Abstract
Thymidine is an important precursor in the production of various antiviral drugs, including azidothymidine for the treatment of AIDS. Since thymidine-containing nucleotides are synthesized only by the de novo pathway during DNA synthesis, it is not easy to produce a large amount of thymidine biologically. In order to develop a host strain to produce thymidine, thymidine phosphorylase, thymidine kinase, and uridine phosphorylase genes were deleted from an Escherichia coli BL21 strain to develop BLdtu. Since the genes coding for the enzymes related to the nucleotide salvage pathway were disrupted, BLdtu was unable to utilize thymidine or thymine, and thymidine degradation activity was completely abrogated. We additionally expressed T4 thymidylate synthase, T4 nucleotide diphosphate reductase, bacteriophage PBS2 TMP phosphohydrolase, E. coli dCTP deaminase, and E. coli uridine kinase in the BLdtu strain to develop a thymidine-producing strain (BLdtu24). BLdtu24 produced 649.3 mg liter−1 of thymidine in a 7-liter batch fermenter for 24 h, and neither thymine nor uridine was detected. However, the dUTP/dTTP ratio was increased in BLdtu24, which could lead to increased double-strand breakages and eventually to cell deaths during fermentation. To enhance thymidine production and to prevent cell deaths during fermentation, we disrupted a gene (encoding uracil-DNA N-glycosylase) involved in DNA excision repair to suppress the consumption of dTTP and developed BLdtug24. Compared with the thymidine production in BLdtu24, the thymidine production in BLdtug24 was increased by ∼1.2-fold (740.3 mg liter−1). Here, we show that a thymidine-producing strain with a relatively high yield can be developed using a metabolic engineering approach.
Thymidine, which is composed of 2-deoxyribose and a thymine base, is a commercially useful precursor in the chemical synthesis of various antiviral drugs, including stavudine and zidovudine (azidothymidine), the active ingredient in a formulation for the treatment of AIDS (18, 19). Because thymidine is required only in DNA synthesis, intracellular thymidine levels are very low and are tightly controlled (40). For the production of precursors for antiviral drugs, thymidine is either biologically produced in a low yield by a few modified microorganisms or chemically synthesized through a very costly process (17, 33, 48, 49). Thus, there is a need for developing a more efficient strain for thymidine production on a large scale.
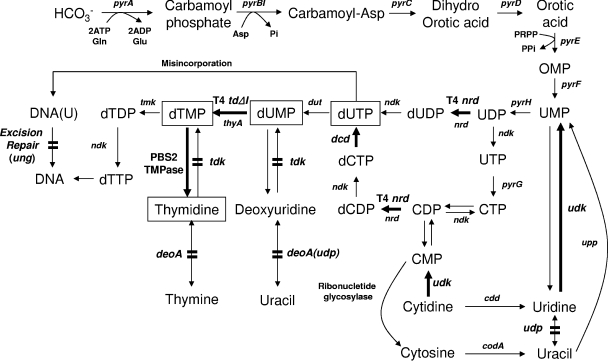
In nature, there are two distinct pathways for dTTP synthesis, the salvage and de novo pathways. The salvage pathway enables the cells to utilize preformed nucleobases and nucleosides for nucleotide synthesis, using thymidine phosphorylase (deoA), uridine phosphorylase (udp), and thymidine kinase (tdk) (Fig. 1) (40).
FIG. 1.
Thymidine biosynthetic pathway. The steps engineered in this study are indicated by the bold arrows and lines. Components of the catabolism are as follows: pyrA, carbamoylphosphate synthase; pyrBI, aspartate-carbamoyl transferase; pyrC, dihydroorotase; pyrD, dihydroorotate oxidase; pyrE, orotate phosphoribosyltransferase; pyrF, OMP decarboxylase; pyrG, CTP synthetase; pyrH, UMP kinase; TMPase, TMP phosphohydrolase; nrd, nucleotide diphosphate reductase; tdΔI, T4 thymidylate synthase (intron deleted); thyA, thymidylate synthase; dcd, dCTP deaminase; udk, uridine kinase; deoA, thymidine phosphorylase; tdk, thymidine kinase; udp, uridine phosphorylase; dut, deoxyribonucleotide triphosphatase; ndk, nucleotide diphosphate kinase; tmk, TMP kinase; ung, uracil-DNA N-glycosylase; upp, uracil phosphoribosyl-transferase; cdd, cytidine deaminase; codA, cytosine deaminase.
As the name indicates, the de novo pathway enables the cells to synthesize nucleobases de novo. The de novo pathway leading to thymidine biosynthesis starts with the condensation of aspartate and carbamoylphosphate, synthesized by carbamoylphosphate synthase (pyrA) (41). This condensation reaction is catalyzed by aspartate-carbamoyl transferase (pyrBI) to produce carbamoyl aspartate, which undergoes several reactions to produce UMP, the common precursor for the synthesis of the pyrimidine ribonucleoside and deoxynucleosides (Fig. 1) (39-41). For thymidine biosynthesis, UMP is converted to UDP in a reaction catalyzed by UMP kinase (pyrH), and UDP is converted to dUDP by ribonucleoside diphosphate reductase (nrdAB), which is regulated by NTP effectors through binding to specific allosteric sites on ribonucleotide diphosphate reductase (nrdA). Escherichia coli can synthesize dUMP from both dCDP and dUDP. The major pathway involves phosphorylation of dCDP to dCTP, deamination of dCTP to dUTP, and hydrolysis of dUTP to dUMP. Only 20 to 30% of the cellular dUMP is supplied by hydrolysis of dUTP (29, 37). The deamination of dCTP (dcd) is located at a branch point in the pyrimidine metabolic pathway. Because of its importance, dcd is regulated by a positive homotropic cooperativity toward dCTP and by a feedback inhibition by dTTP (29, 31, 40).
Deoxyuridine triphosphatase (dUTPase [dut]) is a pyrophosphatase that contains zinc ions (42). dUTPase catalyzes the hydrolysis of dUTP to PPi and dUMP, a substrate for thymidylate synthase (thyA). Generally, the intracellular concentration of dUTP is <10 nmol per 1 g dry cell weight (DCW), and that of dTTP exceeds 500 nmol per 1 g DCW (5, 39, 52). The intracellular dUTP-to-dTTP ratio is increased in dut-deficient mutants, leading to an increased frequency of misincorporation of uracil for thymine in DNA (34). This incorporation is transient only because uracil is removed from DNA via a subsequent excision repair initiated by uracil-DNA N-glycosylase, which is encoded by ung (15, 50). Attempted repair of deoxyuridine residues from DNA without adequate dTTP available to complete the repair reaction can result in multiple single-strand breaks, eventually leading to double-strand breaks (15). Indeed, single- and double-strand breaks accumulate in thymidine-deprived cells (16). In such cells, the loss of uracil glycosylase activity should decrease DNA breaks arising from attempted repair and thereby decrease the toxicity of thymidine depletion.
The synthesis of dTMP from dUMP involves the transfer of a methylene group and two reducing equivalents from 5,10-methylenetetrahydrofolate to dUMP, catalyzed by the dimeric enzyme thymidylate synthase (thyA). Even though ThyA catalyzes the committed step for de novo synthesis of dTTP, neither the activity of the enzyme nor the expression of the thyA gene seems to be regulated (2, 3).
The general strategy used for the development of a thymidine-overproducing strain involves the alleviation of control mechanisms in key pathways. Several different microorganisms have been modified for thymidine production, including E. coli, Brevibacterium helvolum, and Corynebacterium ammoniagenes, by classical mutagenesis methods, and they were selected based on their capacity to grow on toxic thymidine analogues (30, 33, 48, 49). In these studies, feedback inhibition-resistant variants of thymidine biosynthetic enzymes were obtained by random mutation, and high-producing variants were selected. The most optimum B. helvolum strain obtained by this procedure produced 500 mg liter−1 of thymidine by batch fermentation (33). However, engineered B. helvolum and E. coli mutants also produced thymine, deoxyuridine, and uracil, which are unfavorable for thymidine production since it increases costs during the purification process (30, 33, 48, 49). Furthermore, these thymidine-producing strains have residual thymidine degradation activities, resulting in decreased productivities.
Thus, we tried to develop a more efficient thymidine-producing strain by enhancing the de novo pathway leading to thymidine biosynthesis and by disrupting the thymidine salvage pathway. The strategy reported here is based on disrupting genes which encode enzymes involved in thymidine degradation and on expressing foreign genes in the de novo pathway leading to thymidine biosynthesis which encode enzymes that are expected to be less sensitive to feedback inhibition by thymidine than the original enzymes in the host strain. The T4 ribonucleotide diphosphate reductase (nrdAB) operon, T4 thioredoxin (nrdC), T4 thymidylate synthase (td), and PBS2 TMP phosphohydrolase (TMPase) were expressed in an E. coli mutant strain which was modified to block the salvage pathway (deoA, tdk, and udp). In order to increase the influx of dUMP, E. coli dCTP deaminase (dcd), deoxyuridine triphosphatase (dut), and uridine kinase (udk) were expressed with phage-derived genes. We found that the dUTP/dTTP ratio was increased by increasing the level of dUTP in our mutant, leading to the frequent misincorporation of dUTP in DNA. In order to prevent frequent temporary DNA breaks and gaps by excision repair caused by the increased intracellular dUTP/dTTP ratio, uracil-DNA N-glycosylase (ung) was additionally disrupted.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
E. coli strains and plasmids used in this study are listed in Table 1. E. coli BL21 Star (Invitrogen, Groningen, The Netherlands) was used as the host for gene disruption and expression of foreign genes. E. coli XL1-Blue (Stratagene, La Jolla, CA) was used as the cloning host in construction of expression vectors and was grown in Luria-Bertani (LB) medium (5 g liter−1 yeast extract, 10 g liter−1 Bacto tryptone, and 10 g liter−1 NaCl). Conditional replicative oriRγ plasmids were maintained in the pir+ E. coli BW25141 host strain (22). Bacteriophage PBS2 was obtained from the Bacillus Genetic Stock Center (Department of Biochemistry, The Ohio State University). Bacillus subtilis SB19 (ATCC 23856) and bacteriophage T4 (ATCC 11303-B4) were used for the propagation of PBS2 and the amplification of genes, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| BL21 Star(DE3) | F−ompT hsdSB(rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| BW25141 | lacIqrrnBT14 ΔlacZWJ16 ΔphoBR580 hsdR514 ΔaraBADAH33 ΔrhaBADLD78galU95 endABT333uidA(ΔMluI)::pir+recA1 | 22 |
| BLd | BL21 ΔdeoA | This study |
| BLdt | BL21 ΔdeoA Δtdk | This study |
| BLdtu | BL21 ΔdeoA Δtdk Δudp | This study |
| BLdtu1 | BLdtu harboring pETD::Td | This study |
| BLdtu2 | BLdtu harboring pETD::TdNr | This study |
| BLdtu23 | BLdtu2 harboring pACD::DU | This study |
| BLdtu24 | BLdtu2 harboring pACD::DUTm | This study |
| BLdtu24 (dut) | BLdtu2 harboring pACD::DUTmUt | This study |
| BLdtug24 | BLdtu24 Δung | This study |
| Plasmids | ||
| pETDuet | ColE1 replicon, bla | Novagen |
| pACYCDuet | P15A replicon, cat | Novagen |
| pKD3 | Template plasmid, derivative of pANTSγ, FRT-flanked cat | 13 |
| pKD20 | λ Red helper plasmid, derivative of pINT-ts, araC-ParaB and γ β exo DNA fragments | 13 |
| pCP20 | bla and cat, ori(Ts), thermal inducible FRT recombinase | 13 |
| pETD::Td | pETDuet T4 TDΔI expression under tac promoter, bla | This study |
| pETD::TdNr | pETDuet T4 TDΔI, T4 nrdCAB operon expression under tac promoter, bla | This study |
| pACD::DU | pACYCDuet udk-dcd operon expression under tac promoter, cat | This study |
| pACD::DUTm | pACYCDuet udk-dcd operon, PBS2 TMPase expression under tac promoter, cat | This study |
| pACD::DUTmUt | pACYCDuet udk-dcd operon, dut, PBS2 TMPase expression under tac promoter, cat | This study |
The template plasmid pKD3 is a derivative of pANTSγ that contains an FLP recombinase target (FRT) flanking chloramphenicol resistance gene (13, 43). The λ Red helper plasmid, pKD20, is a derivative of pINT-ts which contains araC-ParaB and γ β exo DNA fragments that were PCR generated by using pBAD18 and λ DNA as templates, respectively (24). pCP20 is an ampicillin- and chloramphenicol-resistant plasmid that shows temperature-sensitive replication and thermal induction of FLP synthesis (9).
The E. coli expression vectors pETDuet (Novagen, Madison, WI) and pACYCDuet (Novagen) were used to express foreign genes in E. coli. The pETDuet derivatives contain the origin of ColE1, an ampicillin resistance gene, and foreign genes. The pACYCDuet derivatives contain the origin of P15A, a chloramphenicol resistance gene, and foreign genes.
PCR-mediated gene disruption.
PCR-mediated disruptions of genes were done with slight modifications to the method previously reported (13, 38). PCR fragments which contain a selectable marker flanked by 70-nucleotide (nt)-long oligonucleotides that were comprised of 50-nt-long homology extensions and 20-nt priming sequences for the template, pKD3, were generated. Oligonucleotides used for the generation of gene disruption fragments are shown in Table 2. PCR mixtures containing 1 U thermostable DNA polymerase (Roche Molecular Biomedicals, Basel, Switzerland), 20 mM Tris (pH 8.4), 1.5 mM MgCl2, 1 μM gene disruption primers, 0.2 mM deoxynucleoside triphosphates, and 0.5 μg ml−1 of plasmid pKD3 were incubated at 94°C for 5 min, followed by 30 cycles at 94°C (45 s), 55°C (45 s), and 72°C (1 min), and followed by a final extension time of 10 min at 72°C.
TABLE 2.
Oligonucleotides for gene disruption
| Gene | Direction | Sequencea |
|---|---|---|
| ΔdeoA | Forward | GTTTCAAACCGGCGGGCGGCGTGCGTACTGCGGAAGATGCGCAGAAATATGTGTAGGCTGGAGCTGCTTC |
| Reverse | GACGGGTCAGATTTGGCAGATTGAGCGGGCCTTTACGACCGTTATCAGCTCATATGAATATCCTCCTTAG | |
| Δtdk | Forward | ATGGCACAGCTATATTTCTACTATTCCGCAATGAATGCGGGTAAGTCTACGTGTAGGCTGGAGCTGCTTC |
| Reverse | TTAAGCGTGGCGATGCCTTTCCTGAATAGCCGTTAATGAGTCGACTTGTACATATGAATATCCTCCTTAG | |
| Δudp | Forward | TGATGTTTTTCATCTCGGCCTCACTAAAAACGATTTACAAGGGGCTACGCGTGTAGGCTGGAGCTGCTTC |
| Reverse | GAATTACAGCAGACGACGCGCCGCTTCCACCACGATTTTCACCGCATGGCCATATGAATATCCTCCTTAG | |
| Δung | Forward | TGATGTTTTTCATCTCGGCCTCACTAAAAACGATTTACAAGGGGCTACGCGTGTAGGCTGGAGCTGCTTC |
| Reverse | GAATTACAGCAGACGACGCGCCGCTTCCACCACGATTTTCACCGCATGGCCATATGAATATCCTCCTTAG |
Underlined sequences are priming site sequences for template pKD3.
Transformants carrying a λ Red helper plasmid (pKD20) were grown in 5 ml SOB medium (5 g liter−1 yeast extract, 20 g liter−1 Bacto tryptone, 0.5 g liter−1 NaCl, 0.19 g liter−1 KCl, and 0.95 g liter−1 MgCl2 [pH 7.0]) with 50 mg liter−1 ampicillin and 100 g liter−1 l-arabinose at 30°C to an optical density at 600 nm (OD600) of approximately 0.6 and then were made electrocompetent by concentrating 100-fold and washing three times with ice-cold 10% glycerol. The respective PCR products were purified, digested with DpnI, and then transformed into electrocompetent E. coli BL21 Star carrying a λ Red helper plasmid. Electroporation was done by using a MicroPulser (Bio-Rad Laboratories, Inc., Hercules, CA) with a voltage booster and 0.2-cm chambers using 50 μl of cells and 10 to 100 ng of PCR product, according to the manufacturer's instructions. After electroporation, the cells were grown in 1 ml SOC medium (SOB medium containing 20 mM glucose) to recover the phenotype for 2 h at 37°C, and then one-half was spread onto LB agar plates with 5 μg ml−1 chloramphenicol to select chloramphenicol-resistant transformants. After primary selection, mutants were maintained on medium without antibiotics. To remove helper plasmid, individual colonies were purified nonselectively at 37°C.
Elimination of the antibiotic resistance gene.
Chloramphenicol-resistant mutants selected on plates were transformed with pCP20, and ampicillin-resistant transformants were selected at 30°C, after which a few colonies were nonselectively purified at 43°C and then investigated for loss of all antibiotic resistances. The majority lost the FRT-flanked resistance gene and the FLP helper plasmid, simultaneously (13). Gene disruption and marker elimination were confirmed by using the appropriate antibiotic markers and PCR analysis.
Expression vector construction.
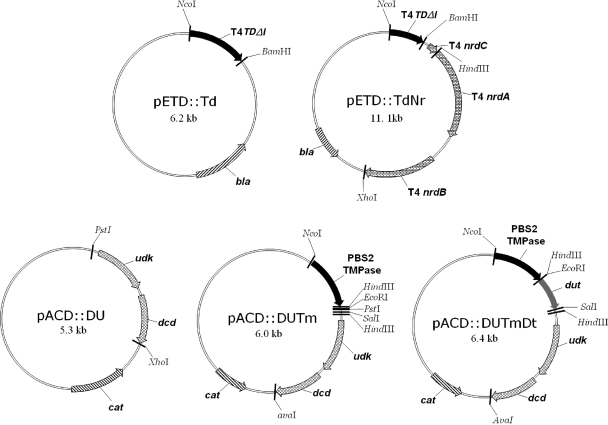
Standard procedures were used for plasmid preparation, restriction enzyme digestions, ligations, transformations, and agarose gel electrophoresis. The T4 thymidylate synthase gene (TD gene) was amplified from the T4 phage by PCR (1,878 bp), and then TDΔI was prepared with deletion of the intron (550 bp to 1,566 bp region) by a gap deletion method for obtaining an active enzyme, since the T4 TD gene is transcribed as preform mRNA and then the intron is deleted by self-splicing (4, 8, 12). The T4 nrdC gene and the T4 nrdA-nrdB operon were amplified by PCR with the flanking primers including 6-nt restriction site extensions for easy cloning and ligated to each other with a ribosome binding site to construct an artificial operon (nrdC-nrdA-nrdB). TDΔI and the nrdC-nrdA-nrdB operon were sequentially ligated to the pETDuet expression vector, according to the standard protocol. The PBS2 TMPase gene, dut, and the udk-dcd operon were amplified by PCR with the flanking primers from each source, and they were sequentially ligated to the pACYCDuet expression vector (Fig. 2). The plasmids pETD::Td, pETD::TdNr, pACD::DU, pACD::DUTm, and pACD::DUTmDt were sequenced by the dideoxy chain termination method with an automatic DNA sequencer to confirm proper cloning. Each expression vector derivative was transformed into BLdtu by the electroporation method, as described in Materials and Methods. These recombinant strains permit transcription from the tac promoter in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside).
FIG. 2.
Maps of plasmids. Plasmid pETDuet was used for the expressions of T4 TDΔI and the T4 nrdCAB artificial operon. Plasmid pACYCDuet was used for the expressions of the udk-dcd operon, the PBS2 TMPase gene, and dut.
Thymidine degradation assay and TMP phosphohydrolase assay.
Equal amounts of transformed cells were harvested by centrifugation and washed with 10 mM Tris (pH 7.4) containing a protease inhibitor mixture (Complete; Amersham Pharmacia, Uppsala, Sweden). Each enzyme solution was prepared by sonication, and its protein concentration was determined by the Lowry assay. For the thymidine degradation assay, 100 μl of 1 mM thymidine was mixed with 100 μl of enzyme solution (1.2 mg ml−1), and the reaction mixture was incubated at 30°C for 1 h, 4 h, and 18 h. The enzyme reaction was subsequently stopped by filtration with polyvinylidene difluoride, and the residual thymidine concentration was measured in the reaction mixture.
For the TMPase assay, the inorganic phosphate released from dTMP was measured by colorimetric assay, as previously described (44). One unit of enzyme catalyzed the hydrolysis of 1 μmol of TMP per minute at 37°C.
RNA isolation and reverse transcription-PCR (RT-PCR).
Total cellular RNA was extracted from mid-log-phase cells with a Qiagen RNeasy mini kit (Qiagen, Hilden, Germany) as described by the manufacturer. RNase-free DNase I (Takara Bio, Shiga, Japan) was treated during the isolation procedure to eliminate possible DNA contamination. Absence of DNA was verified by control PCRs, using the RNA as a template. RNA preparation was diluted to a final concentration of 1 μg μl−1, and reverse transcription was performed with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), using random primer mix, according to the manufacturer's instructions. The sequences of the genes studied were obtained from GenBank, and the primers were designed with the aid of the OLIGO software (version 5.0; Molecular Biology Insights). The PCR was performed under the appropriate condition for each target gene. Relative transcript ratios were quantified using rpoD as an internal control. A negative control was included in all PCR assays.
Culture condition for thymidine production.
A suspension of cells was inoculated into a 250-ml flask containing 50 ml of LB medium and incubated at 37°C and 250 rpm for 8 h. For flask cultures, a 5-ml aliquot of culture broth was transferred to a 500-ml baffled flask containing 50 ml of production medium (60 g liter−1 glycerol as carbon source, 10 g liter−1 CaCO3, 10 g liter−1 yeast extract, 0.4 g liter−1 MgSO4·7H2O, 14.84 g liter−1 soytone, 0.24 g liter−1 phenol red, 50 μg ml−1 chloramphenicol, 50 μg ml−1 ampicillin, and trace elements) and 0.5 mM IPTG for induction and was then incubated at 37°C and 250 rpm for 24 h.
For jar fermenter culture, cells grown in regular LB media in exponential phase were inoculated to the production medium to a final concentration of 10%. All cultivations were performed in a 7-liter jar fermenter (KoBioTech Co., Ltd., Incheon, South Korea) with a volume of 3.5 liters and a stirring rate of 500 rpm for 2 h that was then increased to 800 rpm (two six-blade Rushton-type impellers) at 37°C. The pH was controlled at a minimum of 7.5 with 3 M NH4OH. Thymidine concentration in the medium was determined from the supernatant of cultivation samples. Dissolved oxygen and pH levels were measured with a dissolved-oxygen analyzer (KoBioTech Co., Ltd., Incheon, South Korea) and a pH meter (Mettler Toledo Co., Ltd.).
Analytical methods.
Biomass measured by the OD600 was converted to DCW by using a standard curve (1.0 OD600 = 0.45 g DCW liter−1). The quantitation of bases and nucleosides was analyzed by high-pressure liquid chromatography (Waters 2690; Waters Co., Milford, MA) using an octyldecyl silane MG 5-μm column (4.6 by 250 mm; Shiseido Co., Ltd., Tokyo, Japan) and a UV detector (Waters 2487; Waters Co.) (33). Samples were eluted isocratically with 4% (vol/vol) acetonitrile containing 0.05% (vol/vol) trifluoroacetic acid at a flow rate of 1 ml min−1 and detected at 260 nm. Intracellular dUTP and dTTP levels were measured by high-pressure liquid chromatography as previously reported (20).
RESULTS
Disruption of deoA, tdk, and udp encoding salvage enzymes.
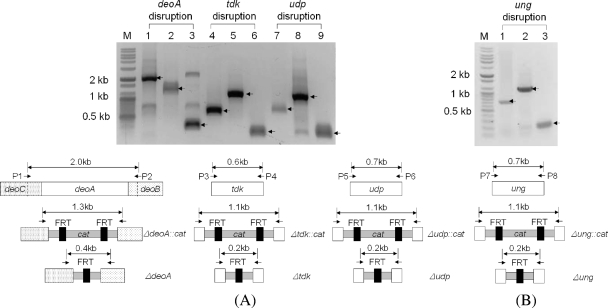
Disruption of deoA was performed using a linear PCR fragment with 50 nt of homology extensions. Using a 1.3-kb fragment, including the 3′ homologous region of deoC, the 5′ homologous region of deoB, and the cat gene, deoA was successfully deleted by gene replacement. Marker elimination was performed using FRT recombinase induced by a temperature shift from 30°C to 42°C in the absence of antibiotic selection. The removal of antibiotic marker was confirmed by PCR amplification of the flanking region of the knocked-out gene. Both tdk and udp were disrupted sequentially by the same method, and the knockout of genes was confirmed by the same method (Fig. 3A).
FIG. 3.
PCR analysis of three disruption mutants. P1 to P8 refer to priming sites. PCR amplification for identifying the deletion of each target gene was performed using each E. coli BL21 mutant genomic DNA as a template. (A) Salvage pathway gene disruptions. M, size marker; lane 1, deoC and deoB region including deoA (2.0 kb); lane 2, ΔdeoC::cat::ΔdeoB (1.3 kb); lane 3, Δ(deoC-deoB) (0.4 kb), represented to ΔdeoA; lane 4, tdk (0.6 kb); lane 5, Δtdk::cat (1.1 kb); lane 6, Δtdk (0.2 kb); lane 7, udp (0.7 kb); lane 8, Δudp::cat (1.1 kb); lane 9, Δudp (0.2 kb). The FRT (black bar)-flanked chloramphenicol-resistant gene was amplified by PCR. The linear disruption PCR fragment (gray bar) was transformed into a strain expressing the λ Red recombinase, and then chloramphenicol-resistant transformants were selected. The selective marker was eliminated by FLP recombinase system. (B) ung disruption. Lane 1, ung (0.7 kb); lane 2, Δung::cat (1.1 kb); lane 3, Δung (0.2 kb).
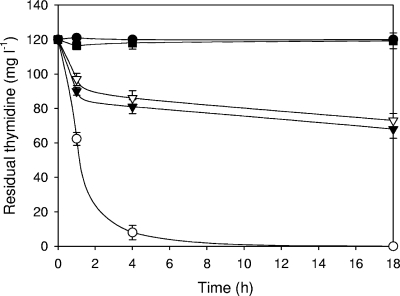
The salvage pathway gene-disrupted strains of E. coli, namely, BLd (ΔdeoA), BLdt (ΔdeoA Δtdk), and BLdtu (ΔdeoA Δtdk Δudp), were prepared and analyzed by in vitro thymidine degradation assays. The thymidine in the wild-type E. coli strain BL21 Star was almost completely degraded to thymine after 4 h (Fig. 4). On the other hand, single disruption of the deoA gene reduced the rate of degradation of thymidine. The residual thymidine concentrations in the BLd strain decreased slowly for 4 h and remained constant at approximately 70% thereafter. The degradation rate of the BLdt strain was almost the same as that of BLd, which is consistent with the fact that Tdk does not participate in thymidine degradation. As shown in Fig. 4, the disruption of deoA alone was not enough to prevent the degradation of thymidine. Since deoA and udp have enough activities to degrade thymidine in E. coli (40), we constructed another strain (BLdtu), in which two genes encoding major enzymes involved in thymidine degradation were disrupted. The result showed that thymidine degradation was completely prevented, not by deoA disruption only but by disruption of both deoA and udp. DeoA and Udp were found to contribute to approximately 60 to 70% and approximately 30 to 40% of thymidine degradation activity after incubation for 18 h, respectively.
FIG. 4.
Thymidine degradation assay. Negative control with thymidine (•), BL21 Star (○), BLd (▾), BLdt (▿), and BLdtu (▪). Values are means ± standard deviations of the results from triplicate experiments.
Expression of de novo thymidine biosynthetic enzymes in a strain with the salvage pathway-deficient background.
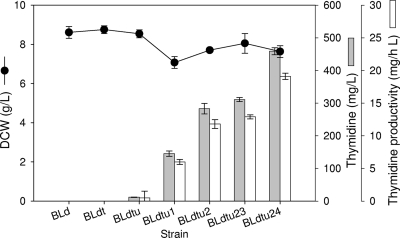
Knocking out three genes (deoA, tdk, udp) involved in the salvage pathway completely abolished thymidine degradation activities, which led to the production of a negligible or small amount of thymidine (Fig. 5). Even though a very small amount of thymidine was produced in the salvage pathway-deficient strain (BLdtu), the inactivation of this pathway can prevent thymidine degradation, so it may be helpful to increase thymidine production, combined with overexpressing foreign genes that are involved in thymidine biosynthesis.
FIG. 5.
Thymidine production by recombinant E. coli strains. Values are means ± standard deviations of the results from triplicate experiments. Thymidine productivity was defined as the produced thymidine concentration per 1 h.
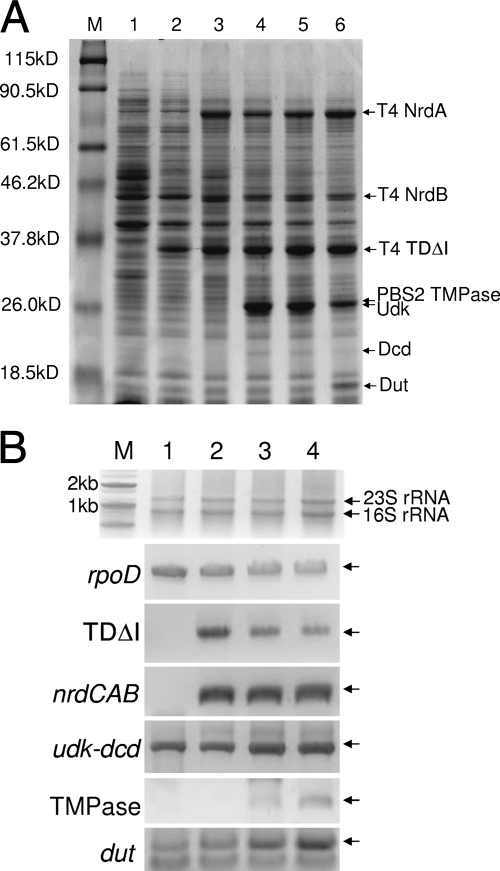
Since ThyA is known to be the committed enzyme to the pyrimidine de novo pathway in E. coli, and since NrdAB is known to be a highly regulated enzyme, increasing the activities of both enzymes could be an effective method to increase thymidine production. Therefore, our strategy to enhance thymidine production involves a combinatorial expression of de novo thymidine biosynthetic pathway genes in a salvage pathway-deficient BLdtu strain. In order to enhance enzyme activities efficiently, the genes encoding intron-deleted thymidylate synthase (TDΔI), ribonucleotide diphosphate reductase (NrdAB), and thioredoxin (NrdC) from T4 phage were cloned and expressed in BLdtu under the control of the tac promoter. The expression of each enzyme was identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 6A). In order to verify whether introduced genes were being reliably expressed, RT-PCR was performed to identify the presence of mRNA transcripts (Fig. 6B).
FIG. 6.
Expression of genes for thymidine de novo biosynthesis in recombinant E. coli strains. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of overexpressing proteins. M, molecular-weight size marker; lane 1, BLdtu; lane 2, BLdtu1; lane 3, BLdtu2; lane 4, BLdtu23; lane 5, BLdtu24; lane 6, Bldtu24 (dut). T4 NrdA, T4 nucleotide diphosphate reductase subunit α; T4 NrdB, T4 nucleotide diphosphate reductase subunit β; T4 TDΔI, T4 thymidylate synthase (intron deleted); PBS2 TMPase, PBS2 TMP phosphohydrolase; Udk, E. coli uridine kinase; Dcd, E. coli dCTP deaminase; Dut, E. coli deoxyribonucleotide triphosphatase. (B) RT-PCR analysis of mRNA transcription. M, size marker; lane 1, BLdtu; lane 2, BLdtu2; lane 3, BLdtu24; lane 4, BLdtu24 (dut).
The expression of T4 TDΔI (BLdtu1) alone triggered extracellular accumulation of a significant amount of thymidine (Fig. 5). In addition, the coexpression of the T4 TDΔI and T4 nrdCAB artificial operons brought about an almost twofold increase in the thymidine titer (BLdtu2), compared with that of BLdtu1. The thymidine concentration of BLdtu2 was further increased by additional expression of T4 nrdCAB, confirming the contribution of nrdCAB to thymidine biosynthesis.
As shown in Fig. 1, the intracellular level of dUMP had a direct effect on thymidine production. In order to increase the influx of dUMP, the udk-dcd operon was cloned from E. coli BL21 and overexpressed in a BLdtu23 strain (Fig. 6A). However, the coexpression of the udk-dcd operon in this strain (BLdtu23) increased the thymidine titer only by approximately 30 mg liter−1, suggesting that there can be other rate-limiting factors (Fig. 5).
To increase the efficiency of thymidine synthesis in vivo, one possible approach is to elevate the activity for TMP phosphohydrolyzation to thymidine by expressing a TMP hydrolysis enzyme (TMPase). Since the hydrolysis of dTMP to thymidine did not seem to occur efficiently (1.3 pmol mg−1 min−1), TMPase from phage PBS2 was cloned and coexpressed in a BLdtu24 strain with other de novo pyrimidine biosynthetic genes (Fig. 6). Corresponding to the elevation in TMP hydrolysis activity by TMPase overexpression (from 1.3 [BLdtu23] to 16.6 pmol mg−1 min−1 [BLdtu24]), the amount of thymidine accumulated using BLdtu24 was 1.5-fold greater than that accumulated using BLdtu23 (Fig. 5). This result clearly indicated the importance of TMP phosphohydrolyzation in thymidine production.
Expression of dut and disruption of ung.
As described in Fig. 5, the coexpression of de novo biosynthetic genes and PBS2 TMPase in BLdtu24 resulted in elevated thymidine levels. However, in BLdtu24, the intracellular concentration of dUTP was increased by sevenfold, whereas dTTP pool concentrations were similar, compared with those of BL21 Star (Table 3). The elevated dUTP/dTTP ratio of BLdtu24 (from 0.005 [BL21] to 0.034 [BLdut24]) may lead to an increased frequency of misincorporation of uracil for thymine in DNA. The resulting excision repair events may result in the consumption of dTTP, thereby reducing thymidine production.
TABLE 3.
Effects on the expression of dut and the disruption of ung
| Strain | Cell growth (g/liter)a | Thymidine (mg/liter)a | Thymidine production per cell (mg/g DCW) | dUTP (nmol/g DCW)b | dTTP (nmol/g DCW)b | dUTP/dTTP ratio |
|---|---|---|---|---|---|---|
| BL21 | 12.5 ± 0.3c | NDd | 0 | 3.1 ± 0.9 | 630.2 ± 10.2 | 0.005 |
| BLdtu24 | 10.9 ± 0.3 | 459.0 ± 10.8 | 42.1 | 21.1 ± 3.2 | 621.3 ± 22.9 | 0.034 |
| BLdtu24 (dut) | 6.5 ± 1.5 | 376.2 ± 9.2 | 57.8 | 9.0 ± 0.2 | 1,317.8 ± 16.9 | 0.007 |
| BLdtug24 | 9.5 ± 0.5 | 550.7 ± 6.2 | 58.0 | 18.5 ± 2.3 | 441.4 ± 2.1 | 0.042 |
These data were obtained from flask cultures for 24 h.
Cultures were harvested at a mid-log phase.
The standard deviations were calculated by using standard propagation of error methods.
ND, not detected.
To determine whether decreasing the dUTP/dTTP ratio in BLdtu24 can boost thymidine production further, dUTPase (dut) from E. coli BL21 was overexpressed in this strain. The transcription level of dut in BLdtu24 (dut) was increased by threefold, compared with that in BLdtu24 (Fig. 6B). Therefore, the intracellular dUTP level was decreased to 43% due to an increase in dut activity, compared with that in BLdtu24. As a result, the dUTP/dTTP ratio of BLdtu24 (dut) decreased from 0.034 to 0.007, and dUTP levels also decreased. At this point, the increase in the Dut expression level was expected to cause an increase in thymidine production, due to the subsequent high activity of TMPase (12.8-fold increase). Even though the overexpression of dut increased thymidine production by 1.4-fold per cell by decreasing intracellular dUTP levels, intracellular dTTP levels were increased, and overall thymidine production was decreased, due to growth retardation (Table 3).
Since the overexpression of dut was not advantageous in overall thymidine production, we tried to enhance thymidine production by preventing cells from consuming additive thymidine nucleotides by suppressing subsequent excision repair. In this approach, ung in BLdtu24 was disrupted without expressing dut (BLdtug24) (Fig. 3B). The disruption of ung brought about an almost 1.2-fold increase in the overall thymidine titer, compared with that in BLdtu24, even though cell growth rate was decreased to some extent (Table 3).
Production of thymidine using BLdtu24 and BLdtug24.
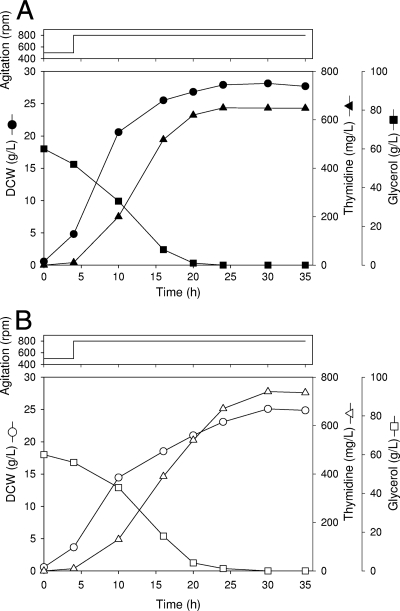
To compare the levels of thymidine production in the BLdtu24 and BLdtug24 strains, aerobic batch cultivation was performed in a 7-liter jar fermenter with thymidine production medium (Fig. 7). In the fermentation of BLdtu24, the carbon source (glycerol) was consumed within 20 h, and the amount of biomass did not increase further after approximately 24 h. The rates of glycerol consumption and cell growth in BLdtug24 were lower than those in BLdtu24. After 30 h of fermentation, BLdtu24 (Fig. 7A) and BLdtug24 (Fig. 7B) reached final biomass concentrations of 28.1 g DCW liter−1 and 25.1 g DCW liter−1, corresponding to final thymidine concentrations of 649.3 mg liter−1 and 740.3 mg liter−1, respectively. Overall thymidine yields for BLdtu24 and BLdtug24 were 10.8 and 12.3 mg of thymidine per 1 g of glycerol, respectively. Neither thymine nor uracil was produced in either strain. These results showed that BLdtug24 is a more effective strain for achieving the overproduction of thymidine than is BLdtu24, even though the growth rate of BLdtug24 was lower than that of BLdtu24.
FIG. 7.
Thymidine production in a 7-liter batch fermenter. (A) Fermentation profile using BLdtu24. (B) Fermentation profile using BLdtug24. • and ○, cell growth; ▵ and ▴, thymidine; ▪ and □, glycerol; and solid line, agitation speed.
DISCUSSION
Generally, de novo biosynthesis of pyrimidine nucleotides is regulated by intracellular concentrations of various nucleotides through feedback inhibition. The intracellular dUMP level is tightly controlled by intracellular dTTP levels through the feedback inhibition of dCTP deaminase and the control of ribonucleotide diphosphate reductase (28, 29, 32). Previously, other workers found that the incorporation rate of exogenously supplied thymine through the salvage pathway during DNA synthesis is very poor in wild-type cells and that the dTTP concentration is maintained at a normal level and increases only during DNA replication (25, 37). In thyA-defective cells, low-dTTP pools promote dUMP accumulation to increase deoxyribose-1-phosphate production through the degradation of excess dUMP, which in turn stimulates the thymidine phosphorylase reaction in the direction of thymidine synthesis (37). On the other hand, thymidine synthesis through the salvage pathway did not occur in a thymidine-overproducing strain (BLdtug24), due to disruptions of deoA and udp (Fig. 4). Because the expression of PBS2 TMPase (44, 45) in this strain may cause the inhibition of DNA biosynthesis from DNA precursor (dTTP) depletion, it can result in decreased feedback inhibitions of dCTP deaminase and ribonucleotide diphosphate reductase. It has been reported that the response of T4 ribonucleotide diphosphate reductase to allosteric effectors such as dATP is different from that of E. coli ribonucleotide diphosphate reductase, although the complete mechanism is not currently known (23, 26, 31). It seems that the expression of T4 nucleotide diphosphate reductase in our system contributed to the enhancement of thymidine production under complex regulatory conditions (Fig. 5), suggesting that the reduction of UDP to dUDP by T4 nucleotide diphosphate reductase may be less inhibited by allosteric regulators.
In this study, increasing the transcriptional levels of E. coli dCTP deaminase fourfold resulted only in an approximately 1.1-fold increase in thymidine production, although a higher yield was expected (Fig. 5). There are several explanations for this behavior. One possibility is that the amount of dCTP deaminase originally present in our system was sufficient to carry out the deamination of dCTP to dUTP, so that increasing the copy number did not greatly increase thymidine production, due to the presence of other limiting factors. A second possibility is that a fourfold overexpression was not sufficient to increase thymidine production and that further increasing the copy number of dCTP deaminase may have a more positive effect on enhancing thymidine production. A third possibility is that E. coli dCTP deaminase is not a good candidate to increase thymidine production. Therefore, expressing foreign deaminase enzymes could potentially be used to increase the thymidine yield. So far, dCTP deaminase is known to be present exclusively in prokaryotes (28, 29, 51, 53). Both bacteria (such as E. coli and Salmonella enterica serovar Typhimurium) and archaea (such as Methanocaldococcus jannaschii) have been shown to contain this enzyme, which is responsible for delivering up to 70 to 80% of the total dUMP needed for dTTP synthesis (29, 40). In most gram-positive bacteria (such as Bacillus subtilis), bacteriophages, and all eukaryotes, dCMP deaminase plays a similar role in the conversion of cytosine nucleotide into uridine nucleotide as that of the above-mentioned dCTP deaminase (1, 6, 14, 36, 46). Both enzyme reactions are key regulatory points of deoxyribonucleotide metabolism and represent the major pathways for dTTP synthesis. It seems that the regulation of dTTP synthesis by dCMP and dCTP deaminases is a part of the overall regulation of deoxynucleoside triphosphate synthesis, along with the complex regulation of ribonucleotide reductase (29, 32). E. coli dCTP deaminase that was used in this study is a monofunctional enzyme that carries out only the deamination of dCTP to dUTP, which is then hydrolyzed to dUMP by dUTPase. Unlike monofunctional dCTP deaminase, the archaeal enzyme sequentially deaminates dCTP and hydrolyzes dUTP to directly yield dUMP and, thus, is bifunctional (7, 27). Therefore, expressing other enzymes that increase the influx of dUTP or dUMP, such as bifunctional dCMP deaminase or dCTP deaminase from other organisms, is a possibility for further enhancing thymidine production in our system.
dUTPase is known to be highly specific for dUTP so that it does not interact with dUDP, UTP, CTP, dCTP, or dTTP, and nonspecific nucleoside diphosphate kinase is known to be a highly active enzyme (21). Thus, it seems reasonable to assume that the dUMP for thymidine synthase is provided via dUTP from dUDP through combined actions of both enzymes. At this point, we do not know exactly why the expression of dut in BLdtu24 was not effective in thymidine production and caused growth rate retardation. One possible explanation could be that intracellular dUTPase activity in BLdtu24 was already too high to show a positive effect on thymidine production and the cells were burdened with the overexpression of dut.
In BLdtu24, the intracellular concentration of dUTP was increased sevenfold, whereas the concentration of the dTTP pool was not altered, resulting in an approximately sevenfold increase in the dUTP/dTTP ratio, compared with that of the parental BL21 strain (Table 3). The increase in the dUTP/dTTP ratio, which was caused by an increase in the dUTP level, may induce incorporation of uracil into its DNA. Therefore, it is possible that an increase in excision repair could have resulted in a decrease in thymidine production by draining the dTTP pool for DNA repair in BLdtu24. ung was disrupted to prevent the loss of dTTP which is consumed in frequent DNA repair, and thymidine production was increased by 1.2-fold compared with that in BLdtu24 (Fig. 7). However, the disruption of ung can cause a build-up of mutations in the BLtug24 strain, which will eventually lead to an increase in genetic instability. Therefore, it will be desirable to find an approach that increases thymidine production while maintaining genetic stability.
In this study, the expression of several viral genes (encoding T4 thymidylate synthase, T4 deoxyribonucleotide diphosphate reductase, and PBS2 TMPase) significantly enhanced thymidine production (Fig. 5). More often than not, viral enzymes are under different regulations compared with E. coli enzymes. Therefore, expressing other viral enzymes can also be considered possible candidates for enhancing thymidine production. For instance, T4 dihydrofolate reductase is known to be more active with NADH than with NADPH, whereas the opposite is true for E. coli enzymes (35). This T4 enzyme is known to have a different stability and response to inhibitors (31). Thus, the expression of T4 dihydrofolate reductase may have beneficial effects on thymidine overproduction.
Previously, we found that more reduced carbon sources, such as glycerol or sorbitol, were better substrates than were all other tested carbon sources (glucose, fructose, lactose, maltose, galactose, or acetate) (unpublished data). In the EMP pathway, two molecules of glycerol are metabolized to two molecules of glyceraldehyde 3-phosphate while producing two molecules of NADH, whereas one glucose molecule is metabolized to two glyceraldehyde 3-phosphate without producing any reducing equivalent (40). In E. coli, there are two nicotinamide nucleotide transhydrogenases (PntAB and UdhA) with a redox-balancing function that can potentially transfer electrons directly from NADH to NADP+ and vice versa (47). In the thymidine biosynthetic pathway, NADPH is known to be essential for the reduction of nucleotides, and the lack of NADPH may inhibit thymidine overproduction by lowering the activities of ribonucleotide diphosphate reductase and dihydrofolate reductase (37, 40). Therefore, growth on glycerol can increase the level of NADH, which in turn can be converted to NADPH by nicotinamide nucleotide transhydrogenases, and thus can be more advantageous to thymidine production.
In this study, several recombinant strains have been engineered to improve thymidine production (Fig. 5). We found that thymidine production using BLdtug24 was significantly improved compared with that using other strains used in previous studies, while suppressing the formation of undesirable by-products to prevent thymidine loss (33, 48, 49). Foreign de novo biosynthetic genes were expressed to enhance thymidine production, while both the salvage pathway genes and ung were disrupted. We found that the overexpression of viral enzymes enhanced thymidine production (Fig. 5). When T4 TD and T4 NrdCAB were overexpressed in a BLdtu strain (BLdtu2), thymidine production was increased approximately 25-fold (from 12 mg liter−1 to 283 mg liter−1) in a flask culture, compared with that using BLdtu. Moreover, overexpressing PBS2 TMPase in BLdtu24 increased thymidine production approximately twofold, compared with that in BLdtu23, in a flask culture. In addition, blocking the drain of dTTP to cells by disrupting ung in BLdtug24 also enhanced thymidine production, although this disruption can potentially lead to genetic instability. As a consequence of these alterations, it was possible to produce thymidine with considerable productivity by simple batch fermentation (7-liter jar) in BLdtug24 (649.3 to 740.3 mg liter−1). This could in principle be easily improved by fed-batch fermentation to further enhance thymidine production and cell growth.
Acknowledgments
We thank Jeff R. Crawford for helpful comments on the manuscript.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Bannwarth, H., and H. G. Schweiger. 1983. The influence of the nucleus on the regulation of the dCMP deaminase in acetabularia. Cell. Biol. Int. Rep. 7:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Belfort, M., G. Maley, J. Pedersen-Lane, and F. Maley. 1983. Primary structure of the Escherichia coli thyA gene and its thymidylate synthase product. Proc. Natl. Acad. Sci. USA 80:4914-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belfort, M., G. F. Maley, and F. Maley. 1983. Characterization of the Escherichia coli thyA gene and its amplified thymidylate synthetase product. Proc. Natl. Acad. Sci. USA 80:1858-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belfort, M., J. Pedersen-Lane, D. West, K. Ehrenman, G. Maley, F. Chu, and F. Maley. 1985. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell 41:375-382. [DOI] [PubMed] [Google Scholar]
- 5.Bertani, L. E., A. Haeggmark, and P. Reichard. 1963. Enzymatic synthesis of deoxyribonucleotides. II. Formation and interconversion of deoxyuridine phosphates. J. Biol. Chem. 238:3407-3413. [PubMed] [Google Scholar]
- 6.Bianchi, V., E. Pontis, and P. Reichard. 1987. Regulation of pyrimidine deoxyribonucleotide metabolism by substrate cycles in dCMP deaminase-deficient V79 hamster cells. Mol. Cell. Biol. 7:4218-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björnberg, O., J. Neuhard, and P. O. Nyman. 2003. A bifunctional dCTP deaminase-dUTP nucleotidohydrolase from the hyperthermophilic archaeon Methanocaldococcus jannaschii. J. Biol. Chem. 278:20667-20672. [DOI] [PubMed] [Google Scholar]
- 8.Capco, G. R., J. R. Krupp, and C. K. Mathews. 1973. Bacteriophage-coded thymidylate synthetase: characteristics of the T4 and T5 enzymes. Arch. Biochem. Biophys. 158:726-735. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Reference deleted.
- 12.Chiu, J., P. E. March, R. Lee, and D. Tillett. 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Saint Vincent, B. R., M. Dechamps, and G. Buttin. 1980. The modulation of the thymidine triphosphate pool of Chinese hamster cells by dCMP deaminase and UDP reductase. Thymidine auxotrophy induced by CTP in dCMP deaminase-deficient lines. J. Biol. Chem. 255:162-167. [PubMed] [Google Scholar]
- 15.Duncan, B. K., P. A. Rockstroh, and H. R. Warner. 1978. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J. Bacteriol. 134:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.el-Hajj, H. H., L. Wang, and B. Weiss. 1992. Multiple mutants of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 174:4450-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freskos, J. N., and K. P. A. Senaratne. March 1988. Synthesis of beta-thymidine. U.S. patent 4,914,233.
- 18.Furman, P. A., J. A. Fyfe, M. H. St. Clair, K. Weinhold, J. L. Rideout, G. A. Freeman, S. N. Lehrman, D. P. Bolognesi, S. Broder, H. Mitsuya, et al. 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 83:8333-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 20.Garrett, C., and D. V. Santi. 1979. A rapid and sensitive high pressure liquid chromatography assay for deoxyribonucleoside triphosphates in cell extracts. Anal. Biochem. 99:268-273. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg, G. R., and R. L. Somerville. 1962. Deoxyuridylate kinase activity and deoxyuridinetriphosphatase in Escherichia coli. Proc. Natl. Acad. Sci. USA 48:247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haldimann, A., L. L. Daniels, and B. L. Wanner. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson, E., and C. K. Mathews. 1994. Allosteric effectors are required for subunit association in T4 phage ribonucleotide reductase. J. Biol. Chem. 269:30999-31005. [PubMed] [Google Scholar]
- 24.Hasan, N., M. Koob, and W. Szybalski. 1994. Escherichia coli genome targeting. I. Cre-lox-mediated in vitro generation of ori− plasmids and their in vivo chromosomal integration and retrieval. Gene 150:51-56. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, K. F., J. C. Leer, and P. Nygaard. 1973. Thymine utilization in Escherichia coli K12 on the role of deoxyribose 1-phosphate and thymidine phosphorylase. Eur. J. Biochem. 40:345-354. [DOI] [PubMed] [Google Scholar]
- 26.Ji, J. P., R. G. Sargent, and C. K. Mathews. 1991. T4 phage ribonucleotide reductase. Allosteric regulation in vivo by thymidine triphosphate. J. Biol. Chem. 266:16289-16292. [PubMed] [Google Scholar]
- 27.Johansson, E., O. Bjornberg, P. O. Nyman, and S. Larsen. 2003. Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases. J. Biol. Chem. 278:27916-27922. [DOI] [PubMed] [Google Scholar]
- 28.Johansson, E., M. Fano, J. H. Bynck, J. Neuhard, S. Larsen, B. W. Sigurskjold, U. Christensen, and M. Willemoes. 2005. Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J. Biol. Chem. 280:3051-3059. [DOI] [PubMed] [Google Scholar]
- 29.Johansson, E., M. Thymark, J. H. Bynck, M. Fano, S. Larsen, and M. Willemoes. 2007. Regulation of dCTP deaminase from Escherichia coli by non allosteric dTTP binding to an inactive form of the enzyme. FEBS J. 274:4188-4198. [DOI] [PubMed] [Google Scholar]
- 30.Kalirai, S. K., B. Scanlon, S. C. Taylor, and S. I. Ahmad. 2000. Screening of microbes, isolation, genetic manipulation, and physiological optimization of Brevibacterium helvolum to produce and excrete thymidine and deoxyuridine in high concentrations. J. Gen. Appl. Microbiol. 46:217-224. [DOI] [PubMed] [Google Scholar]
- 31.Koerner, J. F. 1970. Enzymes of nucleic acid metabolism. Annu. Rev. Biochem. 39:291-322. [DOI] [PubMed] [Google Scholar]
- 32.Larsson, A., and P. Reichard. 1966. Enzymatic synthesis of deoxyribonucleotides. IX. Allosteric effects in the reduction of pyrimidine ribonucleotides by the ribonucleoside diphosphate reductase system of Escherichia coli. J. Biol. Chem. 241:2533-2539. [PubMed] [Google Scholar]
- 33.Lee, H. C., J. M. Ahn, S. N. Lee, and J. H. Kim. 2004. Overproduction of thymidine by recombinant Brevibacterium helvolum amplified with thymidine monophosphate phosphohydrolase gene from bacteriophage PBS2. Biotechnol. Lett. 26:265-268. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl, T. 1979. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol. 22:135-192. [DOI] [PubMed] [Google Scholar]
- 35.Mathews, C. K. 1967. Evidence that bacteriophage-induced dihydrofolate reductase is a viral gene product. J. Biol. Chem. 242:4083-4086. [PubMed] [Google Scholar]
- 36.McIntosh, E. M., and R. H. Haynes. 1986. Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:1711-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Møllgaard, H., and J. Neuhard. 1983. Biosynthesis of deoxythymidine triphosphate, p. 149-198. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides, and nucleobases in microorganisms. Academic Press, London, United Kingdom.
- 38.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 39.Neuhard, J. 1966. Studies on the acid-soluble nucleotide pool in thymine-requiring mutants of Escherichia coli during thymine starvation. 3. On the regulation of the deoxyadenosine triphosphate and deoxycytidine triphosphate pools of Escherichia coli. Biochim. Biophys. Acta 129:104-115. [DOI] [PubMed] [Google Scholar]
- 40.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 446-473. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium. Cellular and molecular biology, vol. 1. American Society of Microbiology, Washington, DC. [Google Scholar]
- 41.O'Donovan, G. A., and J. Neuhard. 1970. Pyrimidine metabolism in microorganisms. Bacteriol. Rev. 34:278-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmén, L. G., K. Becker, L. Bulow, and J. O. Kvassman. 2008. A double role for a strictly conserved serine: further insights into the dUTPase catalytic mechanism. Biochemistry 47:7863-7874. [DOI] [PubMed] [Google Scholar]
- 43.Pósfai, G., M. Koob, Z. Hradecna, N. Hasan, M. Filutowicz, and W. Szybalski. 1994. In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res. 22:2392-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, A. R. 1978. Deoxythymidylate phosphohydrolase from PBS2 phage-infected Bacillus subtilis. Methods Enzymol. 51:285-290. [DOI] [PubMed] [Google Scholar]
- 45.Price, A. R., and S. M. Fogt. 1973. Deoxythymidylate phosphohydrolase induced by bacteriophage PBS2 during infection of Bacillus subtilis. J. Biol. Chem. 248:1372-1380. [PubMed] [Google Scholar]
- 46.Sargent, R. G., and C. K. Mathews. 1987. Imbalanced deoxyribonucleoside triphosphate pools and spontaneous mutation rates determined during dCMP deaminase-defective bacteriophage T4 infections. J. Biol. Chem. 262:5546-5553. [PubMed] [Google Scholar]
- 47.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 48.Song, K. H., D. Y. Kwon, S. Y. Kim, J. K. Lee, and H. H. Hyun. 2005. Thymidine production by Corynebacterium ammoniagenes mutants. J. Microbiol. Biotechnol. 15:477-483. [Google Scholar]
- 49.Tsen, S. 1994. Chemostat selection of Escherichia coli mutants secreting thymidine, cytosine, uracil, guanine, and thymine. Appl. Microbiol. Biotechnol. 41:233-238. [Google Scholar]
- 50.Tye, B. K., P. O. Nyman, I. R. Lehman, S. Hochhauser, and B. Weiss. 1977. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc. Natl. Acad. Sci. USA 74:154-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, L., and B. Weiss. 1992. dcd (dCTP deaminase) gene of Escherichia coli: mapping, cloning, sequencing, and identification as a locus of suppressors of lethal dut (dUTPase) mutations. J. Bacteriol. 174:5647-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner, H. R., B. K. Duncan, C. Garrett, and J. Neuhard. 1981. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 145:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss, B., and L. Wang. 1994. De novo synthesis of thymidylate via deoxycytidine in dcd (dCTP deaminase) mutants of Escherichia coli. J. Bacteriol. 176:2194-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]