Abstract
Molecular fingerprinting techniques offer great promise for analyzing changes in microbial community structure, especially when dealing with large number of samples. However, a serious limitation has been the lack of quantification offered by such techniques since the relative abundances of the identified operational taxonomic units (OTUs) in the original samples are not measured. A quantitative fingerprinting approach designated “qfingerprinting” is proposed here. This method involves serial dilutions of the sample of interest and further systematic fingerprinting of all dilution series. Using the ultimate dilutions for which OTU are still PCR amplifiable and taking into account peak size inaccuracy and peak reproducibility, the relative abundance of each OTU is then simultaneously determined over a scale spanning several orders of magnitude. The approach was illustrated by using a quantitative version of automated ribosomal intergenic spacer analysis (ARISA), here called qARISA. After validating the concept with a synthetic mixture of known DNA targets, qfingerprinting was applied to well-studied marine sediment samples to examine specific changes in OTU abundance associated with sediment depth. The new strategy represents a major advance for the detailed quantitative description of specific OTUs within complex communities. Further ecological applications of the new strategy are also proposed.
Central questions in microbial ecology are related to ecological processes and to spatiotemporal patterns of microbial communities over various scales (29, 36, 39). Despite the persisting difficulty in accurately measuring the vast microbial diversity, microbial ecologists seek to determine the distribution and abundance of microbes, compare the structure of microbial communities, and explain their patterns where contextual parameters are available (37). Although microbial diversity surveys are generally based on the direct sequencing of rRNA genes from environmental samples to assess community composition, this strategy is practically limited by the number of samples and sequences that need to be simultaneously processed to answer medium- to large-scale ecological questions. As an alternative to systematic sequencing, molecular fingerprinting techniques such as denaturing gradient gel electrophoresis (30), terminal restriction fragment length polymorphism (T-RFLP) (24), amplified ribosomal intergenic spacer analysis (ARISA) (8), or single-strand conformation polymorphism (SSCP) (43), which are culture independent, highly reproducible, and robust, have proven useful for time-efficient sample processing and comparative analysis of microbial community structure (1, 25). However, these techniques cannot answer questions about operational taxonomic unit (OTU) richness or about OTU relative abundances (1).
When using community fingerprinting methods, the sizes and the fluorescence intensities of labeled DNA fragments can be analyzed by using an automated sequencer (e.g., in T-RFLP, SSCP, and ARISA). DNA fragments of different sizes are thereafter commonly classified into OTUs (see, for example, references 1, 29, and 44). Often the relative peak areas, i.e., relative to the sample total peak area (54), are used as an indicator of OTU abundance, although it is known that PCR-based fingerprinting methods cannot yield a true representation of the original DNA ratios because of preferential and nonspecific amplifications (11, 46, 51) and of heterogeneity in rrn copy number (7). However, it is generally accepted that the intersample variation in relative peak areas should not be affected by the biased PCR conditions because they similarly apply to all OTUs and samples (2, 42). However, using peak areas as an indicator for absolute abundance has been confirmed to be a fruitless exercise, even when using simplified microbial communities (4, 10, 55). However, when experiments are performed under well-defined laboratory conditions or on simplified communities whose rrs copy numbers per genome are known, quantitative estimates of each target organism may be successfully obtained via community fingerprinting methods (27, 41). A corollary of the lack of quantitativeness in community fingerprinting methods is that they may not be appropriate to assess the richness of microbial communities or to be used with diversity metrics because of their underestimation of the actual richness and because of their limited detection threshold and dynamic range (reviewed in reference 1). These methods are merely strategies designed to screen samples for OTU presence, in contrast to rrs sequencing approaches that are meant to actually sample microbial diversity.
The goal of the present study was to describe a new strategy, called qfingerprinting (for “quantitative fingerprinting”), which provides quantitative estimates of each detected OTU over a scale of several orders of magnitude. In brief, the strategy may be understood as a combination of two techniques, i.e., dilution endpoint PCR (a variant of most-probable-number-PCR [MPN-PCR]) (38) and traditional community fingerprinting analysis. First, the development and subsequent validation of the strategy were performed on a model community composed of a mixture of known sequences. Second, the approach was applied to environmental microbial communities from deep-sea sediment samples for which quantitative estimates of microbial communities were already available (20). Changes in OTU abundance as a function of depth, as well as rare, dominant, and common OTUs, were then determined. Standard ARISA and quantitative ARISA (qARISA) were compared regarding both overall community patterns and the distribution of individual OTUs. Further ecological applications of the new strategy are also proposed.
MATERIALS AND METHODS
Standard ARISA protocol.
The ARISA-PCR mixture (50 μl) contained 1× PCR buffer (Promega, Madison, WI); 2.5 mM MgCl2 (Promega); 0.25 mM deoxynucleoside triphosphate mix (Promega); bovine serum albumin (3 μg μl−1); 20 ng of extracted DNA (quantified by a ND-1000 Nanodrop; Peqlab Biotechnology, Erlangen, Germany); 400 nM universal primer ITSF (5′-GTCGTAACAAGGTAGCCGTA-3′) and eubacterial ITSReub (5′-GCCAAGGCATCCACC-3′) (4), the latter being labeled with the phosphoramidite dye HEX; and 0.05 U of GoTaq polymerase (Promega). PCR primers were chosen based on their ability to yield reproducible and even peaks in the range 100 to 1,000 bp when used in ARISA (unpublished results) compared to other available ARISA primers (8, 40). PCR was carried out in an Eppendorf MasterCycler (Eppendorf, Hamburg, Germany) with an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 90 s, with a final extension at 72°C for 5 min. PCR products were purified utilizing Sephadex G-50 Superfine (Sigma-Aldrich, Germany), and samples were prepared prior to submitting to capillary electrophoresis on a 80-cm-capillary ABI Prism 3130xl genetic analyzer (Applied Biosystems) as follows. A standardized amount of DNA (100 ng of DNA, as determined spectrophotometrically) was added to a separation cocktail containing 0.5 μl of internal size standard Map Marker 1000 ROX (50-1000 bp) (BioVentures, Inc., Washington, DC), 0.5 μl of tracking dye (BioVentures), and 14 μl of deionized Hi-Di formamide (Applied Biosystems, Foster City, CA). The preparation was denatured 3 min at 95°C and kept on ice at least 5 min before being further processed by the sequencer. Separation of the PCR-amplified fragments via capillary electrophoresis was carried out with the following run parameters: 14.6 kV (run voltage), 2.4 kV (injection voltage), 20 s (injection time), and 60°C (oven temperature). Raw profiles were checked for stable baselines and voltage, and peak sizes and absolute areas were then determined by using GeneMapper software v 3.7 (Applied Biosystems) with minimum peak heights of 50 fluorescence units for all dyes. The best-fit size calling curves were built by using a second-order least-squares method (this compensates for any anomalously running fragments in the standard) and the local Southern method. A perfect fit for the calibration curves on the range 100 to 1,000 bp was always checked before further processing the samples.
Binning.
From GeneMapper output tables, conversions to sample-by-fragment tables and subsequently to sample-by-binned-OTU tables were performed by using custom R binning scripts. The algorithm rearranges the data and calculates the relative fluorescence intensity (RFI) of each peak by dividing individual peak areas by the total peak area for the respective sample. All peaks with RFI values of <0.09% were not included in further analyses since they consisted of background peaks. To include the maximum number of peaks while excluding background fluorescence, only fragments above a threshold of 50 fluorescence units and ranging between 100 and 1,000 bp were taken into consideration.
In order to take into account the technical variability in peak size calling (due to computer interpolation of peak sizes) (12), dye migration discrepancies (48), and run-to-run variations (24, 31), all peaks within a range of sizes (a window) can be combined (i.e., binned) into bin window frames, where the window is as wide as the imprecision of the OTU size calling. The window size (WS) must be estimated empirically for each fingerprinting pipeline by running appropriate controls at different times, for instance. Binning has a significant effect on the obtained sample similarities and must thus be accounted for to avoid falsely describing ecological differences that would be only due to technical variability (12). Besides, binning is important because fragment sizes produced by the fingerprinting strategy may later be compared to databases or to sequences from clone libraries (3, 18, 28). In our case, the ARISA imprecision was ±1 bp (i.e., WS = 2 bp) as determined by running known target sequences at different times on the same sequencer under the same analytical conditions (data not shown).
A shifting window size binning strategy was used since it offers the possibility to optimally aligned electrophoretic profiles and to deal with different window starting positions (12). The binning frame that offers the highest similarity among samples is identified out of all binning frames starting at a given position. The distance between two consecutive binning frames is defined as the shift (Sh) value. In the original description of the binning approach (12), only integer values were considered as “true” values to which peaks needed to be assigned to by taking the appropriate technical inaccuracy, resulting in Sh = 1 bp. This would mean that the two bin frames (WS = 2) would start, in our case, at 100 and at 101, respectively. Due to dye migration discrepancies, however, the actual, true (but unknown) size value may be different from integer values (i.e., a decimal value could be also representing a “true” size) and may actually change over the range of sizes being examined due to sequence-specific migration discrepancies (17). For this reason, Sh values must also be variable and a good, but the computing-demanding value may be 0.1 bp. For WS = 2 bp, there would therefore be 20 bin window frames to be calculated and evaluated. The current implementation of the window shifting algorithm enables a user-defined choice of the WS and Sh values, as well as the range size and RFI cutoff value, to calculate the best binning frame for a given data set. The interactive script then reports the best frame among all calculated as the one that maximizes sample similarities.
Another script allows for an automatic calculation of a series of WS values (e.g., 0.5, 1, 2, 3, 4, and 5 bp) for a given Sh value (e.g., 0.1 bp). This enables an optimal determination of the best binning strategy for a data set without a priori knowing the ideal WS value. A compromise between high resolution (low WS) and high similarity among samples (high WS) must be found based on the output of the scanning mode script. The interactive and automatic binning algorithms are implemented in the free, R programming language (The R Foundation for Statistical Computing [http://cran.r-project.org/]) and are available with their respective manuals and examples online (http://www.ecology-research.com). After proper, platform-specific installation of the R base program, the scripts run on Windows, Mac OS, Solaris, and Linux platforms.
Description of the qARISA strategy.
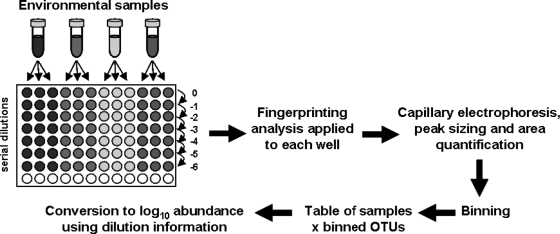
Each DNA sample was serially diluted with sterile PCR-grade water in triplicates, and this is done by transferring few microliters of DNA (typically 1 or 2 μl) from one well to the next by using a single pipette with a new tip each time to avoid carrying DNA over (Fig. 1). Dilutions were directly performed in PCR microtubes each containing, for instance, 9 or 18 μl (in accordance with the volume of DNA transferred) of sterile PCR-grade water if decimal dilutions are chosen. Using a multichannel pipette instead of a single pipette led to more systematic variation due, most likely, to variation in individual volumes that are more difficult to detect when 8 or 12 pipette tips are simultaneously used (unpublished data). Negative controls consisted of PCR-grade water instead of DNA template. The standard ARISA protocol was then applied to a common volume (e.g., 2 μl) from each dilution well, and all reactions were processed at the same time to avoid introducing additional experimental variability between dilution series and replicates. After capillary electrophoresis and peak sizing, binning was also performed on all replicates and dilution series together. Using a combination of the automatic and the interactive binning algorithms was then necessary to obtain the best-fitting binning approach for the data set at hand. The binned table that contained RFI values for the dilution series and for the sample replicates was then converted to a sample-by-OTU table, where OTU abundance values were calculated based on the dilution factor for each sample and on two rules (i.e., the consensus and continuity rules) to determine whether a binned OTU was reliably present. With the consensus rule, a peak is considered present if it appears at least twice among three replicates of the sample for a given dilution level. The continuity rule states that the highest dilution level recorded is the highest one for a continuous series of successful amplifications from undiluted to more diluted DNA preparations. The abundance was then expressed as log10 values for a decimal dilution series. The table conversion that takes into account the dilution levels and the continuity and consensus rules is performed automatically with the qfingerprinting algorithm (provided at http://www.ecology-research.com).
FIG. 1.
qfingerprinting strategy. See the text for details.
To illustrate the continuity rule, we can imagine two OTUs that produce the following series of amplification results, 1101010 and 1100000, with from left to right, PCR success and failure scored as 1 and 0, respectively, for a dilution series from undiluted to 10−6 levels. We may assume for simplicity that a success already satisfies the consensus rule. In those two cases, the calculated log10 values would be 2, i.e., only the highest dilution (i.e., 10−1) that satisfies the consensus rule and that is continuously positive from the undiluted level was kept for estimating abundances. Noticeably, the relative peak areas (RFI) are therefore not taken into consideration but just converted to binary data for the calculations. In our example, up to 7 orders of magnitude in target concentrations may thus be conjointly determined (Fig. 1).
Model community experiments.
The qARISA strategy was applied to a model community consisting of known DNA sequences. Those sequences originate from a sequencing project from deep sea sediments to be reported elsewhere (sequences available upon request). Four plasmids containing cloned partial 16S and ITS sequences were chosen for yielding defined peak sizes of 601, 654, 504, and 681 bp when analyzed by ARISA. They were prepared to obtain the respective decreasing concentrations of 0.2, 0.02, 0.02, and 0.002 ng/μl when mixed together in a total volume of 12 μl. Decimal dilutions were done in triplicates by transferring 1 μl of DNA solution into 9 μl of sterile water until the decimal dilution 10−6 was reached. From each DNA preparation (undiluted and diluted), 2 μl was then used per 50-μl ARISA reaction. After this procedure, the initial amount of DNA for peak 601 bp was 0.4 ng at the undiluted level. The subsequent qARISA steps were as described above. The model community experiment was repeated at three independent times and yielded highly consistent results. Here, the results of only one experiment are presented.
Environmental sample analysis.
Microbiological and geochemical data obtained from the sediment samples of reference core 51-1 used in the present study have been described previously (20). The core was sampled on 1 August 2000 by deploying a video-guided multicorer at the crest of southern Hydrate Ridge (44°34.198′N, 125°08.858′W, 775-m depth) of the Cascadia convergent margin off the coast of Oregon. The core was not enriched in methane in the surface sediments, but parallel cores contained specimens of the chemosynthetic bivalve Acharax, indicating that it was sampled from a site influenced by low seepage below the 15-cm sediment depth (20). DNA was extracted from 1 g of homogenized sediment sample by using an UltraClean soil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. The qARISA was performed as indicated above in triplicate decimal dilution series with a starting DNA concentration of 20 ng per reaction (undiluted level). A total of eight negative controls (PCR water was used instead of DNA) were included in the 2.5 96-well plates processed by the automated sequencer in order to guarantee that no contamination occurred. All samples were analyzed together from DNA extraction to the final capillary electrophoresis separation, so as to limit additional sources of experimental variation.
Statistical analyses.
The nonparametric Kolmogorov-Smirnov two-sample test was used to compare the abundance distributions derived from ARISA and qARISA. The significance of the two-sided test was determined by performing 1,000 permutations of the non-null values of the data set. The Mantel's test was used to determine the level of correlation between the Bray-Curtis similarity matrices among samples inferred from the binned table derived from ARISA and from qARISA. The significance of the correlation value was assessed by 1,000 permutations. Overall, patterns in sample dissimilarity were explored via nonmetric multidimensional scaling (NMDS). NMDS places the samples in a two-dimensional coordinate system so that ranked dissimilarities between the samples are preserved. A stress function assesses the goodness-of-fit of the ordination compared to the original sample ranking (23). Procrustes analysis was used to determine how similar the final sample ordinations deriving from ARISA versus qARISA could be (23). The method estimates the concordance of scores between two ordinations after rotating, translating, and dilating them in order to obtain the best fit. A permutation procedure is then used to test for the significance of the concordance (34).
Detrended correspondence analysis (14) was applied to determine whether linear or unimodal OTU models better fitted the current data set (37). Principal component analysis (PCA) was done on ARISA tables that were first Hellinger transformed (2, 22, 37). To determine whether depth as an environmental parameter could significantly explain the variation in bacterial community structure, a canonical redundancy analysis (RDA) was used, and its significance was assessed by 999 Monte Carlo permutation tests. After a significant RDA, nonparametric Spearman tests were used to detect significant positive or negative correlations of each OTU abundance distribution with depth. All statistical tests and graphics (e.g., heatmap) were produced by using R packages (Stats, Vegan, MASS) and CANOCO for Windows 4.5 (47).
RESULTS AND DISCUSSION
Model community experiment.
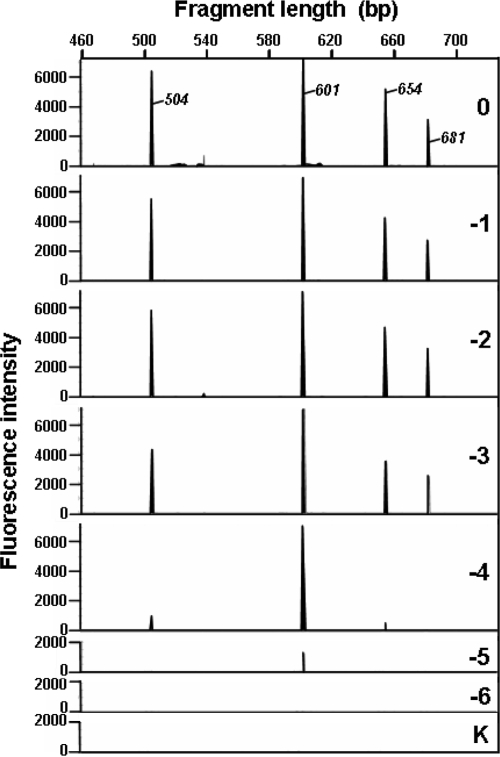
The idea of the qfingerprinting approach is to apply a standard fingerprinting strategy to dilution series of the target samples, in order to identify the highest dilution at which an OTU is still PCR amplifiable (Fig. 1). The qfingerprinting strategy was applied to a mixture of four plasmids harboring cloned ITS inserts whose individual lengths and concentrations (Fig. 2, 3) were known beforehand in order to test the validity of the concept. Capillary electrophoresis separation of the defined samples produced 24 electropherograms (21 dilution samples and 3 negative controls) for the same plasmid mixture (Fig. 2). A clear difference in peak height and a gradual peak loss at dilutions 10−4 to 10−6 were observed as a function of the initial plasmid concentrations.
FIG. 2.
Electropherograms obtained for the qARISA approach performed on a model community. The decimal dilution factor is indicated in each panel by a figure (0 for undiluted, −1 for 0.1, −2 for 0.02, etc.) and the letter K (no DNA control). Only one replicate electropherogram per dilution level is shown (see Fig. 3 for a complete overview). The initial DNA concentrations of individual fragments 504, 601, 654, and 681 bp were 0.02, 0.2, 0.02, and 0.002 ng/μl, respectively.
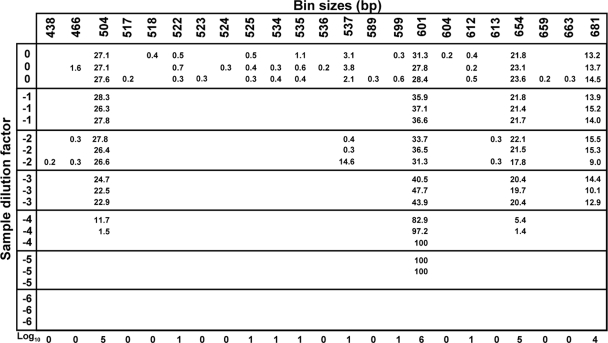
FIG. 3.
Calculations of abundance values for each OTU present in the model community. After binning, the RFI (in percentages of total sample intensity) for each OTU is indicated when present at the corresponding dilution and replicate levels. Using the qARISA strategy, the abundance of each OTU was then calculated by taking the consensus and continuity rules into consideration, as well as the information from all replicates and dilution levels (see text for details). The resulting abundance values for the model community are then expressed in log10 (bottom of the figure).
Following the straightforward binning of the data (WS = 2, Sh = 0.1), the binned table (series of diluted replicated samples by OTUs) was then converted to log10 values by following the consensus and the continuity rules (see Materials and Methods). The consensus rule can be illustrated, for instance, by OTUs 524, 525, and 534 (Fig. 3). Indeed, the corresponding peaks were considered present only for the latter two OTUs, because a peak was identified in two of three replicates at a given dilution level. An example of the continuity rule may be found with OTU 537 for which peaks satisfying the consensus rule were detected at dilution 100 and at 10−2, but not at 10−1. In that case the peak presence was established only for the undiluted sample at 100 (Fig. 3). Using those two rules, OTU presence until the highest dilutions was recorded and transformed into log10 abundance values.
Interestingly, the RFI values did not proportionally change with the dilution factor (Fig. 3). At the undiluted level (i.e., standard ARISA conditions), the RFI values ranging from 13 to 31% did not even reflect the fact that the concentration of each target sequence was initially consisting of 10-fold differences. Indeed, RFI values are obtained by standardizing the individual peak areas per sample (see, for example, references 44 and 54). It is thus not surprising that these values do not follow the dilution trend imposed in the study because their magnitude is directly related to the behavior of the other peaks present in the given sample (see also reference 4). Consequently, the final calculations of abundance values from the dilution series should take the occurrence of the OTUs into account, instead of their RFI values.
On the electropherograms (Fig. 2) and in the resulting sample-by-OTU table (Fig. 3), several OTUs were associated with small peaks, some of which satisfied both the consensus and the continuity rules (e.g., OTUs 522, 525, and 534). Peaks that did not correspond to one of the target sequences in the mixture were always found at low abundances. This may be due to the fact that because few targets were present in the artificial mixture, the background level of false-positive peaks was higher. Thus, unspecific, but weak amplifications were more likely to be detected. When more sequences are present in the DNA mixture, as with environment samples, the small, inconsistent peaks may be diluted out by the presence of more numerous and highly dominant peaks. In a standard ARISA, however, these small peaks would have been considered as real OTUs, whereas here the difference in magnitude reveals that they may most likely be considered PCR artifacts.
By taking the initial insert concentrations and the respective dilution factors into consideration, the theoretical detection limit of the technique can be estimated. By taking the most abundant OTU (601 bp) at the initial concentration of 0.4 ng per reaction at the undiluted level, and assuming 660 g/mol per bp (thus, 6.59 × 10−19 g per fragment), a total of ∼6 × 108 fragment copies of the target sequence were then initially present in the reaction. The highest dilutions where amplifications still occurred were between 10−5 and 10−6 (Fig. 2 and 3), indicating that a minimum of 600 to 6,000 target sequences were needed for a successful amplification to occur under our experimental conditions. This is in accordance with the detection thresholds of 103 cells per ml of sample generally reached when molecular community fingerprinting methods are used (6).
Overall, the qfingerprinting concept and analysis pipeline worked well on the mixture of known ITS sequences. The difference in the magnitude of the initial target ratios was maintained because the derived abundances obtained via qfingerprinting are not based on the final, biased quantitative estimate of PCR success, but rather on the qualitative outcome of the PCR amplification at different dilution levels. The test on plasmids also illustrates the need to apply advanced numerical methods to identify the best binning frames so as to identify the most likely dilution levels for OTU occurrence. Indeed, misclassification of OTUs between replicates at a given dilution level could lead to a false rejection or acceptance of the consensus or continuity rules by altering the continuity of the series of positive amplifications.
Application to environmental samples.
Samples originating from deep marine sediments forming a natural depth gradient were chosen to illustrate the advantages of the new method. A total of nine successive 1-cm sediment layers were subjected to qARISA, thus producing a total of 189 reactions (nine layers × seven dilution levels × three replicates). A total of 15,361 peaks were retained after capillary electrophoresis, which ranged from 100 to 1,000 bp and whose absolute peak areas were over 50 fluorescence units. No peaks satisfying the aforementioned criteria were observed for the eight negative controls. Due to the large number of peaks, using an automatic binning technique was very important because OTU identification and quantification had to satisfy both the consensus and the continuity rules, which could not be performed manually.
After the scanning mode of the binning algorithm was applied, the correlation coefficients between samples were all 0.46 for window sizes 2, 3, 4, and 5 bp and corresponded to total OTU numbers of 438, 298, 225, and 181, respectively. For window sizes of 0.5 and 1 bp, the correlation coefficients were smaller (r = 0.32 and 0.38, respectively), but the OTU numbers were much higher, with 789 and 815 OTUs identified, respectively. These findings are not surprising since increasing the window size leads to lumping more peaks together, hence to a smaller number of OTUs finally identified. As a compromise between high correlation among samples and high OTU number, the binning results of window size 2 were chosen for subsequent analyses. A total of 437 bins (OTUs) were thus obtained out of the 15,361 initial peaks detected when all replicated samples were considered. After the qfingerprinting function was applied to convert the binned table into a log10 abundance table (i.e., by taking the dilution series into account, as well as the consensus and continuity rules), a total of 332 OTUs were finally obtained for the nine environmental samples. For each OTU, an abundance value that ranged from 0 (absent) to 7 (present at abundance levels 7 orders of magnitude higher than at the undiluted level) was obtained, and this produced on average 173 ± 20 (standard deviation) OTUs per sample.
The variability in the quantitative estimates obtained via qfingerprinting was already taken into consideration at the levels of peak size calling and interexperimental variability by using the appropriate binning method and consensus rules. However, the current implementation does not enable the calculation of confidence intervals for those estimates. Alternatively, the MPN (5) approach could be used to analyze the data in a probabilistic framework. MPN-PCR and direct dilution calculations provide slightly different estimations of abundances, but they are mostly linearly related to each other (see, for example, references 35 and 38). The direct dilution calculation was chosen here for its ease of implementation when simultaneously dealing with hundreds of OTUs. It should be noted that the accuracy of the quantification depends on the number of replicates and dilution rates, with more accuracy obtained when more replicates are used per dilution level. The average accuracy has been shown to be almost identical for dilution rates between 2 and 10, but coefficients of variation are more stable and slightly lower for twofold dilution assays (5). The dilution rate may obviously be adapted by taking previous knowledge, specific needs, or available resources into account.
Based on the detection limit calculated in the model community experiment (see above), a specific OTU may be detected if its number of copies is at least in the range 600 to 6,000 copies at the undiluted level, under ideal amplification conditions (i.e., if we assume that DNA from the synthetic community has the same properties as that from environmental communities). This would be indicated by an abundance value of 1 log10 in the result table. Consequently, when values of 7 log10 abundance are obtained (see Fig. 5), this corresponds to at least 6 × 108 target copies detected for the specific OTUs. Noticeably, this number is very close to the reported estimates of highest cell number found in those samples, which ranged from 1.0 × 109 to 6.0 × 109 cells/cm3 (20). Despite this interesting correspondence in abundance estimates, it should be noted that the abundance values obtained via qfingerprinting are difficult to convert to cell densities or biomass because such conversion would require knowledge of the copy number per genome and the genome size, and both numbers can be very variable (9, 19).
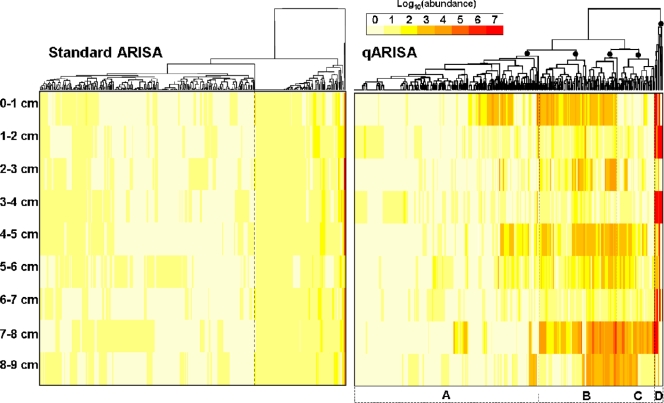
FIG. 5.
Two-way clustering of OTU abundances obtained by standard ARISA and by qARISA. A complete linkage algorithm was used to perform a cluster analysis of the Euclidean-based dissimilarity matrices inferred from RFI values (standard ARISA) and log10 values (qARISA) for the same set of 332 OTUs. Major clusters are indicated by filled circles in the qARISA dendrogram and designated by the letters A to D (see the text for a description of the clusters). This arbitrary cutoff roughly corresponds to 65% similarity.
Comparison of qARISA and standard ARISA.
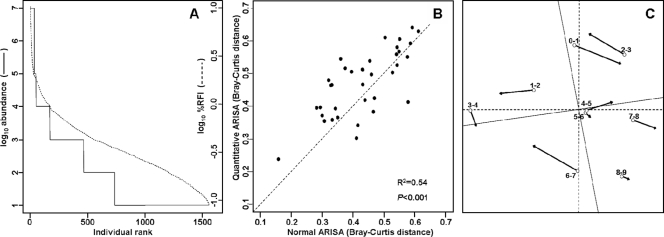
The distributions of RFI values and log10 values obtained from standard ARISA and qARISA, respectively, could not be considered as drawn from the same data distribution (Fig. 4A), as also confirmed by a significant Kolmogorov-Smirnov test (D = 0.8878, P < 0.001; based on 1,560 nonzero values in each case). The latter test takes into account differences in both location and shape of the two empirical cumulative distribution functions. The same conclusions were also obtained if the respective abundance data were first split into frequencies of occurrence per sample and then compared between the standard ARISA and the qARISA approaches (D = 0.77 to 0.98; all P < 0.001).
FIG. 4.
Comparison of the distribution of abundance values and of their effects on sample dissimilarities when ARISA versus qARISA is used on a set of environmental samples. (A) Comparison of all non-null values (1,560 values) from the two distributions (standard versus qARISA) when the two fingerprinting strategies were applied to a gradient of nine environmental samples. (B) Comparison of Bray-Curtis dissimilarities obtained for pairs of samples based either on qARISA (log10 abundance) values or on RFI values obtained from a standard ARISA approach (n = 36 pairwise comparisons). The dotted line corresponds to the case where both approaches provide the same dissimilarities among samples (y = x). (C) Comparison of the ordination results obtained with the two strategies by Procrustean superimposition. Open circles correspond to samples in the ordination derived from the standard ARISA, and the tips of the arrows correspond to the positions of the samples in the ordination when the qARISA procedure is used. The dotted line indicates the axis rotation needed to best superimpose the two ordinations.
At the level of sample similarities, the standard ARISA versus qARISA approaches, however, produced significantly linearly related distance matrices (Mantel r = 0.73, P < 0.001 as determined by 1,000 permutations; Fig. 4B), although a low coefficient of determination (R2 = 0.54) indicated the existence of strong disagreement between the two approaches. As noted from the two abundance distributions, using RFI values from the standard ARISA instead of qARISA-derived values would generally lead to lower dissimilarities among samples, i.e., to concluding that samples are more similar than they really are. The slope coefficient of the linear regression, beta, was estimated to be 0.81 ± 0.129 (standard error), leading to a 95% confidence interval ranging from 0.55 to 1.08 (assuming 34 degrees of freedom for the Student t distribution). This indicates that beta was not significantly different from 1, i.e., that the two methods give related estimates of dissimilarities among samples. Finally, NMDS ordinations based on the two approaches, although displaying substantial deviations from each other (Fig. 4C), were nevertheless significantly correlated (symmetric Procrustes rotation correlation of 0.88; P < 0.001).
In conclusion, using qARISA versus ARISA led to various levels of discrepancies for data distribution, estimation of sample dissimilarities, and ordination results. The significant correlation between sample ordinations implies that the standard ARISA would still be robust for inferring changes in community structure, despite its inability to accurately estimate OTU abundance. This may originate from the DNA normalization steps common to both procedures, i.e., before the initial PCR, but also before separating DNA fragments by capillary electrophoresis. In addition, it has been shown that normalized ARISA peak area may relate well to quantitative relative abundance, as determined by flow cytometry (3), and this also explains why the standard ARISA has been successfully used thus far to describe community dynamics over spatial and temporal gradients in a variety of ecosystems (13, 44, 54). Despite the apparent limited differences between the two approaches, it is not yet known whether the two techniques would (i) still yield comparable sample ordinations when more samples are included in the analyses and (ii) offer the same ecological conclusions when contextual interpretations (i.e., when additional environmental, spatial, or temporal variables are available to explain diversity patterns) of the two sample ordinations are undertaken. Those important points will need particular attention in future qfingerprinting applications.
Validity of the continuity rule.
One of the main assumptions used in the qfingerprinting approach is that, for each OTU, PCR success is monotonically related to the concentration of the OTU target sequence in the sample. It is, however, known that PCR yield may be optimal (i) at high dilution levels of the DNA solution because impurities or PCR inhibitors may be diluted out or (ii) in the middle of a target DNA concentration range because a trade-off between DNA concentration and a low level of contaminants must be found to obtain a successful PCR amplification, thus producing a unimodal distribution of amplification success for a given OTU. The first consideration should not be a major issue if we assume that, in the case of impurities, PCR inhibition would apply to all OTUs at the same time because it is the efficiency of the DNA polymerase that is mostly affected and not a specific OTU amplification. Note that this point could be further debated because it is highly likely that impurities selectively trap DNA sequences based on sequence composition (see, for example, references 45 and 49), but this goes beyond the scope of the present study.
The second hypothesis deserves more attention because it will directly entail a modification of how the algorithm currently assesses OTU presence and calculates the final OTU abundance table. By examining the raw data for each OTU before applying the qfingerprinting calculations, on average 86% ± 10% of the OTUs were present at dilution 100, and for the existing OTUs whose amplification patterns did not start at the undiluted level (i.e., presenting unimodal rather than linear patterns) they consisted more than 61% of the time of OTUs appearing only once at one dilution level, suggesting that they may be considered as unreliable peaks. Thus, it seems that the continuity rule used in OTU quantification represents a valid hypothesis to describe the behavior of OTU amplification across the dilution series. It must be noted that this conclusion, although being valid for this data set, could be inappropriate if more impurities are present (e.g., if different samples or extraction protocols are used) or if particular PCR conditions are more sensitive to suboptimal conditions.
Relationship between OTU distribution patterns and depth.
The environmental samples were coming from nine 1-cm-thick layers from the same core, and it was particularly interesting to look at the OTU distribution patterns and abundance changes across this natural depth gradient. The overall community behavior with respect to depth when determined by detrended correspondence analysis revealed that a short gradient length of 1.74 was associated with the qARISA data set, suggesting that most OTUs were linearly responding to environmental gradients (37). This was confirmed by RDA, indicating that depth significantly explained the variation in the data set (P < 0.001, based on 1,000 permutations of the multivariate model).
A detailed analysis of individual OTU abundance was performed by using two-way cluster analysis and heatmap plot representations (Fig. 5), which allow a display of OTU abundance relationships as a function of depth (the sample order with depth was kept fixed).
To allow for comparisons of abundance classes between RFI and log10 values, RFI values that ranged from 0 to 13% were first converted into a discrete 0 to 7 scale. Therefore, although the ranges of color codes (0 to 7) were the same for standard and quantitative ARISAs, their meaning was different: for standard ARISA they corresponded to fluctuations of ∼1 log10 value difference [log10(13) = 1.11], whereas for qARISA the variation corresponds to 7-order-of-magnitude changes (decimal logarithmic scale). Clustering RFI values from standard ARISA produced overall a very different picture of inter-OTU relationships and of abundance changes with depth compared to the outcome of qARISA log10 abundance values (Fig. 5). Noticeably, OTUs in standard ARISA seemed to fall into two main clusters, whereas four, more variable clusters (designated by the letters A to D) could be identified with qARISA data for a similar cutoff level (Fig. 5). It is important to note that clustering RFI values from standard ARISA could lead to very unpredictable patterns because, as mentioned above, these values cannot truly represent OTU abundances from a sample since they are derived from the overall peak abundances from a given electrophoretic profile. For instance, the two most abundant OTUs in standard ARISA were located in cluster A or B when qARISA was used, i.e., clusters associated with low abundant OTUs.
The four main clusters obtained with qARISA data reflected different patterns of OTU occurrence among samples and different OTU abundance categories, i.e., high, average, or low abundance. Nonparametric Spearman correlation tests were subsequently used to determine significant OTU relationships with depth. Among the 199 OTUs (60% of total OTUs) of cluster A, the abundances of 17 and 2 OTUs significantly decreased and increased with depth, respectively, and contributed to 9.5% of all OTUs in the cluster. Since the highest OTU abundance was 4 log10, cluster A may be qualified as composed of “rare OTUs with low abundance.” Cluster B consisted of 84 OTUs (25% of the whole data set). Among them, two and one OTU, respectively, significantly decreased and increased with depth, representing 3.5% of the OTUs in the cluster. As such, the OTUs in cluster B may be designated as “common, but generally not affected by depth.” Cluster C consisted of 40 OTUs (12% of all OTUs), with 24 OTUs significantly increasing with depth (i.e., 60% of the OTUs of the cluster), leading to the most “depth-associated” cluster. Finally, cluster D with only nine OTUs (3% of all OTUs) consisted of OTUs that were present in several samples and at high abundance (ranging from 5 to 7 log10) but whose abundance overall displayed no trend with depth. This cluster may be designated as that of “common and dominant” OTUs.
Only 27 OTUs (8% of the total OTUs) were present in all nine samples with an average abundance over 2 log10 (mean abundance for all nonzero values), and 20 OTUs (6%) were present in all samples, but with abundances lower than 2. A total of 57 (17%) rare OTUs (i.e., occurring in one sample only) were all found with an average abundance below 2 log10. The large majority (69% or 228 OTUs) of all OTUs were thus not classified as either dominant or rare. It may thus be concluded that qARISA did not only amplify the most common OTUs present in the samples, since the most abundant only consisted of 8% of all OTUs detected. The large majority (85%) was found to be composed of rare or average OTUs with generally low abundance and with weak relationships with depth. It must be noted that the finding of well-represented OTU categories from our environmental samples has a lot to do with the choice of the primer pairs. The ARISA primers in the present study were chosen for their ability to evenly amplify different OTUs in the same sample compared to other primer sets (4; unpublished results). Therefore, the finding of various OTU categories should not directly be attributed to the qfingerprinting approach but to the choice of PCR primers and amplification conditions.
The finding of a substantial proportion of rare or average OTUs is contrary to the common belief that fingerprinting methods only detect the most abundant microbes in a sample (reviewed in reference 1) and suggests that qfingerprinting may also be useful for studying the dynamics of the “rare biosphere” (16, 33). It is, however, difficult to attribute the very rare types either to PCR artifacts, as could be suggested by the low-abundance peaks detected in our model community experiments, or to a true representation of the community composition. It has previously been suggested that rare OTUs (often associated with small peak sizes) may be artifacts of fingerprinting methods (2), thus resulting in fallacious description of microbial community structure (see, for example, reference 55). Using the consensus rule as a conservative way to validate the presence of a given OTU may be one step toward rejecting trivial PCR artifacts. Caution is therefore required at this stage to avoid overinterpreting the presence of rare types in the data, especially when additional molecular proofs are not available, at least for some of the rare OTUs. Alternatively, cutoff abundance values may also be used to remove the low-abundant OTUs if one considers their presence as artifactual.
Inter-OTU relationships.
In most microbial diversity analyses, biotic interactions between components of the microbial community are overlooked, despite the fact that they may be extremely important in determining community dynamics and changes in ecosystem processes, as evidenced from macroecological studies (53). Here, the qfingerprinting approach enabled interrelationships between OTUs to be further examined in contrast to the traditional fingerprinting approaches that only offer a qualitative description of OTU presence or absence. To facilitate the depiction of the relationships, PCA was chosen since it helps identify OTUs whose abundance profiles covary positively, negatively, or neutrally with respect to each other (Fig. 6), insights that cannot be easily deduced from Fig. 5, for instance. In the ordination plot, OTU vectors that are orthogonal to each other may be considered as behaving independently, whereas the ones that are colinear may be seen as positively or negatively covarying, depending on the angles between the vectors being compared (see, for example, references 23 and 37 for further interpretation of PCA ordination plots). Noticeably, individual OTU patterns were clearly spanning all directions in the PCA ordination plot (Fig. 6) and therefore revealed the existence of much more individual variability than what could have been assumed within a single sediment core.
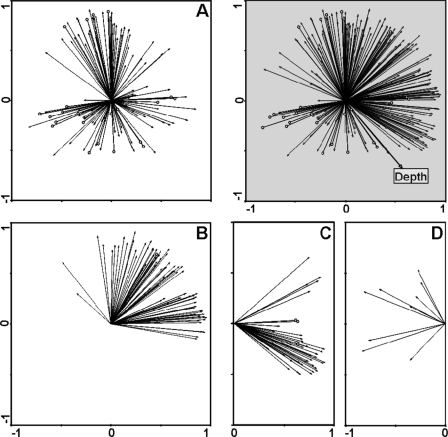
FIG. 6.
PCA of OTU distribution patterns. PCA with scaling focusing on inter-OTU distances for the 332 OTU distribution patterns was chosen to illustrate how inter-OTU relationships could be deduced from qARISA-derived abundance values. The percentages of explained variations for the first two axes were 36 and 20%, respectively (gray panel). Depth is represented as a supplementary variable in the PCA plot. Panels A, B, C, and D represent separate projections of the specific OTUs defined in the four clusters of Fig. 5, respectively. Arrowheads depicted as empty circles indicate OTU distribution patterns that are shared by multiple OTUs (i.e., 22 patterns in panel A, 1 pattern in panel B, and 3 patterns in panel C).
Interestingly, it is also possible to determine the number of OTUs sharing the exact same abundance profile, i.e., being present in the same samples and displaying the same abundance values. The number of OTU patterns occurring only once was 237 (i.e., 90.5% of the 262 abundance patterns), the numbers of those occurring two and three times were 16 (6.1%) and 4 (1.5%), respectively, and those occurring 4, 11, 12, and 20 times were each found at less than 1% of the total patterns (i.e., 1 or 2 patterns) (Fig. 6), thus suggesting that the large majority of OTUs had distinct abundance patterns. Patterns shared by more than two OTUs were mostly associated with low-abundant OTUs found in one or two samples, thus suggesting that the main structure in the data set was created by specific variation in the individual OTU abundances rather by common OTU behaviors. As previously seen (Fig. 5), most OTU distributions did not follow the depth gradient, and this is confirmed here by the finding of only a few OTU vectors being correlated to the depth vector superimposed in the ordination plot (Fig. 6). It therefore seems that some other unmeasured factor(s) may be responsible for the observed patterns, if we opt for a deterministic explanation, and/or that stochastic (random) processes may be at play in the current system.
Advantages and limitations of the quantitative approach.
The present study demonstrates the usefulness of using the qfingerprinting strategy to simultaneously determine the relative abundance of hundreds of OTUs, and this could readily be applied to tens to hundreds of samples. This quantitative approach represents a major improvement for the understanding of microbial diversity patterns at the level of individual OTUs in contrast to standard community fingerprinting methods that merely qualitatively screen for OTU presence or absence in samples. At the level of whole community ordination, i.e., not comparing individual OTUs but sample similarity based on OTU abundances, patterns retrieved by the two approaches were slightly different but still consistent with each other. Hence, if detailed information about each OTU abundance is required in a given study, the new, quantitative strategy should be preferred over the standard strategy. If the study merely focuses on overall patterns of sample similarities and accurate determination of OTU abundance is not needed, then the standard fingerprinting method with the necessary DNA standardization and normalization steps described in the present study should be favored. However, caution must be taken when interpreting diversity patterns based on qualitative measures of β diversity since this has been shown to lead to different ecological insights concerning the factors that structure microbial communities (26).
An additional advantage of the new strategy is the fact that there is no need to design individual PCR primers for each OTU, as would be required for a difficult, multi-OTU quantitative PCR approach. Because it is straightforward, the technique will easily complement the existing applications of ARISA, T-RFLP, and SSCP, and any other fingerprinting methods based on ribosomal or functional genes if the latter are based on high-throughput sorting of the fragments (e.g., via capillary electrophoresis) and if internal size standards are included in every sample. These last conditions are important in order to make sure that a high reliability is associated with peak size calling and that further correction of size calling inaccuracy is possible without losing sensitivity in signal detection.
Despite the considerable advantages of the new strategy, the same weaknesses and limitations as found with fingerprinting methods and more generally with any PCR-based methods may be predicted. For instance, the method also relies on correct DNA extraction methods to obtain a good representation of the original sample diversity and is affected by primer biases, the generation of chimeric sequences, etc., and all of those issues have been extensively reviewed (50). The specificity of the strategy is directly related to the choice of the PCR primers, and this fact may be used to narrow or to expand the range of targeted OTUs (this may be thus seen as a strength or a weakness). Diversity under- or overestimation common to many fragment-based fingerprinting techniques may also be problematic (for a review, see references 1 and 8). Finally, because many PCRs are done in parallel for the dilution series, higher risk of contaminations and cost increase have to be considered. However, because dilution rates can be adapted to the specific needs of the study and as access to sequencing facilities and automatic pipetting devices becomes easier and cheaper, large-scale, high-throughput applications of the technique should become more feasible and affordable in the future.
Foreseen applications.
Future applications of the qfingerprinting strategy should provide datasets that will help establish better mathematical models of intracommunity dynamics and more generally datasets on which microbial ecological theories and community assembly rules can be further developed (15, 21, 39). Detailed analyses of OTU distribution and abundance may also be foreseen when additional contextual parameters are available. For instance, OTU indicator analysis could be performed to determine whether various microbial OTUs are influenced by the same environmental factors or are indicative of specific community status, since microbes may play a major role in detecting and characterizing changes in environmental conditions (32). In addition, density-dependent processes such as quorum-sensing mediated microbial responses (52) may be monitored using qfingerprinting, and this will help us to better understand the relationships between community structure and function in complex ecosystems. Other examples of application should be found in the description of the allelic diversity associated with functional genes so as to quantify specific groups of microorganisms involved in biogeochemical processes, bioremediation, or plant protection and to identify their interactions and behavior under specific environmental conditions.
Acknowledgments
I thank Susanne Menger for excellent technical assistance, Jed A. Fuhrman for helpful discussions about ARISA and binning, and Antje Boetius for critically reviewing the manuscript.
Financial support was provided by the Max Planck Society.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Bent, S. J., and L. J. Forney. 2008. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2:689-695. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood, C. B., T. Marsh, S. H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, M. V., M. S. Schwalbach, I. Hewson, and J. A. Fuhrman. 2005. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 7:1466-1479. [DOI] [PubMed] [Google Scholar]
- 4.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochran, W. G. 1950. Estimation of bacterial densities by means of the most probable number. Biometrics 6:105-116. [PubMed] [Google Scholar]
- 6.Cocolin, L., L. F. Bisson, and D. A. Mills. 2000. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81-87. [DOI] [PubMed] [Google Scholar]
- 7.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 10.Frey, J. C., E. R. Angert, and A. N. Pell. 2006. Assessment of biases associated with profiling simple, model communities using terminal-restriction fragment length polymorphism-based analyses. J. Microbiol. Methods 67:9-19. [DOI] [PubMed] [Google Scholar]
- 11.Hansen, M. C., T. Tolker-Nielsen, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol. Ecol. 26:141-149. [Google Scholar]
- 12.Hewson, I., and J. A. Fuhrman. 2006. Improved strategy for comparing microbial assemblage fingerprints. Microb. Ecol. 51:147-153. [DOI] [PubMed] [Google Scholar]
- 13.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, M. O., and H. G. Gauch. 1980. Detrended correspondence analysis: an improved ordination technique. Vegetatio 42:47-58. [Google Scholar]
- 15.Hubbell, S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton, NJ.
- 16.Huber, J. A., D. Mark Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, C. W., and C. L. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121-125. [DOI] [PubMed] [Google Scholar]
- 18.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knittel, K., A. Boetius, A. Lemke, H. Eilers, K. Lochte, O. Pfannkuche, P. Linke, and R. Amann. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia margin, Oregon). Geomicrobiol. J. 20:269-294. [Google Scholar]
- 21.Koeppel, A., E. B. Perry, J. Sikorski, D. Krizanc, A. Warner, D. M. Ward, A. P. Rooney, E. Brambilla, N. Connor, R. M. Ratcliff, E. Nevo, and F. M. Cohan. 2008. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. USA 105:2504-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legendre, P., and E. D. Gallagher. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271-280. [DOI] [PubMed] [Google Scholar]
- 23.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed. Elsevier Science BV, Amsterdam, The Netherlands.
- 24.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loisel, P., J. Harmand, O. Zemb, E. Latrille, C. Lobry, J. P. Delgenes, and J. J. Godon. 2006. Denaturing gradient electrophoresis (DGE) and single-strand conformation polymorphism (SSCP) molecular fingerprintings revisited by simulation and used as a tool to measure microbial diversity. Environ. Microbiol. 8:720-731. [DOI] [PubMed] [Google Scholar]
- 26.Lozupone, C. A., M. Hamady, S. T. Kelley, and R. Knight. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 30.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial population by DGGE analysis of polymerase chain reaction amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 32.Paerl, H. W., J. Dyble, P. H. Moisander, R. T. Noble, M. F. Piehler, J. L. Pinckney, T. F. Steppe, L. Twomey, and L. M. Valdes. 2003. Microbial indicators of aquatic ecosystem change: current applications to eutrophication studies. FEMS Microbiol. Ecol. 46:233-246. [DOI] [PubMed] [Google Scholar]
- 33.Pedrós-Alió, C. 2007. Dipping into the rare biosphere. Science 315:192-193. [DOI] [PubMed] [Google Scholar]
- 34.Peres-Neto, P. R., and D. A. Jackson. 2001. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129:169-178. [DOI] [PubMed] [Google Scholar]
- 35.Picard, C., C. Ponsonnet, E. Paget, X. Nesme, and P. Simonet. 1992. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl. Environ. Microbiol. 58:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser, J. I., and I. M. Head. 2007. Microorganisms, macroorganisms, and ecology. FEMS Microbiol. Ecol. 62:133-134. [DOI] [PubMed] [Google Scholar]
- 37.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramette, A., and J. M. Tiedje. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197-207. [DOI] [PubMed] [Google Scholar]
- 40.Ranjard, L., F. Poly, J. C. Lata, C. Mougel, J. Thioulouse, and S. Nazaret. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, J. K., B. Konig, and U. Reichl. 2007. Characterization of a three bacteria mixed culture in a chemostat: evaluation and application of a quantitative terminal-restriction fragment length polymorphism (T-RFLP) analysis for absolute and species specific cell enumeration. Biotechnol. Bioeng. 96:738-756. [DOI] [PubMed] [Google Scholar]
- 42.Schütte, U. M. E., Z. Abdo, S. J. Bent, C. Shyu, C. J. Williams, J. D. Pierson, and L. J. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 43.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shade, A., A. D. Kent, R. J. Newton, E. W. Triplett, and K. D. McMahon. 2007. Interannual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol. Oceanogr. 52:487-494. [Google Scholar]
- 45.Stach, J. E. M., S. Bathe, J. P. Clapp, and R. G. Burns. 2001. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 36:139-151. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ter Braak, C. J. F., and P. Smilauer. 2002. CANOCO reference manual and CanoDraw for Windows User's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY.
- 48.Tu, O., T. Knott, M. Marsh, K. Bechtol, D. Harris, D. Barker, and J. Bashkin. 1998. The influence of fluorescent dye structure on the electrophoretic mobility of end-labeled DNA. Nucleic Acids Res. 26:2797-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vettori, C., D. Paffetti, G. Pietramellara, G. Stotzky, and E. Gallori. 1996. Amplification of bacterial DNA bound on clay minerals by the random amplified polymorphic DNA (RAPD) technique. FEMS Microbiol. Ecol. 20:251-260. [Google Scholar]
- 50.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 51.Wang, G. C. Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 53.Weis, J. J., B. J. Cardinale, K. J. Forshay, and A. R. Ives. 2007. Effects of species diversity on community biomass production change over the course of succession. Ecology 88:929-939. [DOI] [PubMed] [Google Scholar]
- 54.Yannarell, A. C., and E. W. Triplett. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, C. P., R. Ahuja, G. Sayler, and K. H. Chu. 2005. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 71:1433-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]