Abstract
Diatoms are very significant primary producers in the world's oceans. Various environmental factors affect the depletion of diatom populations. The importance of viruses as a potential mortality source has recently been recognized. We isolated and characterized a new diatom virus (Chaetoceros socialis f. radians RNA virus [CsfrRNAV]) causing the lysis of the bloom-forming species Chaetoceros socialis Lauder f. radians (Schütt) Proschkina-Lavrenko. The virus infectious to C. socialis f. radians was isolated from water samples collected in Hiroshima Bay. Here we show the physiology, morphology, and genome characteristics of the virus clone. Virions were 22 nm in diameter and accumulated in the cytoplasm of the host cells. The latent period and the burst size were estimated to be <48 h and 66 infectious units per host cell, respectively. CsfrRNAV harbors a single-stranded RNA (ssRNA) genome and encodes at least three polypeptides of 32.0, 28.5, and 25.0 kDa. Sequencing analysis shows the length of the genome is 9,467 bases, excluding a poly(A) tail. The monophyly of CsfrRNAV and other diatom-infecting RNA viruses, Rhizosolenia setigera RNA virus and Chaetoceros tenuissimus RNA virus, was strongly supported by phylogenetic analysis based on the amino acid sequence of the RNA-dependent RNA polymerase domains. This suggested a new ssRNA virus family, Bacillariornaviridae. This discovery of CsfrRNAV may aid in further understanding the ecological dynamics of the C. socialis f. radians population in nature and the relationships between ssRNA diatom viruses and their hosts.
Diatoms (Bacillariophyceae) account for a large part of the marine primary production, up to 35% in oligotrophic oceans and 75% in nutrient-rich systems (13). They play an important role in various marine systems as a food source for zooplankton and animal larvae. Moreover, diatoms are the primary oxygen producers for the atmosphere (25). Therefore, to understand diatom dynamics in nature is significant for biogeochemical science and fisheries studies. Phytoplankton population dynamics are the result of reproduction and losses. Losses include grazing, sinking, and natural mortality. Since the early 1990s, the importance of viruses infectious to microalgae is recognized as one of the principal causes of phytoplankton mortality. The direct evidence for the existence of diatom viruses was reported recently in 2004 (11). Since the discovery of the first diatom virus, the isolation and characterization of new viruses have been conducted. As a result, several new diatom viruses infecting ecologically important diatom members have been successfully isolated and reported.
The first diatom virus, Rhizosolenia setigera RNA virus (RsRNAV), is a small icosahedral virus (32 nm) with a single-stranded RNA (ssRNA) genome at 8,877 nucleotides (nt), excluding a poly(A) tail (11, 15). Thereafter, two Chaetoceros-infecting single-stranded DNA (ssDNA) viruses were isolated and characterized: Chaetoceros salsugineum nuclear inclusion virus (CsNIV), a small (38-nm) virus harboring a covalently closed circular ssDNA (6,000 nt) and a segment of linear ssDNA (997 nt) (12) (H. Mizumoto, unpublished data), and Chaetoceros debilis DNA virus, whose partial genome sequence is highly similar to that of CsNIV (22). The genome analyses of the two ssDNA viruses showed that they are distinctive from previously reported viruses. The isolation of Chaetoceros nuclear inclusion virus (CspNIV) infectious to Chaetoceros cf. gracilis (a Chaetoceros sp. that looks like Chaetoceros gracilis) was also reported (1); however, its nucleic acid type is still unknown. A recent study reports the isolation of the second ssRNA diatom virus infectious to Chaetoceros tenuissimus (CtenRNAV). A phylogenetic analysis showed a putative RNA-dependent RNA polymerase (RdRp) domain from a genome sequence of CtenRNAV is highly similar to RsRNAV but less similar to other marine stramenopile organism viruses (16): Schizochytrium single-stranded RNA virus (SssRNAV) infecting a fungoid protist Aurantiochytrium sp. (formerly Schizochytrium sp.) (19) and Heterosigma akashiwo RNA virus (HaRNAV; Marnaviridae) infecting the bloom-forming raphidophyte H. akashiwo (7, 8). The ssRNA diatom viruses are unlike other known viruses at the family level. These reports suggest that the diatom viruses are an exclusively unique group distinct from previously described viruses where further study of diatom virus biology is significant to understand diatom ecology.
Here we report the isolation and characterization of a new ssRNA virus (Chaetoceros socialis f. radians RNA virus [CsfrRNAV]) infecting Chaetoceros socialis Lauder f. radians (Schütt) Proschkina-Lavrenko, one of the dominant phytoplankton species in the marine environments in especially productive areas during spring blooms; e.g., in the North Water polynya, the maximum concentration of C. socialis was as high as 3.0 × 104 cells ml−1 (2). Here, we also propose a new ssRNA virus family (Bacillariornaviridae), composed of three diatom-infecting ssRNA viruses based on phylogenetic analysis using the RdRp domain and other genomic characters.
MATERIALS AND METHODS
Algal cultures and growth conditions.
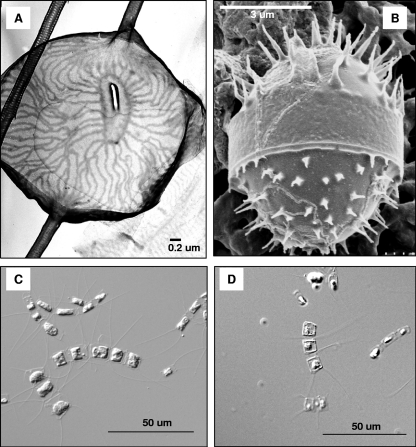
The axenic clonal algal strain used in this study, Chaetoceros socialis f. radians L-4, was isolated from surface water at the Itsukaichi fishing port (34°21.400′N, 132°21.864′E) in Hiroshima Bay, Japan, on 27 April 2005. This algal strain was observed using transmission electron microscopy and scanning electron microscopy (Fig. 1A and B) and identified as C. socialis Lauder f. radians (Schütt) Proschkina-Lavrenko. Algal cultures were grown in modified SWM3 medium enriched with 2 nM Na2SeO3 (3, 5) under a 12/12-h light-dark cycle of ca. 110 to 150 μmol photons m−2 s−1 using cool white fluorescent illumination at 15°C.
FIG. 1.
Chaetoceros socialis f. radians L-4. (A) Transmission electron micrograph of an intact cell, terminal valve with eccentric rimoportula; (B) scanning electron micrograph of a resting spore; (C) optical micrograph of intact cells; (D) optical micrograph of CsfrRNAV-infected cells at 132 hpi.
Isolation of the virus.
Water samples (0.2 m from the bottom) were obtained from Itsukaichi fishing port (ca. 5 m in depth) on 8 July 2005. The samples were immediately filtered through 0.2-μm Dismic-25cs filters (Advantec) to remove eukaryotic microorganisms and most of the bacteria. An aliquot (0.5 ml) of the filtrate was inoculated into an exponentially growing C. socialis f. radians L-4 culture (1.0 ml) and followed by incubation at 15°C using the lighting conditions given above. Algal cultures inoculated with SWM3 served as controls. A C. socialis f. radians L-4 culture inoculated with the filtrate showed the inhibition of algal growth (Fig. 1B). We cloned the responsible pathogen through two extinction dilution cycles (17, 24) using lysed cultures of C. socialis f. radians L-4. The lysate in the highest dilution well of the second assay was made free of bacterial contamination using filtration through a 0.1-μm polycarbonate membrane filter (Nuclepore) and was transferred to another exponentially growing host culture. The resultant lysate was regarded as a clonal virus suspension and was later designated CsfrRNAV01.
Host range.
Interspecies host specificity for the virus CsfrRNAV was tested by adding 5% (vol/vol) aliquots of fresh lysate passed through 0.2-μm filters (Nuclepore) into duplicate cultures of 28 exponentially growing clonal algal strains as follows: Chaetoceros debilis, Chaetoceros salsugineum, C. socialis f. radians, Chaetoceros tenuissimus, Chaetoceros sp., Chaetoceros cf. affinis, Chaetoceros cf. lorenzianus, Chaetoceros cf. pseudocurvisetus, Detonula pumila, Ditylum brightwellii, Eucampia zoodiacus, Rhizosolenia setigera, Skeletonema sp., Stephanopyxis sp. (Bacillariophyceae), Nannochloropsis sp. (Eustigmatophyceae), Teleaulax amphioxeia (Cryptophyceae), Alexandrium catenella, Gymnodinium catenatum, Heterocapsa circularisquama, Heterocapsa triquetra, Karenia mikimotoi, Prorocentrum micans, Scrippsiella sp. (Dinophyceae), Chattonella antiqua, Chattonella marina, Chattonella ovata, Fibrocapsa japonica, and Heterosigma akashiwo (Raphidophyceae). They were cultured under the conditions given above at either 15 or 20°C. Growth and evidence of lysis in each algal culture were monitored by optical microscopy and compared to those in control cultures inoculated with SWM3. The cultures not lysed at 14 days postinoculation were considered to be unsuitable hosts for the viral pathogen.
TEM.
An exponentially growing culture of C. socialis f. radians L-4 was inoculated with CsfrRNAV at a multiplicity of infection of 1.4. As the control, a C. socialis f. radians L-4 culture was inoculated with autoclaved culture medium SWM3. An aliquot of cell suspension was sampled at 48 h postinoculation (hpi). C. socialis f. radians L-4 cells were harvested by centrifugation at 860 × g at 4°C for 10 min and fixed with 1% glutaraldehyde in SWM3 for 4 h at 4°C. The cell pellets were postfixed for 3 h in 2% osmic acid in 0.1 M phosphate buffer (pH 7.2 to 7.4), dehydrated in a graded ethanol series (50 to 100%), and embedded in Quetol 653 resin (Nisshin EM Co., Ltd.). Ultrathin sections were stained with 4% uranyl acetate and 3% lead citrate and observed at 80 kV using a JEOL JEM-1010 transmission electron microscope. CsfrRNAV particles negatively stained with uranyl acetate were also observed using transmission electron microscopy (TEM). Briefly, the virus suspension was mounted on a grid (no. 780111630; JEOL Datum, Ltd.) for 30 s, and excess water was removed using filter paper (no. 1; TOYO Co., Ltd.). Then, 4% uranyl acetate was applied for 10 s, and excess dye was removed using a filter paper. After the grid was dried in a desiccator for 3 h, negatively stained CsfrRNAV particles were observed using TEM at an acceleration voltage of 80 kV. Particle diameters were estimated using the negatively stained images.
Thermal stability.
An exponentially growing culture of C. socialis f. radians L-4 was inoculated with virus CsfrRNAV and incubated for 4 days. The lysate was passed through a 0.2-μm filter (Nuclepore) to remove cellular debris. The titer of the resultant fresh lysate was then estimated using the extinction dilution method, and aliquots of the lysate were stored at 20, 10, 4, −20, −80, or −196°C (in liquid nitrogen) in the dark without the addition of cryoprotectants. After the initial titration, the aliquots were assayed at 50 days after storage to determine the stability of the virus at each temperature.
CsfrRNAV proteins.
The CsfrRNAV suspension was mixed with four volumes of denaturing sample buffer (62.5 mM Tris-HCl, 5% 2-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], 20% glycerol, 0.005% bromophenol blue) and boiled for 5 min. The proteins were then separated using SDS-polyacrylamide gel electrophoresis (80 × 40 × 1 mm, 12.5% polyacrylamide, 150 V) using the XV Pantera system (DRC Co., Ltd.). The proteins were visualized using Coomassie brilliant blue stain. Protein molecular mass standards (Bio-Rad Co., Ltd.) ranging from 6.5 to 200 kDa were used for size calibration.
CsfrRNAV nucleic acids.
A 450-ml exponentially growing C. socialis f. radians L-4 culture was inoculated with 5 ml of virus CsfrRNAV suspension and lysed. The lysate was sequentially passed through 0.8- and 0.4-μm polycarbonate membrane filters (Nuclepore) to remove cellular debris. Polyethylene glycol 6000 (Wako Pure Chemical Industries, Ltd.) was added to the filtrate to a final concentration of 10% (wt/vol), and the suspension was stored at 4°C in the dark overnight. After centrifugation at 57,000 × g at 4°C for 1.5 h, the pellet was washed with 10 mM phosphate buffer (pH 7.2) and centrifuged again at 217,000 × g at 4°C for 4 h to collect the virus particles. They were resuspended in 500 μl of ultrapure water. Nucleic acids were extracted from the pellet using the RNeasy mini kit (Qiagen).
Aliquots (7 μl) of the nucleic acids solution were digested with RNase A (Nippon Gene Co., Ltd.) at 0.025 μg μl−1 in 0.01× SSC or 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M Na3 citrate [pH 7.0]) at 37°C for 1 h (18) and incubated with DNase I (Takara Co, Ltd.) at 0.5 U μl−1 at 37°C for 1 h. Nucleic acid extracts held on ice without any treatment served as controls. The nucleic acid samples thus prepared were electrophoresed in denatured agarose gels (1.5%; SeaKem Gold agarose; BMA, Inc.) at 50 V for 1 h. Nucleic acids were visualized using SYBR gold staining (Molecular Probes, Inc.).
Genome sequencing.
Sequencing of the viral genome was performed as follows: the RNA purified from the virus pellet using the RNeasy mini kit (Qiagen) was reverse transcribed to construct cDNAs with the cDNA synthesis kit M-MLV version (Takara Co., Ltd.) using the random primers according to the manufacturer's recommendation. The 5′ ends of the resultant double-stranded DNA fragments were phosphorylated using T4 polynucleotide kinase (Takara Co., Ltd.); then, the resultant cDNA fragments were electrophoresed on an agarose gel and ∼2-kb fragments were extracted. The fragments were ligated into the HincII-cleaved and dephosphorylated pSTV28 plasmid vector (Takara Co., Ltd.). The ligated double-stranded DNA fragments were transformed into Escherichia coli DH10B competent cells (Invitrogen) and sequenced using the dideoxy method with an ABI 3730xl DNA analyzer (Applied Biosystems). The resultant fragment sequences were reassembled; then, sequencing of the 5′ and 3′ ends was conducted using 5′ and 3′ rapid amplification of cDNA ends following the procedure described previously (10), respectively. The CsfrRNAV genome RNA was fully sequenced. Putative open reading frames (ORFs) were identified using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Automated comparisons were conducted on the CsfrRNAV sequence using BLAST (Basic Local Alignment Research Tool).
Phylogenetic analysis.
We identified a conserved RdRp domain in the assembled contiguous sequence using BLAST. The deduced amino acid sequence of the corresponding region was compared to other virus RdRp domains. They were automatically aligned using ClustalW (20) and manually refined. Phylogenetic trees based on the deduced amino acid sequences of the RdRp domain were constructed using the neighbor-joining (NJ) and maximum likelihood (ML) methods with the Jones-Taylor-Thormton matrix (JTT model) packaged in the Phylip 3.65 program (6). The amino acid sequences used for comparison in the analyses are as follows with the organisms' scientific names and database accession numbers (referring to the NCBI unless otherwise stated): Aichi virus (AB010145), bovine enteric calicivirus (AJ011099), bean pod mottle virus (NC_003496), black queen cell virus (NC_003784), CsfrRNAV (AB469874), CtenRNAV (AB375474), cowpea severe mosaic virus (M83830), cricket paralysis virus (NC_003924), Drosophila C virus (NC_001834), deformed wing virus (NC_004830), HaRNAV (NC_005281), Norwalk virus (M87661), human poliovirus 1 Mahoney (V01149), parsnip yellow fleck virus (D14066), RsRNAV (AB243297), rice turgo spherical virus (AAA66056), sacbrood virus (NC_002066), SssRNAV (BAE47143), Triatoma virus (NC_003783), and Taura syndrome virus (NC_003005).
Growth experiment.
An exponentially growing culture of C. socialis f. radians L-4 (45 ml) was inoculated with CsfrRNAV at a multiplicity of infection of 66. A C. socialis f. radians L-4 culture inoculated with an autoclaved viral suspension served as the control. An aliquot of the cell suspension was sampled from each culture at 0, 12, 24, 36, 48, 60, 72, 84, 108, 120, 156, 180, and 210 hpi, and the number of host cells and lytic agents were estimated. The number of lytic agents was determined using the extinction dilution method (17). The cells at 23 days postinoculation were observed using confocal laser scanning microscope (LSM5 PASCAL) using a 100×/1.4 (Plan-Apochromat) oil immersion objective lens and scanned with a 543-nm helium-neon laser set at 50% intensity with a 560-nm long-pass filter.
RESULTS AND DISCUSSION
Isolation of the viral pathogen.
The isolated algicidal agent retained its lytic activity after filtration through a 0.1-μm filter. The lytic activities were serially transferable to exponentially growing C. socialis f. radians L-4 cultures (data not shown). Cells of C. socialis f. radians L-4 lysed by the virus became pale in color, presumably due to the loss or degradation of photosynthetic pigments (Fig. 1).
Host range.
The host range of CsfrRNAV was tested using 28 phytoplankton strains, including 14 diatom strains. CsfrRNAV caused lysis in only its single host strain and not any other microalgal species tested. This shows that the virus has a high infection specificity (data not shown).
Morphological features.
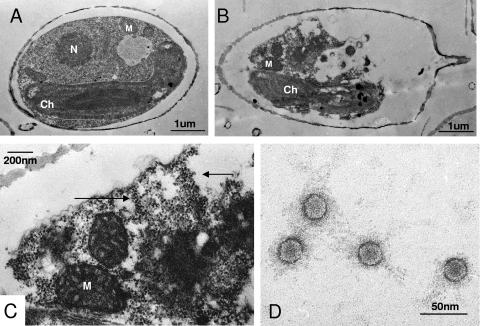
Thin sections of healthy C. socialis f. radians L-4 cells show the cytoplasmic organization and frustules that are typical of these diatoms (Fig. 2A). In contrast, electron micrographs of thin-sectioned C. socialis f. radians L-4 cells at 48 hpi with virus showed the presence of virus-like particles (VLPs) randomly assembled in the host cytoplasm (Fig. 2B and C). No VLPs were found in the healthy control cultures (Fig. 2A). Further, the VLPs were observed in culture lysates using negative staining. They were icosahedral (22 ± 1 nm; n = 50), and lacked a tail or an outer membrane (Fig. 2D). Since (i) the algicidal pathogen was transferable to a fresh algal culture, (ii) the VLPs were observed in the lysed culture, and (iii) the VLPs were not found in healthy cultures, Koch's postulates are fulfilled. We conclude that the VLP observed within the infected cells and in the algal lysates is a previously undescribed virus pathogenic to C. socialis f. radians.
FIG. 2.
Transmission electron micrographs of ultrathin sections of Chaetoceros socialis f. radians L-4 and negatively stained CsfrRNAV particles. (A) Healthy cell. (B, C) Cells infected with CsfrRNAV at 48 hpi. (B) Degraded host cytoplasm. (C) Higher magnification of the VLPs in the cytoplasm. Arrows show the accumulation of VLPs. (D) Negatively stained CsfrRNAV particles in the culture lysate. N, nucleus; Ch, chloroplast; M, mitochondrion.
Thermal stability.
CsfrRNAV suspensions containing 2.1 × 107 infectious units ml−1 were stored at various temperatures. The infectious titers of the virus suspension after 50 days of storage at 20, 10, 4, −20, −80, and −196°C in the dark were 25, 66, 145, 143, 245, and 185% of the initial titer, respectively. Thus, the infectious titers tended to decrease at >4°C. In contrast, the titers seemed to increase when the virus suspensions were preserved at lower temperatures, especially ≤−80°C. This phenomenon has been also reported in other microalgal viruses (16, 23). The virus particles of CsfrRNAV are aggregated in the host cytoplasm (Fig. 2C) and a part of them might form small aggregations in the lysate. The increase in CsfrRNAV titer following cryopreservation is presumably caused by the diffusion of aggregated viruses.
The high thermal stability of CsfrRNAV observed in the present study would be important in revealing its life cycle in nature (9, 21).
Proteins.
The sizes and numbers of structural proteins of the virus particles were determined using SDS-polyacrylamide gel electrophoresis. CsfrRNAV contains at least three major polypeptides at 32.0, 28.5, and 25.0 kDa and one minor polypeptide at 31.0 kDa (Fig. 3). The number of major proteins of CsfrRNAV was the same as those of the ssRNA diatom viruses RsRNAV and CtenRNAV that have three major polypeptides of 41.5, 41.0, and 29.5 kDa, and 33.5, 31.5, and 30.0 kDa, respectively (11, 16).
FIG. 3.
Structural proteins of CsfrRNAV.
Genomic and phylogenetic analysis.
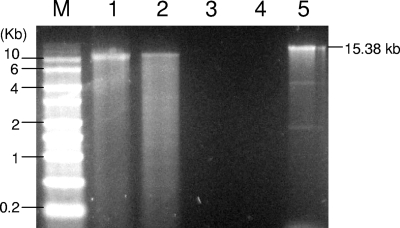
Denaturing agarose gel electrophoresis shows that the nucleic acids extracted from CsfrRNAV were ∼11 kb (Fig. 4). The bands were not sensitive to DNase I, but RNase A at high and low salt treatments completely digested the extracted nucleic acids (Fig. 4). These data suggest that the genome is a ssRNA. Therefore, this new virus was designated as C. socialis f. radians RNA virus (CsfrRNAV) named after its host species and genome type. The full length of the CsfrRNAV was 9,467 nt, excluding a poly(A) tail. Thus, the genome was determined to be smaller than the estimated size by using formaldehyde/agarose gel electrophoresis (Fig. 4), which may reflect the additional length of the 3′ poly(A) tail or the addition of viral genome-linked proteins (VPg); however, we do not entirely understand this. The AU ratio of the CsfrRNAV genome is 60.4%, and this is comparable to that of RsRNAV at 63.7% and CtenRNAV at 61.1% (Table 1). Compared to these RNA viruses, the AU ratios of the other Stramenopile organism-infecting viruses HaRNAV (Marnaviridae) and SssRNAV are much lower at 53.1% and 50.2%, respectively (Table 1). The CsfrRNAV genome includes two ORFs, putative replication-related proteins (∼881 to ∼5,956 nt) and putative capsid proteins (∼6,249 to ∼9,296 nt), and this is the same organization as in RsRNAV and CtenRNAV (Table 1). Therefore, the basic genome structures of CsfrRNAV are identical to those of previously known ssRNA diatom viruses where a BLASTP analysis also shows that the amino acid sequences of CsfrRNAV are highly similar to those of the two diatom viruses (Table 1).
FIG. 4.
Nucleic acids of CsfrRNAV without treatment (lane 1), treatment with DNase I (lane 2), treatment with RNase A in low-salt buffer (lane 3), and treatment with RNase A in high-salt buffer (lane 4). Samples were electrophoresed in a formaldehyde agarose gel with RNA molecular size markers (lane M) and Sendai virus genome RNA (15.3 kb) (lane 5).
TABLE 1.
General features of ssRNA viruses infecting eukaryote microorganisms
| Virus | Particle diameter (nm) | Genome size (nt) | AU ratio (%) | No. of ORFs | BLASTP comparison for ORF-1 of CsfrRNAV
|
BLASTP comparison for ORF-2 of CsfrRNAV
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| E value | No. identical/total no. (%) | No. similar/total no. (%) | E value | No. identical/total no. (%) | No. similar/total no. (%) | |||||
| CsfrRNAV | 22 | 9,467 | 60.4 | 2 | ||||||
| RsRNAV | 32 | 8,877 | 63.7 | 2 | 0.0 | 655/1,652 (39) | 951/1,652 (57) | 2.0E−108 | 287/891 (32) | 446/891 (50) |
| CtenRNAV | 31 | 9,431 | 61.1 | 2 | 0.0 | 651/1,723 (37) | 965/1,723 (56) | 4.0E−126 | 304/896 (33) | 459/896 (51) |
| SssRNAV | 25 | 9,018 | 50.2 | 3 | 4.0E−26 | 147/649 (22) | 265/649 (40) | 2.0E−25 | 92/313 (29) | 149/313 (47) |
| HaRNAV | 25 | 8,587 | 53.1 | 1 | 5.0E−22 | 95/357 (26) | 164/357 (45) | 3.0E−72 | 233/867 (26) | 384/867 (44) |
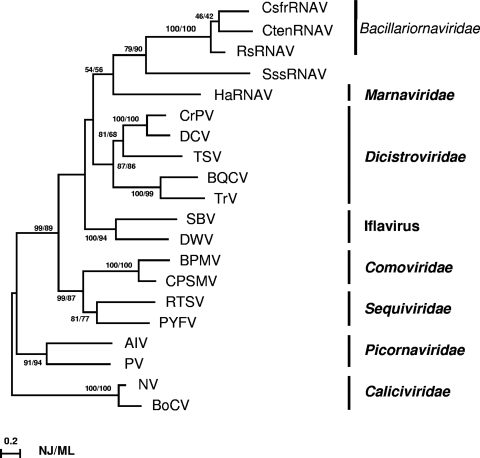
Here, both the NJ and ML methods were used to assess the phylogenetic relationship among positive-sense ssRNA viruses, including CsfrRNAV. Consequently, similar topologies were obtained using the two methods; hence, only the ML phylogenetic tree is shown in Fig. 5. The monophyly of CsfrRNAV, CtenRNAV, and RsRNAV was supported with a bootstrap value of 100% (Fig. 5). This suggests the existence of a diatom-infecting ssRNA virus clade within the positive-sense ssRNA viruses.
FIG. 5.
ML tree based on deduced amino acid sequences of the RdRp whole domain. ML bootstrap values (%) from 100 samples are shown at the nodes, followed by bootstrap values based on the NJ analysis (%) from 100 samples. The ML distance scale bar is shown. CrPV, cricket paralysis virus; DCV, Drosophila C virus; BQCV, black queen cell virus; TrV, Triatoma virus; TSV, Taura syndrome virus; DWV, deformed wing virus; SBV, Sacbrood virus; BPMV, bean pod mottle virus; CPSMV, cowpea severe mosaic virus; RTSV, rice turgo spherical virus; PYFV, parsnip yellow fleck virus; PV, human poliovirus 1 Mahoney; AIV, Aichi virus; BoCV, bovine enteric calicivirus; NV, Norwalk virus.
As a result, a comparison of the genome organization and sequence relationships suggests that diatom-infecting ssRNA viruses are not a member of any currently defined virus family. Therefore, we propose that the ssRNA diatom viruses RsRNAV, CtenRNAV, and CsfrRNAV are members of a new virus family, Bacillariornaviridae. To determine the phylogenetic relationships of ssRNA viruses infecting Stramenopile organisms more precisely, further isolation and characterization of new viruses are essential.
Replication.
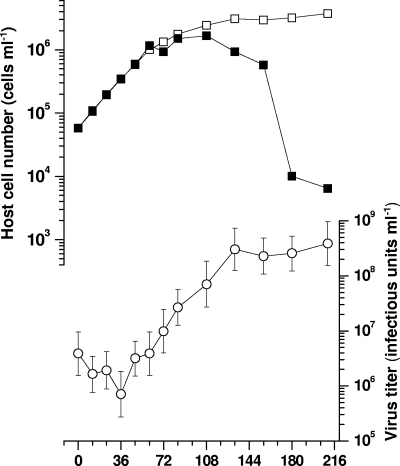
Using a growth experiment, a significant increase in virus abundance was observed after 48 hpi (Fig. 6); thus, the latent period of CsfrRNAV was estimated to be <48 h, whereas the decrease in host cell number was observed after 108 hpi (Fig. 6). If the multiplicity of infection was high enough to make all of the cells exposed to viral attack (i.e., to make all the sensitive cells infected), we presume the viral infection does not necessarily interrupt algal growth in the logarithmic phase. This was also observed with a previously reported ssRNA diatom virus, CtenRNAV (16). One possible explanation may be that the virus sensitivity was highly diverse among cells even in a clonal host culture, i.e., some expressed sensitivity and some did not, and only a portion of the host cells were immediately infected with the virus, lysed, and produced daughter virus particles.
FIG. 6.
Changes in the cell numbers of C. socialis f. radians L-4 used for the growth experiments with (filled square) or without (open square) virus inoculation and the virus titer (open circle). Virus inoculation was performed at 0 h in an exponentially growing host culture using a multiplicity of infection of 66. The error bars in the virus titers indicate the 95% confidential interval.
An estimation of the burst size was considered to be difficult due to the above reasons. The host/virus ratio at 108 to 132 hpi was used to calculate the burst size and was estimated to be 66 infectious units cell−1. Comparing these data to the observed particle numbers found in thin-section views of the infected cells, this value may be underestimated. Possible explanations for the small burst size may be the aggregation of virus particles causing an underestimation of the most probable number, difficulties in distinguishing dead cells and living cells by optical microscopy, or a dominance of defective particles lacking infectivity.
In growth experiments, the cultures did not completely lyse even at 210 hpi. Further, we observed resting spores of C. socialis f. radians L-4 in the culture at 23 days after the virus inoculation. The spores had autofluorescence with 543-nm laser excitation, indicating viability even in the virus suspended culture with 5.1 × 107 infectious units ml−1. The resting spores did not regrow in the infected cultures (data not shown). Similar results have been reported in the relationship between C. debilis and the C. debilis DNA virus (22). The formation of Chaetoceros resting spores are enhanced by nitrogen depletion and other combined factors (4, 14), and they may play an important role in survival in various environments. It is unknown what did affect the resting spore formations in the virus-inoculated culture. Future studies should focus on the viral attacks and the host spores to reveal the survival of C. socialis f. radians in natural environments.
Implications.
The virus CsfrRNAV infectious to C. socialis f. radians was isolated from waters in Hiroshima Bay. The ecological relationships between CsfrRNAV and its host are not understood; however, many other microalgal hosts and their lytic viruses show intimate relationships in nature. Our recent field surveys have shown diatom host-virus relationships in western Japan (unpublished data), e.g., C. debilis (22) and C. tenuissimus (16). One of the foci in the ecological relationships between C. socialis f. radians and CsfrRNAV is to determine the roles of the resting host spores which might be formed in water columns and embedded in sediments. Our prediction is that the resting spores may be significant in protecting the algal population against viral attacks. The lytic viruses infectious to C. socialis f. radians have also been isolated from sediments in Hiroshima Bay (unpublished data); therefore, the hatching of the resting spores from the sediments without viral infection may be important for bloom formation. The relationships between the resting spores and the viral infection should be the focus of further studies to reveal the life cycle of the algae and their viruses in a shallow bay.
The phylogenetic analysis showed that the three diatom viruses had monophyly among the positive-sense ssRNA viruses. Considering the huge biomass and numbers of diatom species, the ssRNA diatom virus group might be a vast virus group in the ocean. The existence of diatom viruses is the result of the interaction with its host diatom. Further studies concerning ssRNA diatom viruses may help to understand diatom biology and virology in the sea.
Acknowledgments
This study was supported by grants-in-aid for scientific research (A)(2)(16208019) from the Ministry of Education, Science, and Culture of Japan and by the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Footnotes
Published ahead of print on 20 February 2009.
REFERENCES
- 1.Bettarel, Y., J. Kan, K. Wang, K. Williamson, S. Cooney, S. Ribblett, F. Chen, E. Wommack, and W. Coats. 2005. Isolation and characterisation of a small nuclear inclusion virus infecting the diatom Chaetoceros c.f. gracilis. Aquat. Microb. Ecol. 40:103-114. [Google Scholar]
- 2.Bootha, B. C., P. Laroucheb, S. Belangerb, B. Kleinc, D. Amield, and Z.-P. Mei. 2002. Dynamics of Chaetoceros socialis blooms in the North Water. Deep-Sea Res. Part II 49:5003-5025. [Google Scholar]
- 3.Chen, L. C. M., T. Edelstein, and J. McLachlan. 1969. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 4.Itakura, S., M. Yamaguchi, and I. Imai. 1993. Resting spore formation and germination of Chaetoceros didymus var. protuberans (Bacillariophyceae) in clonal culture. Nippon Suisan Gakkaishi 59:807-813. [Google Scholar]
- 5.Itoh, K., and I. Imai. 1987. Rafido so (Raphidophyceae), p. 122-130. In A guide for studies of red tide organisms. Japan Fisheries Resource Conservation Association, Shuwa, Tokyo, Japan.
- 6.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 7.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 8.Lang, A. S., and C. A. Suttle. 2008. Marnaviruses, p. 280-285. In B. W. J. Mahy and M. H. V. Van Regenmortel (ed.), Encyclopedia of virology, vol. 5. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 9.Lawrence, J. E., A. M. Chan, and C. A. Suttle. 2002. Viruses causing lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae) are widespread in coastal sediments of British Columbia, Canada. Limnol. Oceanogr. 47:545-550. [Google Scholar]
- 10.Liu, X., and M. A. Gorovsky. 1993. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 21:4954-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagasaki, K., Y. Tomaru, N. Katanozaka, Y. Shirai, K. Nishida, S. Itakura, and M. Yamaguchi. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasaki, K., Y. Tomaru, Y. Takao, K. Nishida, Y. Shirai, H. Suzuki, and T. Nagumo. 2005. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 71:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, D. M., P. Treguer, M. A. Brzezinski, A. Leynaert, and B. Queguiner. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycle 9:359-372. [Google Scholar]
- 14.Oku, O., and A. Kamatani. 1997. Resting spore formation of the marine planktonic diatom Chaetoceros anastomosans induced by high salinity and nitrogen depletion. Mar. Biol. 127:515-520. [Google Scholar]
- 15.Shirai, Y., Y. Takao, H. Mizumoto, Y. Tomaru, D. Honda, and K. Nagasaki. 2006. Genomic and phylogenetic analysis of a single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Assos. UK 86:475-483. [Google Scholar]
- 16.Shirai, Y., Y. Tomaru, Y. Takao, H. Suzuki, T. Nagumo, and K. Nagasaki. 2008. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 74:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suttle, C. A. 1993. Enumeration and isolation of viruses, p. 121-137. In P. F. Kemp, E. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 18.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 19.Takao, Y., K. Nagasaki, K. Mise, T. Okuno, and D. Honda. 2005. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl. Environ. Microbiol. 71:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomaru, Y., N. Hata, T. Masuda, M. Tsuji, K. Igata, Y. Masuda, T. Yamatogi, M. Sakaguchi, and K. Nagasaki. 2007. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 9:1376-1383. [DOI] [PubMed] [Google Scholar]
- 22.Tomaru, Y., Y. Shirai, H. Suzuki, T. Nagumo, and K. Nagasaki. 2008. Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros debilis. Aquat. Microb. Ecol. 50:103-112. [Google Scholar]
- 23.Tomaru, Y., H. Tanabe, S. Yamanaka, and K. Nagasaki. 2005. Effects of temperature and light on stability of microalgal viruses, HaV, HcV and HcRNAV. Plankton Biol. Ecol. 52:1-6. [Google Scholar]
- 24.Tomaru, Y., K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Quantitative and qualitative impacts of viral infection on Heterosigma akashiwo (Raphidophyceae) population during a bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34:227-238. [Google Scholar]
- 25.Werner, D. 1977. Introduction with a note on taxonomy, p. 1-23. In D. Werner (ed.), The biology of diatoms, vol. 13. Blackwell Scientific Publications, Victoria, Australia. [Google Scholar]