Abstract
Industrial production of lactic acid with the current pyruvate decarboxylase-negative Saccharomyces cerevisiae strains requires aeration to allow for respiratory generation of ATP to facilitate growth and, even under nongrowing conditions, cellular maintenance. In the current study, we observed an inhibition of aerobic growth in the presence of lactic acid. Unexpectedly, the cyb2Δ reference strain, used to avoid aerobic consumption of lactic acid, had a specific growth rate of 0.25 h−1 in anaerobic batch cultures containing lactic acid but only 0.16 h−1 in identical aerobic cultures. Measurements of aerobic cultures of S. cerevisiae showed that the addition of lactic acid to the growth medium resulted in elevated levels of reactive oxygen species (ROS). To reduce the accumulation of lactic acid-induced ROS, cytosolic catalase (CTT1) was overexpressed by replacing the native promoter with the strong constitutive TPI1 promoter. Increased activity of catalase was confirmed and later correlated with decreased levels of ROS and increased specific growth rates in the presence of high lactic acid concentrations. The increased fitness of this genetically modified strain demonstrates the successful attenuation of additional stress that is derived from aerobic metabolism and may provide the basis for enhanced (micro)aerobic production of organic acids in S. cerevisiae.
Lactic acid is an organic acid with a wide range of applications. In the food industry, lactic acid has traditionally been used as an antimicrobial as well as a flavor enhancer. Besides having applications in textile, cosmetic, and pharmaceutical industries (5), lactic acid has been applied for the manufacture of lactic acid polymers (11, 40). These polymers have properties that are similar to those of petroleum-derived plastics. Skyrocketing oil prices caused by dwindling fossil fuel reserves coupled with pressures to tackle environmental issues are creating increased demand for bioderived, and often biodegradable, polymers, such as poly-lactic acid.
Current industrial lactic acid fermentations are based on different species of lactic acid bacteria. These bacteria have complex nutrient requirements due to their limited ability to synthesize B vitamins and amino acids (8) and are intolerant to acidic conditions with a pH between 5.5 and 6.5 required for growth (40). Acidification of the growth medium during lactic acid fermentation is typically counteracted by the addition of neutralizing agents (e.g., CaCO3), resulting in the formation of large quantities of insoluble salts, such as gypsum, during downstream processing.
Saccharomyces cerevisiae has received attention as a possible alternative biocatalyst. This organism is relatively tolerant to low pH and has simple nutrient requirements. The production of lactic acid with metabolically engineered S. cerevisiae was achieved by introducing a NAD+-dependent lactate dehydrogenase, leading to the simultaneous formation of both ethanol and lactate (1a, 12, 31, 32, 36). Further improvements were made by constructing a pyruvate decarboxylase-negative (Pdc−) S. cerevisiae strain (1a, 31, 44) that converted glucose to lactic acid as the sole fermentation product.
Although the redox balance and ATP generation in lactic acid fermentation are analogous to those in alcoholic fermentation, engineered homolactic S. cerevisiae strains could not sustain anaerobic growth (44). In addition, the lactate formation rate under anaerobic conditions in the presence of excess glucose was significantly lower than the specific ethanol production rate of the wild-type strain. Moreover, exposure of the anaerobic cell suspension to oxygen immediately led to a 2.5-fold increase in the lactate formation rate. The stimulatory effect of oxygen on lactic acid fermentation may reflect an energetic constraint in lactate fermentation, probably as a consequence of energy-dependent product export (42, 44). In agreement with this hypothesis, intracellular ATP concentrations and the related energy charge decrease rapidly during anaerobic homolactic fermentation by S. cerevisiae (1). Consequently, industrial production of lactic acid with S. cerevisiae may require (micro)aerobic conditions to allow for the generation of sufficient ATP to enable cell growth and, even under nongrowing conditions, maintenance.
The formation of reactive oxygen species (ROS) during cellular respiration is an unavoidable side effect of aerobic life relying on oxygen as the final electron acceptor. At least 2% of oxygen consumed in mitochondrial respiration undergoes only one electron reduction, mainly by the semiquinone form of coenzyme Q, generating superoxide radicals (O2−) (26). In addition, the prooxidant effects of organic acids have been demonstrated using sod mutants (30). An in vitro study by Ali et al. (3) also linked ROS formation to weak organic acids and showed enhanced hydroxy radical (OH) generation in the presence of lactic acid.
Among different ROS, the hydroxy radical that originates from H2O2 in the metal-mediated Fenton/Haber-Weiss reactions is especially reactive. It indiscriminately oxidizes intracellular proteins, nucleic acids, and lipids in the cell membranes (4, 38). Lactate interacts with the ferric ion (Fe3+) to form a stable complex of Fe3+-lactate at a molar ratio of 1:2. This complex then reacts with H2O2 to enhance the OH generation via the Fenton reaction (2, 3). Although a similar in vivo mechanism has not yet been proven, previous research indicates that lactic acid and other weak organic acids enhance oxidative stress of aerobic yeast cultures.
Like other eukaryotic organisms, S. cerevisiae possesses enzymatic defense mechanisms, including several crucial antioxidant enzymes, such as catalase and superoxide dismutase (SOD). SOD removes O2− by converting it to H2O2, which, in turn, can be disproportionated to water by catalase or glutathione peroxidase. Cytosolic catalase, Ctt1p, is thought to play a general role, as CTT1 expression is regulated by various stresses, including oxidative stress, osmotic stress, and starvation (15, 23, 33). More recently, catalase has also been implicated in response to acetic acid tolerance and acetic acid-induced programmed cell death (17, 47).
The goals of the present study were to assess the in vivo relevance of lactate-mediated oxidative stress in S. cerevisiae and to investigate whether its effects could be ameliorated by enhanced expression of catalase.
MATERIALS AND METHODS
Strain construction.
The S. cerevisiae strains used in this work were isogenic members of the CEN.PK family (41). Chromosomal integrations were generated in the genetic background of the uracil-auxotrophic strain CEN.PK 113-5D (MATa ura3 HIS3 LEU2 TRP1 MAL2-8c SUC2). Since this strain is able to consume lactate under aerobic conditions due to the activity of lactate cytochrome c oxidoreductase (cyb2p), the CYB2 gene was knocked out to eliminate lactate consumption and ensure a constant stress level. The cyb2Δ strain was constructed using a disruption cassette obtained with primer pair CYB2-KO-FW/CYB2-KO-RV (Table 1) and pUG6 (18). This strain served as a platform for further genetic modification described below. The genotype of the reference strain is MATa ura3 HIS3 LEU2 TRP1 MAL2-8c SUC2 cyb2::loxP-kanMX-loxP+p426-GPD (empty URA3 vector). The genotype of the CTT1 overexpression strain is MATa ura3 HIS3 LEU2 TRP1 MAL2-8c SUC2 kanMX::loxP-TPI1p::CTT1 cyb2::loxP-URA3-loxP.
TABLE 1.
Primers used in this study
| Primer name | Primer description | Primer sequence (5′-3′) |
|---|---|---|
| CYB2-KO-FW | Knockout of CYB2 | ATCTCGAAGAACTGTGAGGCTGCTATCCTCAGAGCGTCTAAGACTAGATTGTGCGCCTCCAAATCAGAAACAGCTGAAGCTTCGTACGC |
| CYB2-KO-RV | Knockout of CYB2 | CAACACCATTACGACCATAGCATGAGTTCGCATACAAGAATGGTCTACCCGCATAGGCCACTAGTGGATCTG |
| TPI1-Prom-FW | Cloning of TPI1 promoter | GGGGGGCCTATGCGGCCGTGTTTAAAGATTACGGATATTTAACTTAC |
| TPI1-Prom-RV | Cloning of TPI1 promoter | GGGGCCGCGGAGTTTATGTATGTGTTTTTTGTAGTTATAG |
| CTT1-Prom.Repl.-FW | Integration of TPI1 promoter | CGACTCCCTCAACAGGTAGAATTTGTAAGGATTCGACGTAGCCTGGACACCAGCTGAAGCTTCGTACGC |
| CTT1-Prom.Repl.-RV | Integration of TPI1 promoter | CATTTGTGAAGCTGAGCTGATTGATCTTATTGGCATGAGACAAGAGAAGGACACTATAGGGAGACCGGCAG |
Promoter integration cassettes were obtained by amplification of kanMX using pUG6 as a template (18) and specific primers containing sequences homologous to the targeted genes. In order to obtain promoter replacement cassettes containing the TPI1 promoter, this promoter was amplified from pYX222 (R&D Systems Europe Ltd., United Kingdom) using the primers TPI1-Prom-FW and TPI1-Prom-RV (Table 1). The 600-bp PCR product was cloned between the SfiI and SacII sites of pUG6. The resulting PCR template plasmid pUG6-TPI1prom was used for amplification of the kanMX-TPI1 promoter replacement cassettes containing sequences homologous to the upstream region of the targeted open reading frames.
Transformations were performed using a protocol for high-efficiency transformation of yeast (7). After transformation with plasmids, cells were plated on synthetic media. Cultures transformed with integration cassettes were plated on yeast extract-peptone-dextrose containing G418 (200 μg/ml). Confirmation of successful integration was performed by diagnostic PCR.
All stock cultures were grown in shake flasks containing 100 ml synthetic medium with 2% (wt/vol) of d-glucose as a carbon source (as described below). At the end of the exponential phase, 20% (vol/vol) glycerol was added, and 2-ml aliquots were stored at −80°C.
Growth medium and batch cultivation.
Each strain was precultured in synthetic medium containing 5.0 g (NH4)2SO4, 3.0 g KH2PO4, 0.5 g MgSO4·7H2O, and 1 ml trace element solution per liter of demineralized water (45). The initial pH was set to 6.0 using 2 M KOH. After sterilization at 120°C for 30 min, filter-sterilized vitamin solution, as described previously by Verduyn et al. (45), was added. Glucose solution was prepared and sterilized separately (110°C, 20 min) and added to the autoclaved medium to a final concentration of 2% (wt/vol) as a carbon source.
Batch cultivations were conducted in identical synthetic media with the addition of 50 μl liter−1 silicon antifoam (BDH, Poole, United Kingdom) to prevent excessive foaming. Lactic acid was added to a final concentration of 500 mM, and the initial pH was adjusted to 3.0 using KOH pellets. Batches contained 2.5% (wt/vol) glucose as the sole carbon source. Aerobic cultivations with a working volume of 1 liter were sparged with 500 ml min−1 air. The initial batch fermentation was inoculated with an overnight shake flask preculture, and growth was monitored via constant off-gas CO2 measurement. Off-gas was first cooled with a condenser (2°C) and then dried with a Perma Pure dryer (PD-625-12P) before CO2 concentrations in the off-gas were measured with a gas analyzer (NGA 2000; Rosemount). When the concentration of CO2 in the off-gas was ≥1.9% (mid-log to late log phase of growth), a computer-controlled peristaltic waste pump was triggered and about 90% of the culture was removed, leaving approximately 100 ml as an inoculum for the subsequent batch. Fresh growth medium was immediately added with another computer-controlled peristaltic pump connected to an electronic level sensor, which was set to a final volume of 1 liter. The cycle was repeated for a total of six sequential batches for each strain (sequential batch reactor). Specific growth rates for each batch were calculated based on continuous off-gas CO2 measurements.
Enzyme assays.
Cell extracts were prepared with a FastPrep120A machine (Thermo Scientific). Frozen samples (−20°C) were thawed at room temperature, washed, and resuspended in 2 ml ice-cold sonication buffer (100 mM potassium phosphate buffer, 2 mM MgCl2, pH 7.5), with a 1 mM final concentration of 1,4-dithiothreitol. One milliliter of cell suspension was added to safe lock tubes with 0.75 g of cold glass beads (G8772; Sigma). The tubes were placed in a FastPrep120A machine (Qbiogene) and shaken in four bursts of 20 s, at speed 6 (6.0 m/s), for efficient breakdown of the cell membrane. Samples were placed on ice between bursts. The unbroken cells and debris were removed by centrifugation for 20 min at 47,000 × g, 4°C. The supernatant was stored on ice and used for determination of enzyme activities.
Extracts were used to determine the activity of catalase in potassium phosphate buffer (pH 7.5), with hydrogen peroxide as the substrate (46). The rate of H2O2 disappearance was measured spectrophotometrically at 240 nm (U3010; Hitachi). The specific activity of this enzyme was expressed per milligram of soluble protein, and 1 U of enzyme activity corresponded to the decomposition of 1 μmol substrate in 1 min. Protein determination for all cell extracts was carried out by the Lowry method (28).
ROS determination.
Yeast cells were grown overnight in synthetic medium, pH 6.0, with 2% (wt/vol) glucose as a carbon source (as described above). Exponentially growing cells were harvested, transferred into shake flasks containing various stresses, and grown for approximately 18 h. Growth conditions included synthetic medium at pH 3 (negative control) and identical media containing 0.5 mM H2O2 (positive control) or 500 mM lactic acid. Urea was used as a nitrogen source to prevent further acidification of the growth medium (20). After 18 h of growth, the pH of each flask was adjusted to 5.0 with KOH-H2SO4, and 30 ml of each cell suspension was then transferred to 30 ml of potassium phosphate buffer (pH 5.0). Next, 50 μg of the ROS-sensitive fluorescent dye 5-(and -6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA; Invitrogen) dissolved in 100 μl of absolute ethanol was added to each culture (29, 39). Cells were incubated in a shaking incubator at 30°C for 2 h to allow diffusional entry of the dye and reaction with intracellular ROS. After 2 h of loading, 20-ml samples were removed and immediately placed on ice. Cells were harvested by centrifugation at 4,700 rpm, 4°C for 10 min, washed twice with ice-cold demineralized water, and resuspended in 2 ml water. The cell extracts were prepared as described above. Oxidized CM-H2DCFDA in the cell extract was detected using fluorescent spectrophotometry in a 96-well-plate format (Tecan, Austria), with an excitation wavelength of 492 nm and an emission wavelength of 530 nm. Black polystyrene flat-bottomed plates (Corning) were utilized for all measurements. The ROS value was expressed per milligram of soluble protein in the cell extract, as measured by the Lowry method (28).
RESULTS
The effect of lactic acid on the specific growth rate is strongest in aerobic cultures.
The reference strain in this study was derived from the S. cerevisiae CEN.PK family but was altered by disruption of the CYB2 gene to eliminate possible aerobic consumption of lactic acid in order to maintain a consistent level of lactic acid stress at all times. In anaerobic, glucose-grown batch cultures, S. cerevisiae CEN.PK grows with a specific growth rate of 0.34 h−1 (27), while the aerobic growth rate under identical conditions is slightly faster, at 0.37 h−1 (43). Moreover, respiratory dissimilation of glucose generates substantially more ATP than fermentative glucose metabolism. Based on these observations, one would expect that under conditions of lactic acid stress, maintenance of intracellular pH and export of the lactate anion/lactic acid should proceed more easily and the specific growth rate should be higher. However, duplicate aerobic batch cultures containing 500 mM lactic acid at pH 3 exhibited lower growth rates (average of 0.16 h−1) than cultures grown at the same lactic acid concentration and pH under anaerobic conditions (average of 0.25 h−1). The results of these initial batch cultivation experiments were later confirmed in a more tightly controlled and reproducible batch setup (see below).
We hypothesized that the differences in growth rates must be at least partially attributed to the prooxidant effects of weak organic acids (30) and lactic acid in particular (2, 3).
Computer-controlled sequential batch cultures enhance reproducibility of lactic acid stress experiments.
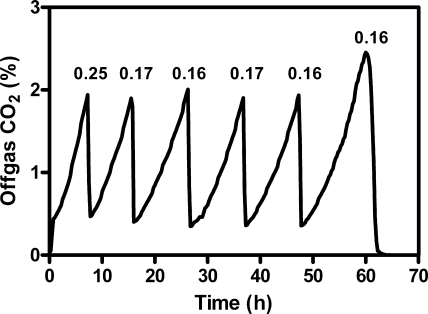
Although initial results using highly standardized shake flask precultivation and single-batch experiments were promising, the reproducibility of the specific growth rate from batch to batch was poor (data not shown). To eliminate the influence of minor variations in cell viability, phase of growth, and residual nutrients that may occur during precultivation, a computer-controlled sequential batch system was utilized (see Materials and Methods). The first batch, grown using an overnight shake flask preculture as the inoculum, consistently grew faster than the subsequent batches. Although we suspected that the initial batch grew faster because of the effects of the precultivation that did not include lactic acid, we did not study these effects any further and omitted these data from calculation of the average growth rate. By use of an automated system enabling inoculation of each batch with a consistent mid-log-phase culture, the standard deviation for the last five batches was very low (Fig. 1; also see Fig. 3), and all reported values are representative of five sequential batch cultures (see Fig. 3).
FIG. 1.
Typical CO2 off-gas profile for fermentations of the reference strain in the sequential batch reactor system. The batch fermentor was emptied and refilled automatically when the CO2 concentration in the off-gas was ≥1.9%. The specific growth rates (h−1) for each batch were calculated from the change in CO2 production and are indicated above each peak.
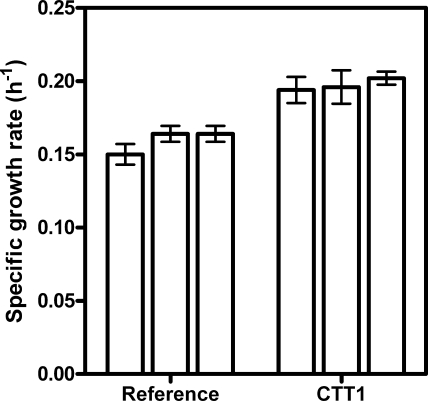
FIG. 3.
Average specific growth rates of aerobic cultivations performed in synthetic medium with glucose containing 500 mM lactic acid and controlled at pH 3. Each bar in this figure indicates the average growth rate of one independent sequencing batch reactor experiment consisting of five sequential batches. Error bars indicate standard deviations. Per strain, a total of 15 batch experiments were performed.
In vivo evidence for lactate-mediated formation of ROS.
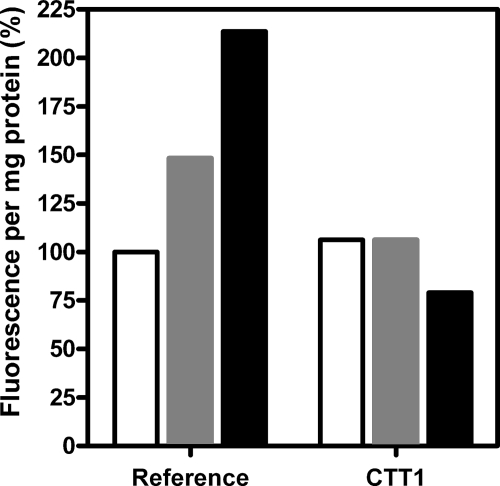
To investigate whether lactate-mediated generation of ROS did indeed occur in aerobic batch cultures, intracellular concentrations of ROS were measured with the fluorescent dye CM-H2DCFDA (see Materials and Methods). Indeed, aerobic cultivation of the reference strain in the presence of lactate led to the enhanced formation of ROS (Fig. 2). The relatively high ROS values obtained upon lactate addition may be attributed to the constant lactate concentrations which were ensured by the cyb2 deletion, which eliminates lactate metabolism. In comparison, hydrogen peroxide may be partially metabolized over time, resulting in a lower signal. It is also possible that the dye interacts differentially with the various forms of ROS (see Table 18.3 in reference 22). Nevertheless, assays performed with the reference strain prove that ROS levels are significantly increased in the presence of peroxide and lactate.
FIG. 2.
Quantification of intracellular ROS in shake flask cultures of S. cerevisiae grown in synthetic medium at pH 3. The fluorescence of CM-H2DCFDA was measured for unstressed control cultures (white bars) and in response to 0.5 mM H2O2 (gray bars) or 500 mM lactic acid (black bars). All values represent the fluorescence per milligram protein and are reported as percentages of values for the reference strain determined under unstressed conditions.
Catalase overexpression partially alleviates aerobic lactic acid stress.
If, as outlined in the introduction, lactate induces oxidative stress via catalysis of the conversion of H2O2 to hydroxyl radicals by lactate-Fe3+ complexes, reduction of intracellular H2O2 concentrations may provide a defense strategy. However, in aerobic batch cultures grown on glucose, either in the absence or in the presence of lactic acid, catalase activity was below 1 U mg protein−1. This result was anticipated based on the severe glucose repression of the CTT1 gene (9, 21, 34), encoding cytosolic catalase.
To investigate whether cytosolic catalase might confer protection against lactate-mediated ROS formation, the promoter of the CTT1 gene was replaced by the strong and constitutive promoter of the glycolytic TPI1 gene. This genetic modification led to high specific activities of catalase (57 U mg protein−1) during batch cultivation in the presence of glucose. The effect of catalase overexpression on lactic acid stress was investigated by the sequential batch reactor system described above. The average specific growth rates from three independent trials of five batches for both the reference strain and the strain containing altered expression of CTT1 showed a small but highly significant positive impact of this engineering strategy (Fig. 3). The overall average growth rate for all 15 batches was 0.16 ± 0.01 h−1 for the reference strain and 0.20 ± 0.01 h−1 when CTT1 was overexpressed. This represents an increase of approximately 25% in the specific growth rate, and the low standard deviations indicate that this change is statistically significant (t test, P < 0.0001).
To prevent the formation of the hydroxy radical, it is obviously important to limit the buildup of the immediate precursor hydrogen peroxide. Therefore, we expected that formation of the hydroxy radical, which is enhanced by lactic acid, should be limited as well and thereby result in decreased ROS levels in the presence of lactic acid. Although the fluorescent dye CM-H2DCFDA cannot discriminate between different ROS species (22), the total intracellular ROS concentration in the catalase overexpression strain was lower than that of the reference strain when exposed to lactic acid (Fig. 2).
DISCUSSION
The increased levels of intracellular ROS in lactic acid-stressed aerobic cultures (Fig. 2) and the significant positive effect of cytosolic catalase overexpression (Fig. 3) prove that lactic acid induces oxidative stress in S. cerevisiae. These results are in agreement with work performed with sod mutants, which showed aerobic sensitivity to organic acids that was relieved under anaerobic conditions (30). Although genes involved in ROS scavenging and detoxification have previously been implicated in tolerance to organic acids (6, 25, 35, 47), to our knowledge this is the first direct proof that aerobic organic acid toxicity and the resulting decreased aerobic growth rates are at least partially mediated by ROS formation.
The protective effect of catalase overexpression is fully consistent with the proposal, based on in vitro research, that lactate-Fe3+ complexes catalyze the conversion of hydrogen peroxide into hydroxy radicals via the Fenton reaction (2, 3). However, since the method used for measuring intracellular ROS levels did not discriminate between different ROS species, we cannot rule out the possibility that lactic acid increases the formation of, for instance, superoxide or hydrogen peroxide via other mechanisms. Although catalase does not act directly on superoxide, the equilibrium between hydrogen peroxide and superoxide combined with decreased levels of hydrogen peroxide may effectively pull the SOD reaction toward hydrogen peroxide formation. Overexpression of SOD appears to provide a logical approach to investigate the role of superoxide in lactate-mediated oxidative stress and to protect S. cerevisiae against such possible effects. However, in contrast to catalase, SOD is present at high levels in nitrogen-limited, glucose-grown aerobic chemostat cultures (37) and even in glucose-grown aerobic batch cultures (13). Moreover, the effects of SOD overexpression in S. cerevisiae are ambiguous and dependent on copper concentrations and/or the activity of the copper chaperone Ccs1p (19). Attempts in our laboratory to achieve combined overexpression of CTT1 and different SOD constructs were unsuccessful, as we could not confirm significantly enhanced SOD activity (data not shown).
In a recent study, Branduardi et al. (6) reported a protective effect of ascorbic acid, produced via the integration of a heterologous pathway, on lactate stress in S. cerevisiae. While their results are fully consistent with those reported in the present study, the introduction of a heterologous pathway may have some disadvantages in industrial applications compared with the overexpression of native defense mechanisms (such as CTT1) against oxidative stress. In addition to the increased complexity, the production of ascorbic acid as a stress protectant may lead to a loss of substrate carbon for its formation and for redox balancing.
The present study has clear implications for metabolic engineering and process design of S. cerevisiae for the production of lactic acid and other organic acids. It provides a strong incentive for developing strains that are capable of anaerobic lactic acid production. At the same time, it identifies oxidative stress as a key factor in metabolic engineering and process design for yeast-based production of lactic acid. Further efforts to quantitate the role of individual oxygen radicals and defense mechanisms will contribute to directing further metabolic engineering. Such studies should take into account that ROS detoxification in S. cerevisiae is a complex and often redundant network of enzymatic and nonenzymatic reactions (38) that can influence cellular processes ranging from metal homeostasis (10), redox metabolism (14), and cell signaling (24) to programmed cell death (16, 17). Thus, altered expression of the enzymes involved may have far-reaching effects and unexpected outcomes.
Acknowledgments
We acknowledge Pervin Aktug, who contributed to this work as a part of her M.S. studies. We are also grateful to Jean-Marc Daran for helpful advice with strain construction.
We thank Tate & Lyle Ingredients Americas, Inc., for financial support. The Kluyver Centre for Genomics of Industrial Fermentation is supported by The Netherlands Genomics Initiative.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Abbott, D. A., J. van den Brink, I. M. K. Minneboo, J. T. Pronk, and A. J. A. van Maris. Anaerobic homolactate fermentation with Saccharomyces cerevisiae results in depletion of ATP and impaired metabolic activity. FEMS Yeast Res., in press. [DOI] [PubMed]
- 1a.Adachi, E., M. Torigoe, M. Sugiyama, J. I. Nikawa, and K. Shimizu. 1998. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. J. Ferment. Bioeng. 86:284-289. [Google Scholar]
- 2.Ali, M. A., and T. Konishi. 1998. Enhancement of hydroxyl radical generation in the Fenton reaction by alpha-hydroxy acid. Biochem. Mol. Biol. Int. 46:137-145. [DOI] [PubMed] [Google Scholar]
- 3.Ali, M. A., F. Yasui, S. Matsugo, and T. Konishi. 2000. The lactate-dependent enhancement of hydroxyl radical generation by the Fenton reaction. Free Radic. Res. 32:429-438. [DOI] [PubMed] [Google Scholar]
- 4.Beckman, J. S., J. Chen, H. Ischiropoulos, and J. P. Crow. 1994. Oxidative chemistry of peroxynitrite. Methods Enzymol. 233:229-240. [DOI] [PubMed] [Google Scholar]
- 5.Benninga, H. A. 1990. The history of lactic acid making. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 6.Branduardi, P., T. Fossati, M. Sauer, R. Pagani, D. Mattanovich, and D. Porro. 2007. Biosynthesis of vitamin C by yeast leads to increased stress resistance. PLoS ONE 2:e1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-38. [DOI] [PubMed] [Google Scholar]
- 9.Cross, H. S., and H. Ruis. 1978. Regulation of catalase synthesis in Saccharomyces cerevisiae by carbon catabolite repression. Mol. Gen. Genet. 166:37-43. [DOI] [PubMed] [Google Scholar]
- 10.Culotta, V. C., H. D. Joh, S. J. Lin, K. H. Slekar, and J. Strain. 1995. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J. Biol. Chem. 270:29991-29997. [DOI] [PubMed] [Google Scholar]
- 11.Datta, R., S. P. Tsai, P. Bonsignore, S. H. Moon, and J. R. Frank. 1995. Technological and economic potential of poly(lactic acid) and lactic acid derivatives. FEMS Microbiol. Rev. 16:221-231. [Google Scholar]
- 12.Dequin, S., and P. Barre. 1994. Mixed lactic acid-alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei L(+)-LDH. Bio/Technology (New York) 12:173-177. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 14.Drakulic, T., M. D. Temple, R. Guido, S. Jarolim, M. Breitenbach, P. V. Attfield, and I. W. Dawes. 2005. Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 5:1215-1228. [DOI] [PubMed] [Google Scholar]
- 15.Franca, M. B., A. D. Panek, and E. C. A. Eleutherio. 2005. The role of cytoplasmic catalase in dehydration tolerance of Saccharomyces cerevisiae. Cell Stress Chaperones 10:167-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guaragnella, N., L. Antonacci, S. Passarella, E. Marra, and S. Giannattasio. 2007. Hydrogen peroxide and superoxide anion production during acetic acid-induced yeast programmed cell death. Folia Microbiol. 52:237-240. [DOI] [PubMed] [Google Scholar]
- 17.Guaragnella, N., L. Antonacci, S. Giannattasio, E. Marra, and S. Passarella. 2008. Catalase T and Cu, Zn-superoxide dismutase in the acetic acid-induced programmed cell death in Saccharomyces cerevisiae. FEBS Lett. 582:210-214. [DOI] [PubMed] [Google Scholar]
- 18.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, N., M. Bachler, V. Costa, M. Mollapour, P. Moradas-Ferreira, and P. W. Piper. 2005. Overexpressed Sod1p acts either to reduce or to increase the lifespans and stress resistance of yeast, depending on whether it is Cu2+-deficient or an active Cu, Zn-superoxide dismutase. Aging Cell 4:41-52. [DOI] [PubMed] [Google Scholar]
- 20.Hensing, M. C. M., K. A. Bangma, L. M. Raamsdonk, E. Dehulster, J. P. van Dijken, and J. T. Pronk. 1995. Effects of cultivation conditions on the production of heterologous α-galactosidase by Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 43:58-64. [Google Scholar]
- 21.Hörtner, H., G. Ammerer, E. Hartter, B. Hamilton, J. Rytka, T. Bilinski, and H. Ruis. 1982. Regulation of synthesis of catalases and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur. J. Biochem. 128:179-184. [DOI] [PubMed] [Google Scholar]
- 22.Invitrogen. 2008. Probes for reactive oxygen species, including nitric oxide. The handbook—a guide to fluorescent probes and labeling technologies. Invitrogen, Carlsbad, CA. http://www.invitrogen.com/site/us/en/home/References/Molecular-Probes-The-Handbook/Probes-for-Reactive-Oxygen-Species-Including-Nitric-Oxide.html.
- 23.Izawa, S., Y. Inoue, and A. Kimura. 1996. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 320:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 25.Kawahata, M., K. Masaki, T. Fujii, and H. Iefuji. 2006. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 6:924-936. [DOI] [PubMed] [Google Scholar]
- 26.Kowaltowski, A. J., and A. E. Vercesi. 1999. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. 26:463-471. [DOI] [PubMed] [Google Scholar]
- 27.Kuyper, M., A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 4:655-664. [DOI] [PubMed] [Google Scholar]
- 28.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 29.Pereira, M. D., R. S. Herdeiro, P. N. Fernandes, E. C. A. Eleutherio, and A. D. Panek. 2003. Targets of oxidative stress in yeast sod mutants. Biochim. Biophys. Acta 1620:245-251. [DOI] [PubMed] [Google Scholar]
- 30.Piper, P. 1999. Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic. Biol. Med. 27:1219-1227. [DOI] [PubMed] [Google Scholar]
- 31.Porro, D., M. M. Bianchi, L. Brambilla, R. Menghini, D. Bolzani, V. Carrera, J. Lievense, C. L. Liu, B. M. Ranzi, L. Frontali, and L. Alberghina. 1999. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl. Environ. Microbiol. 65:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porro, D., L. Brambilla, B. M. Ranzi, E. Martegani, and L. Alberghina. 1995. Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol. Prog. 11:294-298. [DOI] [PubMed] [Google Scholar]
- 33.Ruis, H., and B. Hamilton. 1992. Regulation of yeast catalase genes, p. 153-172. In J. G. Scandalios (ed.), Molecular biology of free radical scavenging systems. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Rytka, J., A. Sledziewski, J. Lukaszkiewicz, and T. Bilinski. 1978. Haemoprotein formation in yeast. III. The role of carbon catabolite repression in the regulation of catalase A and T formation. Mol. Gen. Genet. 160:51-57. [PubMed] [Google Scholar]
- 35.Schüller, C., Y. M. Mamnun, M. Mollapour, G. Krapf, M. Schuster, B. E. Bauer, P. W. Piper, and K. Kuchler. 2004. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell 15:706-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skory, C. 2003. Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J. Ind. Microbiol. Biotechnol. 30:22-27. [DOI] [PubMed] [Google Scholar]
- 37.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures: combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 38.Temple, M. D., G. G. Perrone, and I. W. Dawes. 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15:319-326. [DOI] [PubMed] [Google Scholar]
- 39.Ueom, J., S. Kwon, S. Kim, Y. Chae, and K. Lee. 2003. Acquisition of heat shock tolerance by regulation of intracellular redox states. Biochim. Biophys. Acta 1642:9-16. [DOI] [PubMed] [Google Scholar]
- 40.Vaidya, A. N., R. A. Pandey, S. Mudliar, M. Suresh Kumar, T. Chakrabarti, and S. Devotta. 2005. Production and recovery of lactic acid for polylactide—an overview. Crit. Rev. Environ. Sci. Technol. 35:429-467. [Google Scholar]
- 41.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. F. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindeløv, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 42.van Maris, A. J. A., W. N. Konings, J. P. van Dijken, and J. T. Pronk. 2004. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab. Eng. 6:245-255. [DOI] [PubMed] [Google Scholar]
- 43.van Maris, A. J. A., J. M. Geertman, A. Vermeulen, M. K. Groothuizen, A. A. Winkler, M. D. W. Piper, J. P. van Dijken, and J. T. Pronk. 2004. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl. Environ. Microbiol. 70:159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Maris, A. J. A., A. A. Winkler, D. Porro, J. P. van Dijken, and J. T. Pronk. 2004. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl. Environ. Microbiol. 70:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeast: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 46.Verduyn, C., J. P. van Dijken, and W. A. Scheffers. 1984. Colorimetric alcohol assays with alcohol oxidase. J. Microbiol. Methods 2:15-25. [Google Scholar]
- 47.Wei, P., Z. Li, P. He, Y. Lin, and N. Jiang. 2008. Genome shuffling in the ethanologenic yeast Candida krusei to improve acetic acid tolerance. Biotechnol. Appl. Biochem. 49:113-120. [DOI] [PubMed] [Google Scholar]