Abstract
Recently, a new type of hybrid resulting from the hybridization between Saccharomyces cerevisiae and Saccharomyces kudriavzevii was described. These strains exhibit physiological properties of potential biotechnological interest. A preliminary characterization of these hybrids showed a trend to reduce the S. kudriavzevii fraction of the hybrid genome. We characterized the genomic constitution of several wine S. cerevisiae × S. kudriavzevii strains by using a combined approach based on the restriction fragment length polymorphism analysis of gene regions, comparative genome hybridizations with S. cerevisiae DNA arrays, ploidy analysis, and gene dose determination by quantitative real-time PCR. The high similarity in the genome structures of the S. cerevisiae × S. kudriavzevii hybrids under study indicates that they originated from a single hybridization event. After hybridization, the hybrid genome underwent extensive chromosomal rearrangements, including chromosome losses and the generation of chimeric chromosomes by the nonreciprocal recombination between homeologous chromosomes. These nonreciprocal recombinations between homeologous chromosomes occurred in highly conserved regions, such as Ty long terminal repeats (LTRs), rRNA regions, and conserved protein-coding genes. This study supports the hypothesis that chimeric chromosomes may have been generated by a mechanism similar to the recombination-mediated chromosome loss acting during meiosis in Saccharomyces hybrids. As a result of the selective processes acting during fermentation, hybrid genomes maintained the S. cerevisiae genome but reduced the S. kudriavzevii fraction.
The genus Saccharomyces consists of seven biological species: S. arboricolus, S. bayanus, S. cariocanus, S. cerevisiae, S. kudriavzevii, S. mikatae, and S. paradoxus (29, 59) and the partially allotetraploid species S. pastorianus (46, 58).
The hybrid species S. pastorianus, restricted to lager brewing environments, arose from two or more natural hybridization events between S. cerevisiae and a S. bayanus-like yeast (7, 16, 28, 46). Recent studies of S. bayanus have also revealed the hybrid nature of certain strains of this species, which has subsequently been subdivided into two groups, S. bayanus var. bayanus, containing a variety of hybrid strains, and S. bayanus var. uvarum, also referred to as S. uvarum, that contains nonhybrid strains (45, 46).
New hybrids of other species from the genus Saccharomyces have recently been described. Hybrid yeasts of S. cerevisiae and S. kudriavzevii have been characterized among wine (6, 20, 33) and brewing yeasts (21); even triple hybrids of S. cerevisiae, S. bayanus, and S. kudriavzevii have been identified (20, 41).
The first natural Saccharomyces interspecific hybrid identified, the lager brewing yeast S. pastorianus (S. carlsbergensis) (42, 57), has become one of the most investigated types of yeast hybrids. The genome structure of these hybrids has been examined by competitive array comparative genome hybridization (aCGH) (5, 16, 28), complete genome sequencing (28), and PCR-restriction fragment length polymorphism (RFLP) analysis of 48 genes and partial sequences of 16 genes (46). The aCGH analyses of several S. pastorianus strains with S. cerevisiae-only DNA arrays (5, 28) revealed the presence of aneuploidies due to deletions of entire regions of the S. cerevisiae fraction of the hybrid genomes. A recent aCGH analysis of S. pastorianus strains with S. cerevisiae and S. bayanus DNA arrays (16) showed two groups of strains according to their genome structure and composition. These groups arose from two independent hybridization events, and each one is characterized by a reduction and an amplification of the S. cerevisiae genome fraction, respectively.
The genetic characterization of the wine S. cerevisiae and S. kudriavzevii hybrids by restriction analysis of five nuclear genes located in different chromosomes, 5.8S-ITS rDNA region and the mitochondrial COX2 gene, revealed the presence of three types of hybrids in Swiss wines, thus indicating the presence of different hybrid genomes (20). In a recent study (21), we identified six new types of S. cerevisiae and S. kudriavzevii hybrids among brewing strains, which were compared to wine hybrids by a genetic characterization based on RFLP analysis of 35 protein-encoding genes. This analysis confirmed the presence of three different genome types among wine hybrids that contain putative chimeric chromosomes, probably generated by a recombination between homeologous chromosomes of different parental origins.
The aim of the present study is to investigate the genome composition and structure of wine hybrids of S. cerevisiae and S. kudriavzevii. This has been achieved by a combined approach based on the RFLP analysis of 35 gene regions from our previous study, comparative genome hybridizations using S. cerevisiae DNA macroarrays, a ploidy analysis by flow cytometry, and gene dose determinations by quantitative real-time PCR. This multiple approach allowed us to confirm the presence of chimeric chromosomes and define the mechanisms involved in their origins.
MATERIALS AND METHODS
Strains.
The yeast strains used in this study are four S. cerevisiae × S. kudriavzevii hybrids. W27 and W46 were isolated from pinot noir must fermentations in Jenins and Stäfa, Switzerland, respectively. They correspond to commercial wine yeasts (Lallemand, Inc., Montreal, Canada). SPG16-91 and 441 were isolated from Muller-Thurgau must fermentations in Wädenswil, Switzerland. The homoploid S. cerevisiae lab strain FY-1679, the haploid S. cerevisiae lab strain S288c, and the S. kudriavzevii type strain IFO 1802 were also used in different experiments performed in the present study.
DNA labeling and competitive genome hybridization.
DNA was extracted from yeast strains grown from single colonies in GPY (2% glucose, 0.5% peptone, 0.5% yeast extract) following the procedure described by Querol et al. (44). Before the labeling reaction, 10 μg of DNA were digested with HinfI (Takara, Japan), according to the manufacturer's instructions, to an average length of 250 bp to 8 kb. The fragmented sample was purified using a QIA-quick gel extraction kit (Qiagen, Germany), heat denatured for 5 min at 100°C, and then cooled on ice. DNA was labeled in 50-μl reaction mixtures by random priming using GE Healthcare Ready-To-Go DNA labeling beads (GE Healthcare Ltd., Amersham Biosciences). Each reaction mix bead contains buffer, FPLCpure Klenow fragment (7 to 12 units), random oligonucleotides (primarily 9-mers), dATP, dGTP, and dTTP. The room-temperature-stable bead is reconstituted with a solution of denatured template (1 μg) and [α-33P]dCTP (40 μC) to a total of 50 μl. The mixture is then incubated at 37°C for 1 to 2 h. Unincorporated nucleotides are removed by filtration through a MicroSpin S-200 HR column (GE Healthcare Ltd., Amersham Biosciences, Germany). Equal amounts of labeled DNA (5 × 106 dpm/ml) were used as probes to hybridize to PCR-amplified open reading frames (ORFs) of homoploid S. cerevisiae FY 1679 DNA spotted onto membrane macroarrays (described in reference 1; see also http://scsie.uv.es/chipsdna/index.html). New macroarrays are pretreated for 30 min at 80°C with 0.5% sodium dodecyl sulfate (SDS). The filters are prehybridized in a rotator oven with 5 ml of prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution, 0.5% SDS, and 100 μg herring sperm DNA/ml) at 65°C for 2 h. The prehybridization solution is replaced with 5 ml of the same solution containing the desired amount of radioactive sample and hybridized for 40 h at 65°C. After the hybridization membranes were washed once at 65°C for 20 min in 2× SSC, 0.1% SDS and twice at 65°C for 30 min in 0.2× SSC, 0.1% SDS. After the washing step, the membranes were exposed to an imaging plate (BAS-MP; Fuji Film, Japan) for 3 days.
Macroarray scanning and data normalization.
Images were acquired using a Fujifilm FLA3000 phosphorimager. The raw data were processed by using Excel spreadsheets. Spot intensities from three replicate hybridizations were measured as artifact-removed density, background, and background-corrected artifact-removed density by using the Array Vision 7.0 software (Imaging Research, Inc., Canada). Poor or inconsistent signals were not considered for further analysis. Only signal intensities higher than 1.5 times the background were considered as valid data and normalized. The normalization process and the measure of the significance level for each ORF were done by using ArrayStat software (Imaging Research, Inc., Canada), considering the data as independent and allowing the program to take a minimum number of two valid replicates in order to calculate the mean and standard deviation values for every gene (only one of the three replicates was allowed to be a removable outlier). Genes with missing values of more than 80% were removed.
Background-corrected DNA macroarray data intensities were normalized to a common reference in another channel by following a global locally weighted scatterplot smoothing (LOWESS) procedure. The common reference used in normalization was generated with respect to the sum of the intensities of all genes. Logarithms were calculated, and the intensity ratio versus the average intensity for dot plots (M-A plots) were represented. The hybridization signal of each ORF from the hybrid and parental strains was normalized to that of the homoploid strain FY1679. Hybridization signals were depicted as the log2 hybridization signal ratio (hybrid/FY1679) with respect to the S. cerevisiae gene order using the program ChARM v1.6 (36).
Flow cytometry.
Hybrid yeasts and the reference homoploid strain were grown in 10 ml of liquid medium under agitation. Early-stationary-phase cells (ca. 106 cells ml−1) were harvested by centrifugation, washed, and fixed in 70% ethanol at 4°C for 5 min. The fixed cells were harvested by centrifugation and resuspended in 0.01 M phosphate-buffered saline buffer (pH 7.2) containing 400 μl of RNase (10 mg ml−1). After incubation at 37°C for 30 min, the cells were harvested by centrifugation and resuspended in 950 μl of 0.01 M phosphate-buffered saline buffer (pH 7.2) containing 50 μl of propidium iodide (0.005%). The samples were analyzed using a flow cytometer FACScan analyzer (Becton Dickinson). Ploidy determination of the hybrid strains was done by comparison against the haploid S288c and diploid FY1679 strains.
Gene copy estimation by qRT-PCR.
General and species-specific oligonucleotide PCR primers for genes located in different chromosome regions were designed (see Table S1 in the supplemental material). These general and species-specific primers were designed according to the available genome sequences of the laboratory strain S. cerevisiae S288c and the Japanese type strain S. kudriavzevii IFO1802, representatives of the parental species. Specificity, efficiency, and accuracy of the primers were tested and optimized by standard PCRs, using DNA from parental and hybrid strains. Primers showing amplification were used in the subsequent quantitative real-time PCR (qRT-PCR) analysis. The amplification of gene fragments from different yeast strains was determined by qRT-PCR using a standard curve method (61). DNA from overnight stationed precultures was extracted in triplicate with a PrepMan kit (PE Applied Biosystems) as described previously (34). qRT-PCR was performed with gene-specific primers (200 nM) in a 20-μl reaction mixture, using the LightCycler FastStart DNA MasterPLUS SYBR green (Roche Applied Science, Germany) in a LightCycler 2.0 system (Roche Applied Science, Germany) device. All samples were processed for melting curve analysis, amplification efficiency, and DNA concentration determination using the LightCycler 2.0 system. For every strain, DNA extracted from 106 CFU and serial dilutions (10−1 to 10−5) were used for a standard curve. The copy number for each gene was estimated by comparing the DNA concentration for S288c (haploid S. cerevisiae strain) or IFO 1802 (diploid S. kudriavzevii strain) with that of the W27 hybrid strain.
Recombination sites in the chimeric chromosomes.
The approximate location of the recombination points in the mosaic chromosomes IV and V was determined from the up and down “jump” locations in the ORF mapping by macroarray analysis of the hybrid yeast genomes.
The recombination site in chromosome IV was approximately placed in the PMT1 gene by amplification with general primers (direct, 5′-CATCTTTGCAAGRGGMGGATGCACRTCCAT-3′, and reverse, 5′-TACCGTTTTGGATTTGGCRAAYTGCATCAT-3′) and subsequent sequencing. Once the recombination region was identified, oligonucleotide primers specific for S. cerevisiae (ScePMT1up, 5′-GACAAACAGGTGCATTGTTAAAGCGAGGT-3′, and ScePMT1down, 5′-CTGTCAGGCCATGCCAAGCCAT-3′) and S. kudriavzevii (SkuPMT1up, 5′-GAGGTGCATTGTGCAATACTTAGGCGAAAC-3′, and SkuPMT1down, 5′-GTCTGGCCAGGCGAGGCTGC-3′) were designed. PCR amplification with different combinations of the species-specific primers allowed us to corroborate the presence of chimeric chromosomes IV, and the subsequent sequencing revealed the recombination site located at the 5′ end of the PMT1 gene.
The PCR was prepared with rTaq, buffer, and deoxynucleoside triphosphate mix (Takara, Japan) according to the manufacturer's instructions as well as 4 μl of DNA (10 to 100 ng) and 1 μl of each oligonucleotide to a final volume of 50 μl. The PCRs were performed in a G-STORM thermocycler (Gene Technologies Ltd., United Kingdom).
The PCR amplification of DNA upstream and downstream of the PMT1 gene was as follows: 45 cycles of 94°C for 45 s, 62°C for 35 s, and 72°C for 1 min, with a final extension of 10 min at 72°C. The PCR products were purified using the PerfectPrep kit (Eppendorf, Germany) and directly sequenced by using the BigDye Terminator v3.1 cycle sequencing kit (PE Applied Biosystems), following the manufacturer's instructions, in an automatic DNA sequencer, model ABI 3730 (PE Applied Biosystems).
Nucleotide sequence accession numbers.
The PMT1 sequences from this study have been submitted to the EMBL database under accession numbers FM211146 to FM211153, and aCGH data were submitted to Gene Expression Omnibus under accession no. GSE12774.
RESULTS
Comparative genome analysis of hybrid strains based on DNA macroarrays.
To determine the genome composition and structure of wine hybrids of S. cerevisiae and S. kudriavzevii, we performed a comparative genome hybridization analysis with S. cerevisiae DNA macroarrays. Of the seven wine S. cerevisiae × S. kudriavzevii hybrids described (20), we selected four strains (W27, W46, SPG16-91, and 441), representative of the three genome types characterized by González et al. (21).
The analysis of the presence of chromosomal rearrangements among Saccharomyces species (17), as well as the complete genome sequencing of S. kudriavzevii (9, 10), showed that this species contains a colinear (syntenic) genome with respect to that of S. cerevisiae, and hence, a similar position of the genes under analysis is expected in the hybrid's chromosomes coming from the S. kudriavzevii parent.
Relative gene copy numbers for the hybrid strains were determined by comparative genome hybridization with respect to the S. cerevisiae homoploid strain FY1679. This strain contains the same genetic background as the haploid strain S288C, whose complete genome sequence is known (19) and is the basis of the DNA macrochip synthesis (1).
The hybridization signal of each ORF from the hybrid strains was normalized to that of FY1679 and shown as the log2 signal ratio (hybrid/FY1679) with respect to the S. cerevisiae gene order in each chromosome (see the supplemental material). Nucleotide divergences among the genomes of the parental species S. cerevisiae and S. kudriavzevii are on average ∼30%. Due to the DNA hybridization stringency conditions used (<10% nucleotide divergence), most genes from the S. kudriavzevii genome fraction of the hybrids did not hybridize to the S. cerevisiae macroarray, and hence, the hybridization differences correspond mainly to the hybrid genome fraction coming from the S. cerevisiae parent.
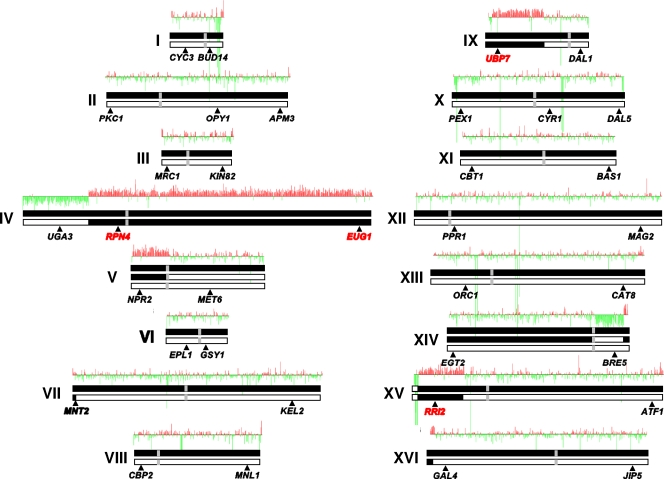
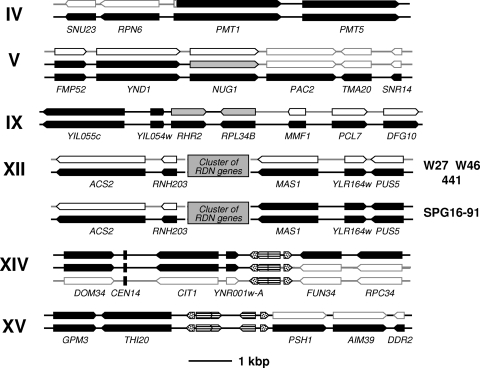
As an example, Fig. 1 shows the hybridization ratios versus the chromosomal gene order for the wine hybrid strain W27. When the log2 hybridization ratios for the hybrid yeast genes were arranged according to the S. cerevisiae gene order, we observed up and down “jumps.” Up jumps correspond to those genes that are overrepresented, i.e., more copies than average, in the hybrid genome fraction coming from S. cerevisiae, and down jumps correspond to genes either absent or underrepresented, with fewer copies than average. Jumps involving large chromosomal regions were observed in chromosomes IV, V, IX, XIV, and XV for all four hybrids and also in chromosome XII for hybrid SPG16-91 (Fig. 2a). Other jumps involved shorter subtelomeric regions in several chromosomes or interdispersed regions of a few genes.
FIG. 1.
Chromosome structures in the S. cerevisiae × S. kudriavzevii hybrid strain W27 as deduced from the DNA array analysis. These putative structures were deduced from the plots of the log2 hybridization ratios (W27 hybrid gene signals divided by those of the homoploid S. cerevisiae FY1679 genes) with respect to the S. cerevisiae gene order of each chromosome. Abrupt changes in the hybridization ratios of some chromosome regions are due to the presence of chimeric recombinant chromosomes generated by the nonreciprocal recombination between homeologous chromosomes. Chromosomes and chromosome regions coming from the S. cerevisiae parent are represented as black bars and those coming from the S. kudriavzevii parent as white bars; vertical gray bars correspond to centromeres. These chromosome structures are congruent with the results of a previous study on the presence/absence of parental genes based on the RFLP analysis of 35 gene regions (21). The locations of these gene regions are indicated above each chromosome, and the results of presence/absence of the parental alleles are summarized as follows: when both parental genes are present, the gene name is in black, but when only the S. cerevisiae gene is present, the gene name is in red. The presence of a S. kudriavzevii gene was not observed.
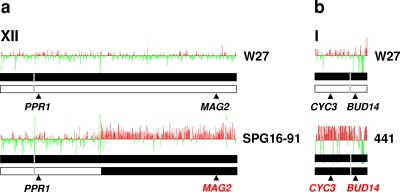
FIG. 2.
Variable chromosome structures in S. cerevisiae × S. kudriavzevii hybrids. Putative structures, as deduced from the results of the macroarray analysis, of those chromosomes of S. cerevisiae × S. kudriavzevii hybrids differing from the structures of strain W27 chromosomes. (a) Structure differences in chromosome XII of hybrid SPG16-91 with respect to that of hybrid W27; (b) structure differences in chromosome I of hybrid 441 with respect to that of hybrid W27. The representation of the chromosomes, plots, and presence/absence of parental genes is the same as described in the legend to Fig. 1.
Large over- or underrepresented chromosomal regions can be interpreted as indicative of regional duplications and deletions, respectively. But, in the case of the hybrid strains, the simplest explanation of the ratios of hybridization data are that the jump locations represent regions where the homeologous (i.e., homologous from different parental species) chromosomes have undergone interchromosomal nonreciprocal recombination (4). This way, the different points of recombination between homeologous chromosomes appear as abrupt changes in the hybridization ratios because the extra copies of some of the hybrid genes coming from the S. cerevisiae parent are in the resulting chimeric chromosomes. This is the case with chromosomes IV, V, IX, XIV, and XV in all hybrids (Fig. 1), as well as chromosome XII in strain SPG16-91 (Fig. 2a). As an example, SPG16-91 genes, coming from S. cerevisiae that are located in the left arm and the first third of the right arm of chromosome XII, show log2 hybridization ratios between −0.5 and 0.5, but those located in the rest of the right arm exhibit log2 ratios around 1. These results indicate that the hybrid contains two copies of the genes in the second region per each copy of the genes in the first, which is compatible with the presence in the SPG16-91 hybrid genome of complete and partial or chimeric forms of S. cerevisiae chromosome XII as indicated in Fig. 2a.
In a previous study (21), we characterized the genome structures of hybrids according to the presence/absence of the parental alleles for 36 gene regions located in the different chromosomal arms. This analysis, summarized in Fig. 1 and 2, demonstrated the absence of the S. kudriavzevii alleles for some genes and their presence for other genes of the same chromosome. These results were already interpreted as evidence of the presence of chimeric chromosomes in the hybrid genomes. As can be seen, these results are congruent with the aCGH analysis, indicating the presence of chimeric forms of chromosomes IV, IX, and XV according to the chromosome structures depicted in Fig. 1, as well as of chromosome XII in the case of hybrid SPG16-91 (Fig. 2a). Both analyses are also congruent for the short subtelomeric rearranged region located in the left arm of chromosome VII, according to the position of gene MNT2 (Fig. 1). Nonetheless, there seems to be a discrepancy between both studies with respect to chromosomes V and XIV. On the one hand, the aCGH analysis of all hybrids shows up and down jumps in the log2 hybridization ratio of genes located in the left arm of chromosome V and in the right arm of chromosome XIV, respectively, which is indicative of either a regional duplication in chromosome V and a deletion in chromosome XIV or the presence of chimeric chromosomes. On the other hand, the RFLP analysis of genes located in these chromosomal regions showed the presence of the two parental alleles. However, both kinds of results are compatible with the presence in the hybrid genome of three different structural forms of chromosomes V and XIV—an intact S. cerevisiae chromosome, an entire S. kudriavzevii chromosome, and a rearranged form, chimeric chromosome, as depicted in Fig. 1.
The remaining genes of the S. cerevisiae genome fraction of the hybrids, located in chromosomes II, III, VI, VII, VIII, X, XI, XIII, XVI, and XII for all hybrids except SPG16-91, in general showed relative hybridization ratios oscillating between −0.5 and 0.5. These results indicate that they have similar relative copy numbers. In our previous analysis of hybrid genome composition, we could observe that genes located on these chromosomes contain two different alleles, each one coming from a different parent. The only exceptions are genes located in chromosome I for strain 441 (Fig. 2b) that exhibit hybridization ratios between 0.5 and 1, which is indicative of the presence of an extra copy of the S. cerevisiae chromosome I in this hybrid genome. Moreover, the RFLP analyses of genes located in this chromosome (21) indicated the absence of S. kudriavzevii chromosome I in the hybrid genome of this strain.
Hybrid ploidy and chromosome copy numbers.
Due to the normalization procedure, the hybridization ratios derived from macroarray analysis show the relative proportions of each gene with respect to the average in the reference strain. Although this method allows those over- and underrepresented regions to be identified, it does not permit the absolute copy numbers of each S. cerevisiae chromosomal region present in the hybrid genome to be determined. The RFLP analysis of gene regions located in the different chromosomes complements the aCGH analysis, providing only qualitative information about the presence/absence of S. cerevisiae and S. kudriavzevii chromosomal regions.
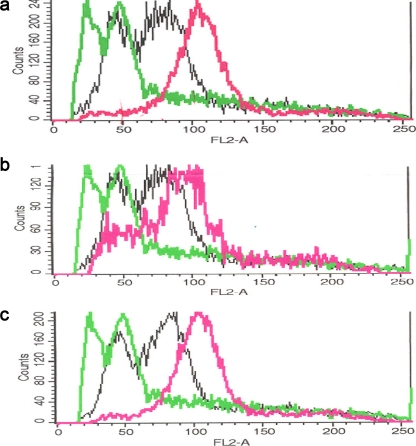
Therefore, we used two additional approaches to decipher the precise genome structure and composition of wine hybrids. First, we used flow cytometry to estimate hybrid ploidies. The comparison of the distribution of DNA content in hybrid cells (depicted in Fig. 3) to those obtained for the reference haploid (S288C) and diploid (FY1679) strains rendered the approximate ploidies of the hybrid strains. Wine hybrids of S. cerevisiae and S. kudriavzevii displayed an estimated ploidy slightly higher than diploidy (n = 2.2 to 2.3), which is indicative of a low level of aneuploidy. This estimated ploidy is compatible with a hybrid genome composition of two copies for most chromosomes and three for chromosomes V and XIV, corresponding to an expected ploidy of 2.11.
FIG. 3.
Flow cytometry analysis of the DNA content per cell in the S. cerevisiae × S. kudriavzevii hybrid strains. The DNA content per cell was measured by flow cytometry for the following S. cerevisiae × S. kudriavzevii hybrid strains: W27 (a), SPG16-91 (b), and 441 (c). The signal of the haploid reference strain S288c is depicted in light green, that of the reference diploid strain FY1679 in black, and those of the hybrids under study in purple.
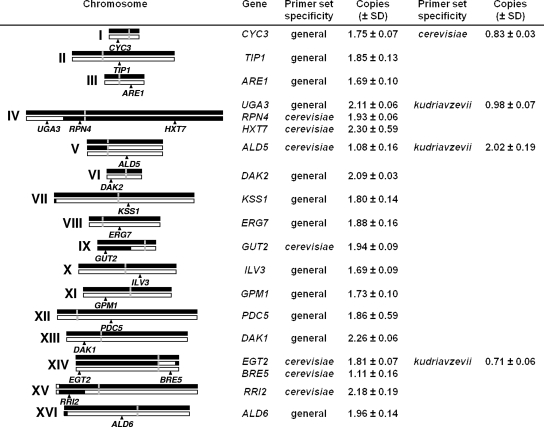
To estimate the copy numbers of the different chromosomes present in the hybrid genomes, we performed a qRT-PCR analysis of several genes, located in the different chromosome regions defined by the aCGH analysis. Since hybrids exhibited the same ploidy and very similar genome composition, this analysis was done only for all gene regions of hybrid strain W27. Figure 4 summarizes the copy numbers estimated for 19 gene regions amplified with general primers (designed and tested for S. cerevisiae and S. kudriavzevii) and species-specific primers, either for S. cerevisiae or for S. kudriavzevii. These primers were designed according to the available genome sequences of the strains S. cerevisiae S288c and S. kudriavzevii IFO1802T, also used for qPCR amplification testing and the generation of the copy number standard curve by the serial DNA dilution method. However, these sequences may differ from the allele sequences present in hybrids, especially in the case of the S. kudriavzevii alleles, according to the sequences available for some other genes (20, 21, 51). This putative sequence divergence explains differences in the amplification efficiency of DNA from hybrids that are responsible for the biases observed in the gene copy number estimates. However, gene copies should correspond to natural numbers and values smaller or higher than expected can be rounded to the closest integer. For example, a value of 1.75 ± 0.07 obtained for CYC3 with general primers is not indicative of a non-sense value of 1¾ copies of the gene present in the hybrid but of two copies amplified with lower efficiency due to nucleotide divergence.
FIG. 4.
Chromosome copy numbers in the S. cerevisiae × S. kudriavzevii hybrids. Copy numbers of chromosomes from hybrid W27 were deduced from the gene dose analysis by the qRT-PCR of 19 gene regions. The gene copy estimations were performed, as indicated, with general primers designed and tested for S. cerevisiae and S. kudriavzevii and/or with species-specific primers, either for S. cerevisiae or for S. kudriavzevii. The genes under analysis are located, as depicted, in the different chromosome regions defined by the aCGH analysis. Chromosomes and chromosome regions from the S. cerevisiae parent are represented as black bars and those from the S. kudriavzevii parent as white bars; the positions of the centromeres are shown as vertical gray bars.
Taking this into consideration, these results support the hypothesis that the hybrid genome of wine strain W27 contains three homeologous forms of chromosomes V and XIV, two of them corresponding to complete S. cerevisiae and S. kudriavzevii chromosomes and the third to a chimeric chromosome. Chromosomes IV, IX, and XIV are present in two forms, one corresponding to a complete S. cerevisiae chromosome and the other to a chimeric chromosome, with one end coming from S. cerevisiae and the other from S. kudriavzevii. Finally, the remaining chromosomes (I to III, VI to VIII, X to XIII, and XVI) are also present in two versions, one coming from S. cerevisiae and the other from S. kudriavzevii, with the exception of some rearranged subtelomeric regions.
With respect to the genome composition of the other hybrids, W46 exhibits the same ploidy and identical chromosome structure as W27. Hybrid 441 differs from W27 only in the composition of chromosome I. The qRT-PCR quantification of the copy numbers of gene CYC3, located in chromosome I, indicates that strain 441 contains two copies of the gene (total copies per cell, 2.07 ± 0.11, estimated using general primers), but both come from S. cerevisiae (S. cerevisiae copies per cell, 1.95 ± 0.10, estimated with species-specific primers). Finally, hybrid SPG16-91 differs only in the structure of chromosome XII, which contains two copies (total copies of gene PDC5, 1.80 ± 0.15), but one corresponds to a complete S. cerevisiae chromosome and the other to a chimeric chromosome.
Recombination sites in chimeric chromosomes.
In the aCGH analysis of hybrid genomes, jumps in the log2 hybridization ratios of genes, located in large chromosomal regions, are due to the presence of chimeric chromosomes generated by the nonreciprocal recombination between homeologous chromosomes. These abrupt changes are observed between the genes flanking (or located at the vicinities of) the recombination points where crossing-over occurred. In this way, we could identify in the chimeric chromosomes those regions where the putative recombination sites are located. The conformation of these regions in the hybrid genomes are described in Fig. 5.
FIG. 5.
Location of the putative recombination sites in the chimeric chromosomes of S. cerevisiae × S. kudriavzevii hybrids. The location of these recombinant regions was deduced from the abrupt changes in the hybridization ratios depicted in Fig. 1 and 2. In the case of chromosome XII, the recombinant form is present only in hybrid SPG16-91 but not in the other, as depicted. The hybrid genes from the S. cerevisiae parent are indicated as black arrows and those from the S. kudriavzevii parent as white arrows, and those genes where the putative recombination site is located are indicated as gray arrows. A gray box represents the large cluster of 100 to 200 tandem repeats containing the highly conserved rRNA genes (RDN genes), where the putative recombination site in the chimeric chromosome XII of strain SPG16-91 is located. A vertical black bar denotes the position of the centromere in chromosome XIV. Dotted and striped arrows represent tRNA and LTRs (delta, sigma, or tau) from Ty retrotransposons, respectively, which could be involved in the recombination events. The recombination site located within PMT1 was confirmed by sequencing (see Fig. 6).
The recombination sites in the chimeric chromosomes XIV and XV are located between genes flanking large regions containing Ty1 delta, Ty3 sigma, and Ty4 tau elements and tRNA genes, according to the genome sequences of S. cerevisiae, S. kudriavzevii, and other Saccharomyces species (available at the Saccharomyces Genome Database [http://db.yeastgenome.org/]).
The recombination site in the chimeric chromosome XII, present only in the hybrid genome of strain SPG16-91, is located within the large cluster of 100 to 200 tandem repeats containing the highly conserved rRNA (RDN) genes.
Finally, recombination sites in chimeric chromosomes IV, V, and IX appear as located within protein-coding genes. The recombination site in chromosome IV is located in the region between genes RPN6 and PMT1, coding, respectively, for a regulatory subunit of the 26S proteasome required for its assembly and activity and an O-mannosyltransferase involved in glycosylation, respectively. The recombination site in chromosome IX is located in the vicinity of the highly conserved gene RPL34B, encoding a ribosomal protein of the large ribosomal subunit (60S). This region has been identified as a meiotic recombination hot spot in the S. cerevisiae genome (18). Finally, the site in chromosome V is apparently located in the region around NUG1, which encodes a GTPase required for the export of 60S ribosomal subunits from the nucleus.
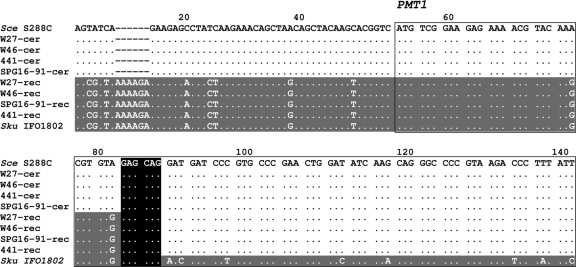
The definite demonstration of the presence of chimeric chromosomes in the hybrid genome would require the sequencing of at least one of the recombinant regions. Thus, we decided to amplify by PCR and sequence one of these putative regions. Recombinant sites located in the chimeric chromosomes XII, XIV, and XV were discarded because they are located within large potential regions. Among those hypothetical recombinant sites placed in the vicinities of protein-coding sequences, the one located in the chimeric chromosome IV was the more feasible. DNAs from the different hybrid strains were used for the PCR amplifications with general primers (described in the supplemental material) of a DNA fragment spanning the RPN6 and PMT1 genes and the region between them. The direct sequencing of the DNA fragments with an automatic sequencer allowed us to determine the location of the recombination region. This region was identified, in the resulting chromatogram, as the nucleotide positions where a transition from double peaks, due to the overlapping S. cerevisiae and S. kudriavzevii sequences, to single peaks, corresponding to the S. cerevisiae sequence, is observed. From these sequences, species-specific primers were designed. The PCR amplification of a 297-bp DNA fragment using S. cerevisiae-specific primers (described in Materials and Methods) pinpointed the presence of nonrecombinant S. cerevisiae chromosome IV. As expected, no amplifications were obtained with both S. kudriavzevii-specific primers (see Materials and Methods), indicative of the absence of a nonrecombinant S. kudriavzevii chromosome IV. But the PCR amplification of a 303-bp DNA fragment, using the suitable combination of S. cerevisiae- and S. kudriavzevii-specific primers (ScePMT1up plus SkuPMT1down [see Materials and Methods]), unequivocally demonstrated the presence of a chimeric chromosome IV in the hybrid genomes.
The subsequent sequencing of the recombination region of the chimeric chromosome IV from the different hybrid strains showed that they share the same recombination event, which occurred in a short region of microhomology located within the PMT1 gene at codons 11 to 12 (Fig. 6). The coding regions of the two PMT1 alleles from hybrids differ only by two synonymous substitutions, and hence, hybrids contain two alleles encoding the same protein but under the control of divergent promoters of different parent origin.
FIG. 6.
Alignment of the partial sequences of the nonrecombinant (cer; S. cerevisiae origin) and recombinant (rec) PMT1 alleles found in S. cerevisiae × S. kudriavzevii hybrid strains and those of the reference strains of the parent species, S. cerevisiae (Sce) S288c and S. kudriavzevii (Sku) type strain IFO1802. A dot indicates nucleotides identical to that from the reference PMT1 sequence of S. cerevisiae S288c. Regions in the hybrid alleles that exhibit a higher similarity to S. cerevisiae sequences are indicated by a lack of shading, and those with a higher similarity to S. kudriavzevii sequences are indicated by gray shading. A continuous rectangle highlights the 5′ end of the PMT1 coding region. The black box indicates the crossing-over site involved in the nonreciprocal recombination between homeologous chromosomes V of the common ancestor of the hybrids.
DISCUSSION
Natural hybridization among Saccharomyces yeasts seems to be more common than previously suspected (53). The best known example of Saccharomyces hybrids are those of the lager yeasts, hybrids of S. cerevisiae and S. bayanus (28, 46). S. cerevisiae × S. bayanus var. uvarum hybrid strains have also been found in wines (2, 30, 35). A new type of hybrids resulting from the hybridization between S. cerevisiae and S. kudriavzevii, the subject of the present study, was first described for wine strains (6, 20, 33) and has recently been described for brewing yeasts (21).
The presence of S. cerevisiae × S. bayanus hybrids is easily explainable, since both species coexist in the same fermentation processes, such as brewing (46), cider production (11, 40, 55), and winemaking (2, 13, 49, 56). Moreover, Le Jeune et al. (30) have recently found S. cerevisiae × S. bayanus var. uvarum hybrids and their putative parents in the same winery.
However, in the case of S. cerevisiae × S. kudriavzevii hybrids, it is not so easy to understand how this hybridization occurred and how these hybrids colonized central European wine fermentations (21, 53). Although the parental species S. cerevisiae is the dominant yeast species in most wine fermentations, S. kudriavzevii has never been found so far in fermentative environments, which suggest that hybridizations likely took place in wild environments. The first strains of S. kudriavzevii were isolated from decayed leaves and soil in Japan (38), and recently, new strains were found on the bark of oak trees in several locations in Portugal (51). Phylogenetic analyses based on some gene sequences showed that hybrids are closer to the Portuguese S. kudriavzevii strains than the Japanese strains (20, 51), which is indicative that hybridization probably occurred in Europe.
The high similarity in the genome structure of the S. cerevisiae × S. kudriavzevii hybrids under study clearly indicates that they share a common ancestor. This is supported by the geographical origin of the strains (eastern Switzerland) and the similar fermentation conditions from which they were isolated. Besides, these hybrids have shown similar tolerance to several stress conditions that are important in wine fermentation (3, 22). In a previous study, we observed that some S. cerevisiae × S. kudriavzevii hybrids isolated in Belgium share a similar genome constitution with Swiss wine strains (21). All these observations support that S. cerevisiae × S. kudriavzevii hybrids were originated by a limited number of hybridization events.
The hybridization of different Saccharomyces species, by the conjugation of haploid spores or cells probably mediated by insects (43, 50), by the rare mating of a vegetative diploid cell and a haploid spore or cell, or by the rare mating of vegetative diploid cells (12), produces a hybrid genome consisting of different copies of the parental genomes. Hybrid diploids are sterile (37), and can only be maintained by asexual reproduction. Only allotetraploid (or amphidiploid) hybrids are fertile, producing diploid hybrid spores (39), but S. cerevisiae × S. kudriavzevii wine hybrids are almost diploid and sterile.
Meiotic segregation in diploid hybrids is known to be ineffective, and the resulting spores are nonviable (<1%) due to the high frequency of aneuploidies (25). Previous studies (24, 32) have shown that reproductive isolation between the Saccharomyces species is due primarily to sequence divergence acted upon by the mismatch repair system (MRS), which contributes to sterility in an interspecific hybrid by preventing successful meiotic crossing-over leading to aneuploidy (25).
Chambers et al. (8) demonstrated that MRS reduces meiotic homeologous recombination in artificial S. cerevisiae × S. paradoxus partial hybrids. When recombination between divergent regions is aborted by the MRS, the absence of functional interchromosomal crossovers may result in aberrant chromosome segregation (meiosis II nondisjunction). However, when recombination is initiated in a region with high homology, the MRS stimulates the loss of one partner of the recombination event in the hybrids and the fixation of the other, a chimeric recombinant chromosome. This mechanism, called recombinant-dependent chromosome loss (RDCL), was demonstrated during meiosis and explains the sterility of hybrids and the correlation between genome divergence and postzygotic reproductive isolation observed in Saccharomyces (32). If a similar mechanism is acting during the less-frequent mitotic recombination, it could explain the chromosome rearrangements observed in the S. cerevisiae × S. kudriavzevii hybrids in the present study, as well as those described for different S. cerevisiae × S. bayanus hybrids (5, 16, 28).
The MRS-mediated chromosome unstability observed in hybrids could also explain differences in the frequency of hybridization among Saccharomyces species. In the case of an RDCL mechanism acting during mitosis, a direct correlation between hybrid genome stability and divergence would be expected. This way, the frequency of homeologous recombination will be lower in a hybrid containing divergent homeologous chromosomes than in a hybrid from closely related species, and hence, the genome stability will be higher in the former than in the latter. Natural hybrids among Saccharomyces species have only been described among the three more distantly related species of this genus, i.e., S. cerevisiae, S. bayanus, and S. kudriavzevii (29), but not among closely related species, such as S. cerevisiae and S. paradoxus, which coexist in the same habitats (48, 54). Although Liti et al. (31) postulated putative S. cerevisiae × S. paradoxus hybrids from the distribution of telomeric repetitive sequences and transposable elements, these strains do not exhibit a hybrid genome (unpublished results from our laboratory), they instead correspond to S. paradoxus strains introgressed with some S. cerevisiae subtelomeric genome regions (32). S. cerevisiae strains introgressed with several S. paradoxus genes have also been described recently (14, 60). All these introgressions are clear indications of ancient unstable hybridization events between these species.
The RDCL model predicts that chimeric chromosomes are generated when the homeologous recombination is initiated in a region of high homology. In the case of S. cerevisiae × S. kudriavzevii hybrids, we have demonstrated that the homeologous recombinations, involved in the generation of chimeric chromosomes, occurred in highly conserved regions, such as defective Ty elements (long terminal repeat [LTR] regions), rRNA regions, and conserved coding genes. Similarly, lager S. cerevisiae × S. bayanus hybrids contain chromosomal rearrangements generated by the recombination between homeologous chromosomes in regions positioned at or close to tRNAs, transposon-related sequences, and origins of replication, well-known sites associated with meiosis-induced double-strand breaks (5, 16). Even though there seems to be no clear association between homologous recombination hot spots and repetitive DNA elements (18), the comparative analysis of Saccharomyces chromosomes and genomes (15, 17, 26, 27) showed that conserved paralogous genes, transposons, and tRNAs are located at the rearrangement breakpoints generated by ectopic recombination initiated in these repetitive and highly conserved regions.
After hybridization, the hybrid genome suffers random genomic rearrangements that, under the strong selective conditions prevailing during wine fermentation (nutrient depletion, osmotic stress, fermenting temperature, increasing levels of ethanol, etc.), are selected. Natural Saccharomyces hybrids have been found so far to be associated with fermentation processes in temperate areas of Europe, regions of continental climate located in the northern limit of grapevine distribution such as northern Spain, France, Germany, Switzerland, Austria, Hungary, etc., where they can also predominate (2, 20, 30, 33). Under such circumstances, however, hybrids may have clear advantages over the parental species (3, 52), because although they are generally less suited than the parents to specific environmental conditions, hybrids can be better adapted to intermediate or fluctuating conditions that can be present in these peripheral areas. This is due to the acquisition of physiological properties from both parents, which provide a mechanism for the natural selection of hybrids (22, 23, 35, 62). For example, S. cerevisiae × S. kudriavzevii hybrids acquired the physiological properties of both parents, a good alcohol and glucose tolerance and a fast fermentation performance from S. cerevisiae and a better adaptation to low and intermediate temperatures, as well as a higher production of glycerol and aroma compounds, from S. kudriavzevii (3, 22).
As a result of these selective processes, a trend to maintain the S. cerevisiae genome and to reduce the S. kudriavzevii fraction is observed in S. cerevisiae × S. kudriavzevii hybrids (21). On the contrary, a group of lager S. cerevisiae × S. bayanus hybrids exhibit an opposite tendency to preserve the S. bayanus-like genome and to reduce the S. cerevisiae fraction (16, 28), while another maintains both genomes (16). However, both types of natural hybrids contain the non-S. cerevisiae mitochondrial genomes, S. kudriavzevii-like in S. cerevisiae × S. kudriavzevii hybrids (21) and S. bayanus-like in S. cerevisiae × S. bayanus hybrids (47), which may impose restrictions to the loss of some non-S. cerevisiae genes from the nuclear hybrid genome involved in mitochondrial functions.
Supplementary Material
Acknowledgments
This work was supported by CICYT grants AGL2006-12703-CO2-01/ALI and -02/ALI from the Spanish Government to A.Q. and E.B., respectively. C.B. gratefully acknowledges an I3P postdoctoral fellowship from the Spanish Research Council (CSIC).
Hybrid strains were kindly provided by Antonio Palacios, Lallemand, Inc., and Jürg Gafner, Swiss Federal Research Station, Wädenswil, Switzerland.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alberola, T. M., J. García-Martínez, O. Antúnez, L. Viladevall, A. Barceló, J. Ariño, and J. E. Pérez-Ortín. 2004. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7:199-206. [PubMed] [Google Scholar]
- 2.Antunovics, Z., L. Irinyi, and M. Sipiczki. 2005. Combined application of methods to taxonomic identification of Saccharomyces strains in fermenting botrytized grape must. J. Appl. Microbiol. 98:971-979. [DOI] [PubMed] [Google Scholar]
- 3.Belloch, C., S. Orlić, E. Barrio, and A. Querol. 2008. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int. J. Food Microbiol. 122:188-195. [DOI] [PubMed] [Google Scholar]
- 4.Bond, U., and A. Blomberg. 2006. Principles and applications of genomics and proteomics in the analysis of industrial yeast strains, p. 175-214. In A. Querol and G. H. Fleet (ed.), Yeasts in food and beverages. Springer-Verlag, Berlin, Germany.
- 5.Bond, U., C. Neal, D. Donnelly, and T. C. James. 2004. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr. Genet. 45:360-370. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury, J., K. Richards, H. Niederer, S. Lee, P. Rod Dunbar, and R. Gardner. 2006. A homozygous diploid subset of commercial wine yeast strains. Antonie van Leeuwenhoek 89:27-37. [DOI] [PubMed] [Google Scholar]
- 7.Casaregola, S., H. V. Nguyen, G. Lapathitis, A. Kotyk, and C. Gaillardin. 2001. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int. J. Syst. Evol. Microbiol. 51:1607-1618. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, S. R., N. Hunter, E. J. Louis, and R. H. Borts. 1996. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 16:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliften, P. F., R. S. Fulton, R. K. Wilson, and M. Johnston. 2006. After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics 172:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliften, P. F., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Coton, E., M. Coton, D. Levert, S. Casaregola, and D. Sohier. 2005. Yeast ecology in French cider and black olive natural fermentations. Int. J. Food Microbiol. 108:130-135. [DOI] [PubMed] [Google Scholar]
- 12.de Barros Lopes, M., J. R. Bellon, N. J. Shirley, and P. F. Ganter. 2002. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1:323-331. [DOI] [PubMed] [Google Scholar]
- 13.Demuyter, C., M. Lollier, J. L. Legras, and C. Le Jeune. 2004. Predominance of Saccharomyces uvarum during spontaneous alcoholic fermentation, for three consecutive years, in an Alsatian winery. J. Appl. Microbiol. 97:1140-1148. [DOI] [PubMed] [Google Scholar]
- 14.Doniger, S. W., H. S. Kim, D. Swain, D. Corcuera, M. Williams, S. P. Yang, and J. C. Fay. 2008. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 4:e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn, B., R. P. Levine, and G. Sherlock. 2005. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, B., and G. Sherlock. 11 September 2008. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed]
- 17.Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405:451-454. [DOI] [PubMed] [Google Scholar]
- 18.Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown, and T. D. Petes. 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6,000 genes. Science 274:546-567. [DOI] [PubMed] [Google Scholar]
- 20.González, S. S., E. Barrio, J. Gafner, and A. Querol. 2006. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6:1221-1234. [DOI] [PubMed] [Google Scholar]
- 21.González, S. S., E. Barrio, and A. Querol. 2008. Molecular characterization of new natural hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii from brewing. Appl. Environ. Microbiol. 74:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González, S. S., L. Gallo, M. D. Climent, E. Barrio, and A. Querol. 2007. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int. J. Food Microbiol. 116:11-18. [DOI] [PubMed] [Google Scholar]
- 23.Greig, D., E. J. Louis, R. H. Borts, and M. Travisano. 2002. Hybrid speciation in experimental populations of yeast. Science 298:1773-1775. [DOI] [PubMed] [Google Scholar]
- 24.Greig, D., M. Travisano, E. J. Louis, and R. H. Borts. 2003. A role for the mismatch repair system during incipient speciation in Saccharomyces. J. Evol. Biol. 16:429-437. [DOI] [PubMed] [Google Scholar]
- 25.Hunter, N., S. R. Chambers, E. J. Louis, and R. H. Borts. 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15:1726-1733. [PMC free article] [PubMed] [Google Scholar]
- 26.Infante, J. J., K. M. Dombek, L. Rebordinos, J. M. Cantoral, and E. T. Young. 2003. Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics 165:1745-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellis, M., N. Patterson, M. Endrizzi, B. W. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 28.Kodama, Y., M. C. Kielland-Brandt, and J. Hansen. 2005. Lager brewing yeast, p. 145-164. In P. Sunnerhagen and J. Piškur (ed.), Comparative genomics: using fungi as models. Springer-Verlag, Berlin, Germany.
- 29.Kurtzman, C. P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233-245. [DOI] [PubMed] [Google Scholar]
- 30.Le Jeune, C., M. Lollier, C. Demuyter, C. Erny, J. L. Legras, M. Aigle, and I. Masneuf-Pomarède. 2007. Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 7:540-549. [DOI] [PubMed] [Google Scholar]
- 31.Liti, G., A. Peruffo, S. A. James, I. N. Roberts, and E. J. Louis. 2005. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22:177-192. [DOI] [PubMed] [Google Scholar]
- 32.Liti, G., D. B. H. Barton, and E. J. Louis. 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopandić, K., H. Gangl, E. Wallner, G. Tscheik, G. Leitner, A. Querol, N. Borth, M. Breitenbach, H. Prillinger, and W. Tiefenbrunner. 2007. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 7:953-965. [DOI] [PubMed] [Google Scholar]
- 34.Martorell, P., A. Querol, and M. T. Fernández-Espinar. 2005. Rapid identification and enumeration of Saccharomyces cerevisiae cells in wine by real-time PCR. Appl. Environ. Microbiol. 71:6823-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masneuf, I., J. Hansen, C. Groth, J. Piškur, and D. Dubourdieu. 1998. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64:3887-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers, C. L., M. J. Dunham, S. Y. Kung, and O. G. Troyanskaya. 2004. Accurate detection of aneuploidies in array CGH and gene expression microarray data. Bioinformatics 20:3533-3543. [DOI] [PubMed] [Google Scholar]
- 37.Naumov, G. I. 1996. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 17:295-302. [Google Scholar]
- 38.Naumov, G. I., S. A. James, E. S. Naumova, E. J. Louis, and I. N. Roberts. 2000. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50:1931-1942. [DOI] [PubMed] [Google Scholar]
- 39.Naumov, G. I., E. S. Naumova, I. Masneuf, M. Aigle, V. I. Kondratieva, and D. Dubourdieu. 2000. Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: auto- and allotetraploidy. Syst. Appl. Microbiol. 23:442-449. [DOI] [PubMed] [Google Scholar]
- 40.Naumov, G. I., H. V. Nguyen, E. S. Naumova, A. Michel, M. Aigle, and C. Gaillardin. 2001. Genetic identification of Saccharomyces bayanus var. uvarum, a cider-fermenting yeast. Int. J. Food Microbiol. 65:163-171. [DOI] [PubMed] [Google Scholar]
- 41.Naumova, E. S., G. I. Naumov, I. Masneuf-Pomarède, M. Aigle, and D. Dubourdieu. 2005. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast 22:1099-1115. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson-Tillgren, T., C. Gjermansen, M. C. Kielland-Brandt, J. G. L. Petersen, and S. Holmberg. 1981. Genetic differences between Saccharomyces carlsbergensis and S. cerevisiae. Analysis of chromosome III by single chromosome transfer. Carlsberg Res. Commun. 46:65-76. [Google Scholar]
- 43.Pulvirenti, A., C. Zambonelli, A. Todaro, and P. Giudici. 2002. Interspecific hybridisation by digestive tract of invertebrates as a source of environmental biodiversity within the Saccharomyces cerevisiae. Ann. Microbiol. 52:245-255. [Google Scholar]
- 44.Querol, A., E. Barrio, and D. Ramon. 1992. A comparative study of different methods of yeast-strain characterization. Syst. Appl. Microbiol. 15:439-446. [Google Scholar]
- 45.Rainieri, S., C. Zambonelli, J. E. Hallsworth, A. Pulvirenti, and P. Giudici. 1999. Saccharomyces uvarum, a distinct group within Saccharomyces sensu stricto. FEMS Microbiol. Lett. 177:177-185. [DOI] [PubMed] [Google Scholar]
- 46.Rainieri, S., Y. Kodama, Y. Kaneko, K. Mikata, Y. Nakao, and T. Ashikari. 2006. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 72:3968-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rainieri, S., Y. Kodama, Y. Nakao, A. Pulvirenti, and P. Giudici. 2008. The inheritance of mitochondrial DNA in lager brewing strains. FEMS Yeast Res. 8:586-596. [DOI] [PubMed] [Google Scholar]
- 48.Redžepović, S., S. Orlić, A. Majdak, and I. S. Pretorius. 2002. Identification and characterization of Saccharomyces cerevisiae and Saccharomyces paradoxus strains isolated from Croatian wines. Lett. Appl. Microbiol. 35:305-310. [DOI] [PubMed] [Google Scholar]
- 49.Rementería, A., J. A. Rodríguez, A. Cadaval, R. Amenábar, J. R. Muguruza, F. L. Hernando, and M. J. Sevilla. 2003. Yeast associated with spontaneous fermentations of white wines from the “Txakoli de Bizkaia” region (Basque Country, North Spain). Int. J. Food Microbiol. 86:201-207. [DOI] [PubMed] [Google Scholar]
- 50.Reuter, M., G. Bell, and D. Greig. 2007. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 17:R81-R83. [DOI] [PubMed] [Google Scholar]
- 51.Sampaio, J. P., and P. Gonçalves. 2008. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74:2144-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serra, A., P. Strehaiano, and P. Taillandier. 2005. Influence of temperature and pH on Saccharomyces bayanus var. uvarum growth; impact of a wine yeast interspecific hybridization on these parameters. Int. J. Food Microbiol. 104:257-265. [DOI] [PubMed] [Google Scholar]
- 53.Sipiczki, M. 18 March 2008. Interspecies hybridisation and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed]
- 54.Sniegowski, P. D., P. G. Dombrowski, and E. Fingerman. 2002. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 1:299-306. [DOI] [PubMed] [Google Scholar]
- 55.Suárez-Valles, B., R. Pando-Bedriñana, N. Fernández-Tascón, A. Querol, and R. Rodríguez-Madrera. 2007. Yeast species associated with the spontaneous fermentation of cider. Food Microbiol. 24:25-31. [DOI] [PubMed] [Google Scholar]
- 56.Torriani, S., G. Zapparoli, and G. Suzzi. 1999. Genetic and phenotypic diversity of Saccharomyces sensu stricto strains isolated from Amarone wine. Antonie van Leeuwenhoek 75:207-215. [DOI] [PubMed] [Google Scholar]
- 57.Vaughan-Martini, A., and C. P. Kurtzman. 1985. Deoxyribonuclease-acid relatedness among species of the genus Saccharomyces sensu stricto. Int. J. Syst. Bacteriol. 35:508-511. [Google Scholar]
- 58.Vaughan-Martini, A., and A. Martini. 1998. Saccharomyces Meyen ex Reess, p. 358-371. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts: a taxonomic study. Elsevier, Amsterdam, The Netherlands.
- 59.Wang, S. A., and F. Y. Bai. 2008. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 58:510-514. [DOI] [PubMed] [Google Scholar]
- 60.Wei, W., J. H. McCusker, R. W. Hyman, T. Jones, Y. Ning, Z. Cao, Z. Gu, D. Bruno, M. Miranda, M. Nguyen, J. Wilhelmy, C. Komp, R. Tamse, X. Wang, P. Jia, P. Luedi, P. J. Oefner, L. David, F. S. Dietrich, Y. Li, R. W. Davis, and L. M. Steinmetz. 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104:12825-12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilhelm, J., A. Pingoud, and M. Hahn. 2003. Real-time PCR-based method for the estimation of genome sizes. Nucleic Acids Res. 31:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zambonelli, C., P. Passarelli, S. Rainieri, L. Bertolini, P. Giudici, and L. Castellari. 1997. Technological properties and temperature response of interspecific Saccharomyces hybrids. J. Sci. Food Agric. 74:7-12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.