Two RNA samples of sample 86 (86-1 and 86-2) were processed independently and hybridized to two Affymetrix arrays. The RMA-normalized data of all 14,554 probe sets are plotted in a scatter plot. (B) Total RNA from two independent noninduced control fermentations (sorbitol), 96 and 52, are hybridized, and RMA normalized-data are plotted. The lines above and below the diagonal line indicate twofold difference relative to the diagonal line.
A transcriptome analysis revealed 24 genes that are significantly differentially expressed in d-xylose-induced cultures compared to sorbitol-grown cultures (Table 5). This gene list contains both xlnB and xlnD, the two genes used in the qPCR measurements to assess the induction of the xylanolytic system, as well as an additional eight enzymes that are known to be under XlnR control (34). The presence of four sugar transporter-encoding genes, as well as two genes encoding glycosyl hydrolases, is in accordance with the view that d-xylose initializes a response to degrade complex carbohydrates, such as hemicellulose. In addition, two genes that encode enzymes of the Leloir pathway, the classical pathway of d-galactose catabolism, are upregulated in the induced samples. An02g03590, the ortholog of the Saccharomyces cerevisiae GAL7 protein, encodes the second enzyme of the Leloir pathway, while the required glucose epimerase activity is encoded by An11g10890, the ortholog of S. cerevisiae GAL10. The first step of this pathway, the galactokinase that phosphorylates d-galactose to d-galactose 1-phosphate, is encoded by An16g04160. This gene is induced about twofold by d-xylose, but a P value of 0.604 excludes this gene from our list of statistically differentially expressed genes. Finally, for six genes in our list of differentially expressed genes, their roles in the cellular response toward d-xylose are difficult to assess.
TABLE 5.
Differentially expressed genes on d-xylose induction
| A. niger gene | Signal sequence predicted | Description | Motifa |
|---|---|---|---|
| An01g00780 | Yes | xlnB (xynB); xylanase B | −216 |
| An01g09960 | Yes | xlnD; β-xylosidase | (−250), −147 |
| An14g05800 | Yes | aguA; α-glucuronidase | −277, (−183) |
| An09g00120 | Yes | faeA; ferulic acid esterase A | −265, (−121) |
| An18g03570 | Yes | bglA; β-glucosidase | |
| An14g02760 | Yes | eglA; endoglucanase A | |
| An09g03300 | Yes | Glycosyl hydrolase, family 31 | −138 |
| An03g00500 | Glycosyl hydrolase, family 30 | ||
| An15g01500 | Sugar transporter | (−417) | |
| An11g01100 | Sugar transporter | ||
| An03g01620 | Yes | Sugar transporter | −407 |
| An06g00560 | Sugar transporter | −415 | |
| An01g10920 | ladA; l-arabitol dehydrogenase | −334 | |
| An01g03740 | xyrA; d-xylose reductase XyrA | (−311) | |
| An12g00030 | xdhA; xylitol dehydrogenase | −333 | |
| An07g03140 | xkiA; d-xylulokinase | (−672), −482 | |
| An02g03590 | Galactose-1-phosphate uridylyltransferase (similar to S. cerevisiae GAL7p) | (−371) | |
| An11g10890 | UDP-glucose 4-epimerase (similar to S. cerevisiae GAL10p) | (−577) | |
| An07g03160 | talB; transaldolase | −195 | |
| An08g01740 | Aldehyde reductase | −765 | |
| An02g13980 | Yes | Conserved hypothetical protein (mono-oxygenase?); cytochrome P450 domain | −540, (+26) |
| An06g00830 | Weak similarity to hypothetical transcription regulatory protein | ||
| An01g11180 | Hypothetical protein, FAD/FMN, containing dehydrogenase | (−329) | |
| An11g05340 | Yes | Fructosamine oxidase |
The location of the cis-acting motif 5′-GGNTAAA-3′ is indicated in base pairs counted upstream of the ATG start codon. Values in parentheses indicate the reverse direction of this motif.
DISCUSSION
Experimental variation.
Although, in theory, a protocol is a rigid and established code that describes all proceedings relating to an experiment, in practice, protocol steps are subject to interpretation by different experimenters. A precise description of how to handle given steps and the removal of unclear or ambiguous sentences decrease the necessity to interpret the exact meaning of a protocol phrase, which in turn leads to more-reproducible experiments. The effects of our optimization and quantitative determination of the variation in our laboratory setup were determined by qPCR, a cost-effective yet powerful technique to study transcript abundance.
Important parameters for high-quality qPCR measurements are the specificity of a primer pair for its target, amplification efficiency, and reproducibility (5, 20). Typically, when a newly designed primer pair is tested prior to use in quantification, a range of different annealing temperatures or primer concentrations is examined to identify conditions where only a single DNA fragment is amplified. In this study, a multitude of target genes were measured using a single PCR profile and primer concentration. The possibility that this approach affects the robustness of our qPCR measurements was assessed by the examination of the variation that is introduced in the amplification efficiency values. Within-gene variation is minimal, even when the amplification efficiency per gene varies. The most variable amplification efficiency values are for gene An02g04120, with a mean value of 1.72 and a standard deviation of 0.04 (n = 326). The reproducibility of this generalized method can be also derived from the analysis of variance while looking at the variance component titled “qPCR measurement” (Table 3), which includes pipetting and the actual measurement. The median relative contribution is 6.1% of the total variation (Table 3). Transcript An02g04120 again shows the highest variation, 20.1%, in this step. When this percentage is placed in its biological context, 95% of all cycle threshold values will range between 21.3 ± 0.9, which translates to 1.6-fold differences. We conclude that our standardized qPCR method is both precise and accurate.
Before the actual determination of the sources of variation in our experiment, we hypothesized that most variation is introduced by differences in the day-to-day handling and processing. The next largest variation was anticipated to be the use of the individual fermentor vessels. The results of the analysis of variance components comply with our initial hypothesis: day-to-day variation contributes to about 70% of the total variation (Table 3). This step includes the growth and harvesting of spores, the preparation of fermentation media, and the assembly of fermentors. The large effect that the day-to-day variation has on the total variation can effectively be excluded by examination of the expression ratio. This ratio presents the relative change in a gene's transcript level between preinduced and postinduced fermentation conditions. As this ratio is calculated from expression data of samples taken from the same fermentor, this result effectively cancels out the contribution of day-to-day variation. When fermentor cultures are compared by this gene expression ratio, the analysis of variance components of the ratio-derived data shows that the three steps of fermentation, cDNA synthesis, and actual qPCR measurement each contribute about equally in the case of the four endogenous reference genes (Table 3).
The effect that the individual fermentation vessels have on the variation in transcript level is 60% and 80% for the two d-xylose-induced genes, xlnB and xlnD, respectively. For the malate synthase-encoding gene, the effect is 75%. Different amounts of fungal cells present in a fermentor cannot explain this effect, as only a weak correlation of cycle threshold values with the biomass concentration is found (i.e., for xlnB, R2 = 0.35). One explanation is that small differences between fermentor headplates and vessels result in unique mixture characteristics for each fermentor. These fermentor-specific effects are reflected in small physiological differences between cultures, which may account for the observed differences in transcript levels. Since samples are taken 1 h after induction with d-xylose, such differences may affect the actual d-xylose concentration per fermentor, leading to a high reproducibility of a gene's expression within a fermentor but variation of its expression between fermentors. For example, xlnD induction is on average 240-fold higher for fermentors with headplate “2” but only 100-fold higher for fermentors with headplate “4.”
d-Xylose-induced genes assessed by DNA microarray analysis.
The qPCR measurements clearly showed elevated transcript levels of xlnB and xlnD after induction with d-xylose. For xlnB, the average increase for all 15 fermentations was by 16-fold compared to that of noninduced conditions, while for xlnD, the average increase was by 138-fold. For the six DNA microarrays analyzed, the average increase is by 6-fold for xlnB and by 110-fold for xlnD after induction with d-xylose. The results for a gene measured by both methods are in good agreement; for instance, the Spearman rank correlation coefficient for xlnD measurements obtained by qPCR and microarray technologies is 0.90.
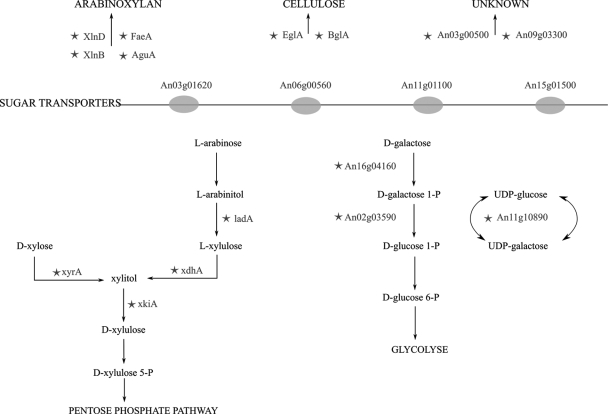
Comparison of the noninduced and d-xylose-induced samples that were hybridized onto microarrays revealed 24 genes that are statistically differentially expressed after induction with d-xylose (Table 5). Degradation of complex polysaccharide substrates starts with the uptake of signal molecules such as d-xylose that activate specific induction pathways, resulting in the expression and secretion of enzymes necessary to degrade and metabolize the polysaccharide. However, since d-xylose is rarely found by itself under natural conditions, it is likely that A. niger interprets the presence of d-xylose as proxy for the availability of complex carbohydrate polymers, such as (hemi)cellulose. Not only is this heterogenic response reflected in the induction of secreted enzymes but also in the activation of multiple metabolic pathways (Fig. 4). For instance, genes encoding the second step in l-arabinose metabolism and enzymes of the classical d-galactose catabolic route are significantly upregulated as well.
FIG. 4.
Enzymes induced by d-xylose. Schematic of the enzymes encoded by significantly differentially expressed genes after induction by d-xylose. (Top) Extracellular enzymes acting on complex polysaccharides, arabinoxylan, and cellulose. (Middle) Sugar transporters. (Bottom) Metabolic routes of the degradation of d-xylose, l-arabinose, and d-galactose. The enzymes encoded by significantly differentially expressed genes are indicated by stars.
Five genes appear to encode enzymes that might well be related to (hemi)cellulose degradation, but of which the exact function has not been elucidated. An08g01740 encodes an aldehyde reductase. Reduction of the carboxyl group of carbohydrates by aldehyde reductases, such as the reduction of d-xylose to xylitol, is a first step in the catabolism of many monosaccharides, and the product of this gene might well play a role in the catabolism of monosaccharide substrates that are the products of (hemi)cellulose degradation. Products of d-xylose or l-arabinose catabolism are channeled into the nonoxidative branch of the pentose phosphate pathway. This pathway in turn is linked to glycolysis by the enzymes transaldolase and transketolase (30). While the talB transaldolase is upregulated by sixfold in d-xylose-induced samples, both transketolase-encoding genes as well as the major transaldolase-encoding gene of A. niger have no differential expression levels under induced and noninduced conditions.
The transcriptional regulator XlnR activates the transcription of (hemi)cellulose-degrading enzymes (34) as well as the transcription of genes encoding metabolic enzymes (34, 33). A cis-acting element in the promoter region of d-xylose-induced genes, described as 5′-GGCTAAA-3′ (34) and proposed to be generalized to 5′-GGNTAAA-3′ after comparative transcriptome analysis of three Aspergillus species (2), was detected in all but five statistically significant differentially expressed genes (Table 5), and also, the GAL1p ortholog An16g04160 has this consensus motif present 153 bp upstream in its promoter region.
In this and other studies (2), it is observed that nonstrictly d-xylose metabolism-related genes have elevated transcript levels upon d-xylose induction. The most likely explanation for this is that the fungus fine-tunes its response toward the diversity of complex substrates it encounters by coordinated action of partly overlapping regulatory systems. Expression of the XlnR-controlled ferulic acid esterase A is greater when A. niger is induced with both d-xylose and ferulic acid relative to induction by d-xylose or ferulic acid alone (7). Aromatic compounds such as ferulic acid are not only part of arabinoxylan but are also part of pectin. No XlnR-related motif is present in the promoter region of the polysaccharide-degrading enzymes acting on cellulose, BglA, and EglA, nor is that present in the promoter region of one of the uncharacterized family 30 glycosyl hydrolases. The induction of these genes in the absence of the consensus motif suggests the action of other transcriptional activators besides the xylanolytic activator XlnR. Interestingly, one of the genes of unclear function encodes a hypothetical transcription regulatory protein, and its expression patterns correlate strongly with the three genes of the Leloir pathway.
Andersen and coworkers have described a conserved set of 23 genes for which transcription is elevated on d-xylose medium (2). Nine of these d-xylose-responsive genes are not present in our gene list, including three genes encoding sugar transporters, three glycosyl hydrolases, and three metabolic enzyme-encoding genes (encoding an aldose 1-epimerase, a short-chain dehydrogenase, and an aldehyde reductase). The difference between the two gene lists can be explained by the experimental approach chosen: Andersen and coworkers have grown A. niger with either d-glucose or d-xylose as the sole carbon source and have sampled at d-xylose levels around 12 mM. In this study, sorbitol-grown cultures were induced with minute concentrations of d-xylose only.
In conclusion, the work presented in this study has resulted in an improved method for the generation of high-quality qPCR and microarray data from fermentations of the filamentous fungus Aspergillus niger. The decreased variation improves data quality and eases the use of data analysis, which is an essential prerequisite to study transcript profiling and gene regulation.
This work provides new insights into the mechanisms following d-xylose induction. The data link xylose metabolism not only to l-arabinose metabolism but also to d-galactose metabolism.
Supplementary Material
Acknowledgments
We thank J. Thissen of the Biometris expert group of Wageningen UR for performing the variance components analysis and DSM Food Specialties for providing aid in fermentor optimization and use of the DSM A. niger microarrays.
Footnotes
Published ahead of print on 20 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Affymetrix. 2004. GeneChip expression analysis technical manual. Affymetrix, Santa Clara, CA.
- 2.Andersen, M. R., W. Vongsangnak, G. Panagiotou, M. P. Salazar, L. Lehmann, and J. Nielsen. 2008. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. USA 105:4387-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 4.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, S. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 6.Carlile, M. 1995. The success of the hypha and mycelium. In N. Gow and G. Gadd (ed.), The growing fungus. Chapman & Hall, London, United Kingdom.
- 7.de Vries, R. P., and J. Visser. 1999. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl. Environ. Microbiol. 65:5500-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries, R. P., J. Visser, and L. H. de Graaff. 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150:281-285. [DOI] [PubMed] [Google Scholar]
- 9.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehinger, A., S. H. Denison, and G. S. May. 1990. Sequence, organization and expression of the core histone genes of Aspergillus nidulans. Mol. Gen. Genet. 222:416-424. [DOI] [PubMed] [Google Scholar]
- 11.Etienne, W., M. H. Meyer, J. Peppers, and Meyer, R. A., Jr. 2004. Comparison of mRNA gene expression by RT-PCR and DNA microarray. BioTechniques 36:618-620, 622, 624-626. [DOI] [PubMed] [Google Scholar]
- 12.Huggett, J., K. Dheda, S. Bustin, and A. Zumla. 2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6:279-284. [DOI] [PubMed] [Google Scholar]
- 13.Imbeaud, S., and C. Auffray. 2005. ‘The 39 steps’ in gene expression profiling: critical issues and proposed best practices for microarray experiments. Drug Discov. Today 10:1175-1182. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry, R., B. Bolstad, F. Collin, L. Cope, B. Hobbs, and T. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jafari, P., and F. Azuaje. 2006. An assessment of recently published gene expression data analyses: reporting experimental design and statistical factors. BMC Med. Inform. Decis. Mak. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, G. W., P. Hooley, S. M. Farrington, S. G. Shawcross, L. A. Iwanejko, and P. Strike. 1999. Cloning and characterisation of the sagA gene of Aspergillus nidulans: a gene which affects sensitivity to DNA-damaging agents. Mol. Gen. Genet. 261:251-258. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita, K., M. Takano, T. Koseki, K. Ito, and K. Iwano. 1995. Cloning of the xynB gene encoding xylanase B from Aspergillus niger and its expression in Aspergillus kawachii. J. Ferment. Bioeng. 79:422-428. [Google Scholar]
- 18.Lee, S., M. Jo, J. Lee, S. S. Koh, and S. Kim. 2007. Identification of novel universal housekeeping genes by statistical analysis of microarray data. J. Biochem. Mol. Biol. 40:226-231. [DOI] [PubMed] [Google Scholar]
- 19.Lin, S. J., and V. C. Culotta. 1996. Suppression of oxidative damage by Saccharomyces cerevisiae ATX2, which encodes a manganese-trafficking protein that localizes to Golgi-like vesicles. Mol. Cell. Biol. 16:6303-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutfalla, G., and G. Uze. 2006. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol. 410:386-400. [DOI] [PubMed] [Google Scholar]
- 21.Morey, J. S., J. C. Ryan, and F. M. Van Dolah. 2006. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 8:175-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murone, M., and V. Simanis. 1996. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 15:6605-6616. [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson, H. D., and R. Thompson. 1971. Recovery of inter-block information when block sizes are unequal. Biometrika 58:545-554. [Google Scholar]
- 24.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. Macdonald, and A. W. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 26.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 27.Salit, M., A. Kimmel, and O. Brian. 2006. Standards in gene expression microarray experiments. Methods Enzymol. 411:63. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder, A., O. Mueller, S. Stocker, R. Salowsky, M. Leiber, M. Gassmann, S. Lightfoot, W. Menzel, M. Granzow, and T. Ragg. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. [DOI] [PubMed] [Google Scholar]
- 30.Stryer, L. 1995. Biochemistry, 4th ed. W. H. Freeman and Company, New York, NY.
- 31.Tichopad, A., M. Dilger, G. Schwarz, and M. Pfaffl. 2003. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res. 31:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuma, R. S., M. P. Beaudet, X. Jin, L. J. Jones, C. Y. Cheung, S. Yue, and V. L. Singer. 1999. Characterization of SYBR Gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators. Anal. Biochem. 268:278-288. [DOI] [PubMed] [Google Scholar]
- 33.vanKuyk, P. A., M. J. de Groot, G. J. Ruijter, R. P. de Vries, and J. Visser. 2001. The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on D-xylose and L-arabinose. Eur. J. Biochem. 268:5414-5423. [DOI] [PubMed] [Google Scholar]
- 34.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Peij, N. N., J. Brinkmann, M. Vrsanská, J. Visser, and L. H. de Graaff. 1997. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur. J. Biochem. 245:164-173. [DOI] [PubMed] [Google Scholar]
- 36.Wei, C., J. Li, and R. E. Bumgarner. 2004. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics 5:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, S., M. McIntyre, D. R. Berry, and B. McNeil. 2002. The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 22:1-14. [DOI] [PubMed] [Google Scholar]
- 38.Wosten, H. A., S. M. Moukha, J. H. Sietsma, and J. G. Wessels. 1991. Localization of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 137:2017-2023. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.