Abstract
Prokaryotic diversity was investigated near the inlet and outlet of a plug-flow reactor. After analyzing 800 clones, 50 bacterial and 3 archaeal phylogenetic groups were defined. Clostridia (>92%) dominated among bacteria and Methanoculleus (>90%) among archaea. Significant changes in pH and volatile fatty acids did not invoke a major shift in the phylogenetic groups. We suggest that the environmental filter imposed by the saline conditions (20 g liter−1) selected a stable community of halotolerant and halophilic prokaryotes.
The anaerobic digestion of organic wastes constitutes a major research focus due to the global needs for waste recycling and renewable energy production. Currently, the linkage between digester performance and the diversity and dynamics of anaerobic prokaryotes is still not fully understood (2). Bacterial diversity in anaerobic reactors has always been judged to be greater than archaeal diversity (9, 13, 30). This probably reflects the metabolic flexibility of bacteria and the range of available substrates in complex input materials. However, several recent discoveries pose the question as to whether archaeal diversity and physiological versatility are greater than currently thought: that is, the huge diversity of yet-to-be cultured archaea (4, 6), the detection of energy metabolisms not known previously in archaea (e.g., chemoorganotrophy [1]), and the unexpected predominance of archaeal groups among prokaryotes in unstressed environments, such as ammonia oxidizers in soils (19).
Several surveys have investigated the shifts in prokaryotic diversity occurring with waste maturation or under different reactor operating conditions. Some evidence demonstrates bacterial phylogenetic stability under constant operation conditions (18). Generally, however, the dominant bacterial communities are very dynamic, showing chaotic shifts even with stable reactor performance (9, 32). Hypothetically, this is due to the functional redundancy among diverse phylogenetic groups allowing oscillations of their populations with no effects on the reactor function (2). Archaeal communities are less dynamic than bacterial communities (32), their shifts being related to changes in reactor performance (6) and correlated with important process parameters such as volatile fatty acids (VFAs) (13, 16).
We aimed to analyze the change in prokaryotic diversity in a plug-flow reactor associated with the maturation of biowastes. In a previous study, stable bacterial and archaeal denaturing gradient gel electrophoresis patterns were found in the sludge collected close to the outlet over a year of unstable reactor performance (23). This temporal pattern contradicts the general idea of extremely dynamic bacterial communities proliferating in bioreactors. Here, we investigated the phylogenetic identity of the organisms in sludge samples collected near the inlet and outlet pipes after a period of stable operation and performance in terms of pH and biogas production.
Operation of the bioreactor and sampling of sludges.
Samples were taken from the thermophilic (50 to 55°C) anaerobic plug-flow reactor located in Roppen (Tirol, Austria), which has a capacity of 750,000 liters. At the time of sampling (October 2006), the reactor had been operating stably for several years. That is, it had been fed daily with 20 to 25 Mg biowaste (i.e., source-separated organic household waste mixed with garden residues in a season-dependent ratio) with continuous radial stirring and a directional axial flow from the inlet to the outlet, with no backward flow possible, and a 3-week hydraulic retention time. The biogas production had been stable for several months, averaging 120 m3 h−1, with 62% CH4 and 38% CO2. Fresh and mature sludges were collected from two 1-m-depth sampling ports located on top of the reactor near the inlet and outlet pipes (ca. 30 m away from each other). Five 1-liter samples were collected per sampling port.
Physical and chemical sludge properties.
Table 1 shows the physical and chemical parameters of fresh and mature sludges. The C and N contents of oven-dried (60°C) ground sludge samples were measured in an automated CNHS analyzer (TruSpec; LECO). VFAs were analyzed by high-pressure liquid chromatography, after injection of 20 μl of solution (i.e., supernatant after centrifuging 2 ml of 1:4 sludge-water suspensions) into a Shimadzu LC-20A prominence high-pressure liquid chromatograph. VFAs were separated with an Aminex HPX-87H column (300 by 7.8 mm) and detected photometrically (220 nm).
TABLE 1.
Physical and chemical properties of the fresh and mature sludges
| Parameter | Result for sludgea
|
|
|---|---|---|
| Fresh | Mature | |
| Total solids (%) | 29.1 (1.0) | 27.1 (1.8) |
| Volatile solids (%) | 57.3 (2.2) | 54.8 (2.2) |
| pH at 1:5 (wt/vol) | 7.93 (0.05) | 8.65 (0.03)* |
| Electrical conductivity at 1:10 wt/vol (mS cm−1) | 3.97 (0.05)* | 3.75 (0.10) |
| C (%) | 30.07 (2.50)* | 24.72 (1.50) |
| N (%) | 2.00 (0.08) | 1.92 (0.11) |
| C/N ratio | 15.0 (0.5)* | 12.9 (0.2) |
| Acetate (mM) | 10.02 (1.01)* | 1.01 (0.20) |
| Propionate (mM) | 7.16 (0.73) | 6.69 (0.49) |
| Butyrate (mM) | 3.31 (0.40)* | 0.06 (0.04) |
| i-Valeriate (mM) | 1.03 (0.11)* | 0.52 (0.10) |
| i-Butyrate (mM) | 0.57 (0.03)* | 0.28 (0.03) |
| Valeriate (mM) | 0.37 (0.05) | 0.35 (0.24) |
Standard deviations are given in parentheses (n = 5). Asterisks indicate significant differences at P < 0.001 after Student's t test or multivariate one-way analysis of variance (for VFAs) with SPSS 15.0.
The fresh and mature sludges differed significantly in their main chemical properties (Table 1). The high content of organic substances in the fresh sludge resulted in the production of a large amount of short-chain fatty acids. This explains the significantly lower pH and higher electrical conductivity of the fresh sludge compared with the mature sludge. As expected, the recalcitrance of the organic matter in the biowastes increased together with the process of anaerobic digestion, as indicated by the decrease in both the C/N ratio and the levels of VFAs, which dropped from 22.5 mM in the fresh sludge to 8.9 mM in the mature sludge. All VFAs underwent a significant decrease during the digestion, except for propionate, which remained at high and constant levels, and valeriate, which was present at low levels in both sludges.
Biogas production from the fresh and mature sludges.
Biogas production was measured in a batch incubation assay. Within 5 h of sampling, 2.5 g sludge was transferred to 250-ml Schott bottles flushed with 100% N2 and incubated in the dark at 50°C with continuous stirring. The pressure within the test bottles was measured for 17 days using a digital manometric system (Sensomat; Aqualytic, Germany). Nonlinear regression analysis was used to estimate the kinetic parameters of biogas production using SigmaPlot v9.01.
The reduction in substrate concentration during the digestion invoked a decrease in the rate of biogas production. After 17 days of incubation, biogas production decreased from 19.2 ml biogas g−1 fresh sludge to 5.6 ml biogas g−1 mature sludge (wet weight). Both curves fitted well (R2 = 0.995) to a first-order kinetic equation, and the rate constant in the fresh sludge was twofold that of the mature sludge (0.18 and 0.09 day−1).
DNA extraction and PCR amplification.
DNA was extracted from 0.25 g sludge within 5 h of sampling using the PowerSoil DNA isolation kit (MO BIO Laboratories). DNA was subjected to PCR amplification of the 16S rRNA genes with universal bacterial and archaeal primers (Table 2). PCR amplifications were performed in a Thermo Hybaid PCR Express Thermalcycler (Fisher Scientific) in 25-μl volumes, with each reaction mixture containing 1× reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]) (Applied Biosystems), 200 μM each deoxynucleoside triphosphate (dNTP) (Genxpress, Austria), 0.2 μM each primer, 1× enhancer (Peqlab, Germany), 0.4 mg ml−1 bovine serum albumin, 10 mM tetramethylammonium chloride, 1.25 mM MgCl2, 0.63 U AmpliTaq Gold DNA polymerase LD (Applied Biosystems), and sterile water. Two microliters of extracted DNA (diluted 1/10 for archaea and 1/50 for bacteria) was applied to the PCR mix. Bacterial DNA was amplified with an initial denaturation at 95°C for 5 min, 25 amplification cycles (45 s at 95°C, 45 s at 52°C, and 4 min at 72°C), and a final elongation at 72°C for 10 min. Archaeal DNA was amplified as described for bacteria, but amplification cycles consisted of 1 min at 95°C, 1 min at 55°C, and 3 min at 72°C. PCR products obtained for the five independent replicates were pooled and purified with the GenElute PCR clean-up kit (Sigma).
TABLE 2.
Primers used in this study
| Target | Primer | Sequence (5′ → 3′) | E. coli position | Reference |
|---|---|---|---|---|
| Bacteria | 8F | AGAGTTTGATYMTGGCTC | 8-27 | 21 |
| 1492R | GGYTACCTTGTTACGACTT | 1492-1507 | ||
| Archaea | 109F | ACKGCTCAGTAACACGT | 109-125 | 12 |
| 934R | GTGCTCCCCCGCCAATTCCT | 915-934 | ||
| Methanoculleusa | 298F | GGAGCAAGAGCCCGGAGT | 298-315 | 10 |
| 586R | CCAAGAGACTTAACAACCCA | 586-606 | ||
| Methanothermobactera | 410F | CTCTTAACGGGGTGGCTTTT | 410-440 | 10 |
| 667R | CCCTGGGAGTACCTCCAGC | 667-686 | ||
| Uncultured groupa | 195F | AAAACTCCGGTGCCTTAGGATT | 195-230 | 10 |
| 330R | CCCGTAGGGCCTGGACTCA | 330-348 | ||
| Methanosaetaa | MS1b | CCGGCCGGATAAGTCTCTTGA | 585-606 | 28 |
| SAE835R | GACAACGGTCGCACCGTGGCC | 835-851 | ||
| Methanosarcinaa | 240F | CCTATCAGGTAGTAGTGGGTGTAAT | 240-264 | 10 |
| 589R | CCCGGAGGACTGACCAAA | 589-606 |
Source DNA for preparing standards: clone 2FA36 (AM947508), Methanoculleus bourgensis; DSM 2970, Methanothermobacter wolfeii; clone 1FA28 (AM947509), uncultured archaea; DSM 2139, Methanosaeta concilii; DSM 800, Methanosarcina barkeri. All pure cultures from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany.
Clone library construction and analysis.
A total of four clone libraries were constructed from the DNA extracted from the fresh (F) and mature (M) sludges. DNA amplified with both the bacterial (-B) and archaeal (-A) primers was subjected to cloning with the TOPO TA cloning kit using One Shot TOP10 competent cells (Invitrogen). To analyze the clones for inserts, thermal cycling was performed in 20-μl volumes, with each reaction mixture containing 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 0.01% Tween 20], 200 μM each dNTP, 0.2 μM T3 (5′-ATTAACCCTCACTAAAGGGA) and T7 (5′-TAATACGACTCACTATAGGG) primers, 0.45 U BioTherm DNA polymerase (GeneCraft, Germany), and sterile water. A small amount of the colony was directly added to the PCR mix. The initial denaturation (94°C, 5 min) was followed by 30 cycles of amplification (50 s at 94°C, 50 s at 50°C, and 2 min at 72°C) and a final extension (10 min at 72°C).
DNA from insert-containing clones was analyzed by restriction digestion analysis. Ten microliters of PCR product was digested in a 10-μl reaction mixture containing 0.5 U HaeIII, 1× Tango buffer (Fermentas, Germany), and sterile water. Products digested at 37°C for 3 h were separated in 2.5% agarose gels (1× Tris-borate-EDTA buffer, 50 V, 60 min) and stained (0.1% [vol/vol] ethidium bromide). Analysis of the restriction digestion patterns was carried out with GelCompar 3.1 (Applied Maths, Belgium). Ward's method was applied to calculate dendrograms, after which the phylotypes (i.e., clones with the same restriction digestion patterns) were defined.
Sequencing and phylogenetic analysis.
DNA from at least one representative of each phylotype was amplified as described above. The DNA was purified with the NucleoSpin Extract II kit (Macherey-Nagel, Germany), and forward and reverse sequencing reactions were performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). PCR products were analyzed using a 3130 Genetic Analyzer (Applied Biosystems) at the BioSeq facilities of the University of Innsbruck, Innsbruck, Austria. Full-length sequences were NAST-aligned with Greengenes (7) and checked for anomalies using Bellephoron (15). The phylogenetic affiliation of error-free sequences was determined with the ARB software package (22). Neighbor-joining trees of nearly complete 16S rRNA gene bacterial and partial 16S rRNA gene archaeal sequences and their corresponding similarity matrices were generated by the neighbor-joining distance method. Operational taxonomic units (OTUs) were defined as phylotypes having 16S rRNA gene sequences with ≥97% identity. Bootstrap values were calculated based on 1,000 replications.
Quantitative RT-PCR.
Sludge DNA was subjected to real-time PCR (RT-PCR) amplification with specific primers for several groups of methanogens (Table 2) as described in reference 10. Amplifications were conducted using the Quantimix Easy SYG kit (Biotools, Spain) in a Rotor-Gene 6000 (Corbett Life Sciences, Australia) in 20-μl volumes including 2 μl sludge DNA (1/70 diluted). Standard curves were constructed with PCR-amplified 16S rRNA genes using the source DNA and methanogen-specific primers in Table 2. Spiking experiments were performed to check for PCR efficiency reduction due to the presence of inhibitors in the sludge matrix. The amplification efficiency of the unspiked standard curves was similar to that of the standard curves spiked with sludge DNA (data not shown). Thus, the effect of inhibitors in the sludge DNA was considered to be negligible.
Bacterial community structure.
Bacterial clone libraries were constructed from the fresh and mature sludges (F-B and M-B), and 400 clones were screened. The number of phylotypes indicated a higher bacterial diversity in the mature sludge, with 46 phylotypes detected in F-B and 70 in M-B (Table 3). However, the numbers of OTUs were similar: 31 and 37 OTUs were defined in F-B and M-B, respectively, 18 of which were common to both libraries. Firmicutes (97.7%), Bacteroidetes (1.3%) and Thermotogae (1.0%) were the bacterial phyla detected in the bioreactor (Fig. 1). The same phyla have been reported to dominate a lab-scale thermophilic reactor fed with organic household waste (20).
TABLE 3.
Distribution of clones in the fresh and mature sludge bacterial clone libraries
| Putative division | No. of clones (no. of phylotypes)
|
Closest cultured species or environmental clone (accession no.)a | % Sequence identity | |
|---|---|---|---|---|
| F-B | M-B | |||
| Firmicutes | ||||
| Clostridiales | 69 (9) | 82 (15) | Clone TIBH10 (DQ887962.1) | 90-99 |
| 1 (1) | 11 (3) | Clone MBA02 (AB114312.1) | 98-99 | |
| 19 (6) | 12 (5) | Clone GMB4D08 (DQ887933.1) | 90-99 | |
| 0 | 4 (3) | Clone GMB4H01 (DQ887946.1) | 88-97 | |
| 2 (1) | 2 (2) | Clone MRE50b01 (AY684075.1) | 92 | |
| 9 (2) | 10 (4) | Clone GMB4C10 (DQ887942.1) | 98 | |
| 2 (1) | 7 (4) | Clone TIBA10 (DQ887956.1) | 93-98 | |
| 11 (4) | 4 (4) | Carboxydocella ferrireducens (EF092457.1) | 83-87 | |
| 4 (3) | 1 (1) | Catabacter hongkongensis (AY574991.1) | 86-88 | |
| 9 (2) | 6 (2) | Tepidimicrobium quinonicus (AY656718.1) | 93-94 | |
| 6 (2) | 6 (3) | Clostridium cellulolyticum (X71847.1) | 91 | |
| 6 (1) | 1 (1) | Clostridium piliforme (D14639.1) | 86 | |
| 1 (1) | 0 | Clostridium stercorarium (AJ310082.1) | 90 | |
| 0 | 2 (1) | Clostridium ultunense (Z69293.1) | 94 | |
| 0 | 1 (1) | Syntrophomonas erecta (AY536889.1) | 90 | |
| 0 | 1 (1) | Syntrophomonas wolfei (M26492.1) | 94 | |
| 0 | 1 (1) | Pelotomaculum isophthalicicum (AB232785.1) | 94 | |
| Halanaerobiales | 40 (6) | 30 (9) | Halocella cellulosilytica (X89072.1) | 91-92 |
| Bacillales | 9 (3) | 4 (4) | Geobacillus caldoxylosilyticus (AF067651.1) | 82-87 |
| Thermotogae | 3 (2) | 1 (1) | Petrotoga mobilis (Y15479.1) | 77-91 |
| Bacteroidetes | 2 (2) | 2 (2) | Bacteroides uniformis (AB050110.1) | 84 |
| 0 | 1 (1) | Clone GMB4C12 (DQ887931.1) | 99 | |
Environmental clones are given for sequences not closely related to any cultured species.
FIG. 1.
Neighbor-joining tree of partial 16S rRNA gene sequences depicting the phylogenetic relationships of 16S rRNA gene bacterial clones from fresh (F) and mature (M) sludge DNA. Numbers at nodes represent bootstrap values based on 1,000 replications. Escherichia coli was used as the outgroup of the tree. Sequences from this study are shown in bold.
The class Clostridia dominated both clone libraries (92.7% in F-B and 95.8% in M-B), a finding reported previously for cellulolytic environments (5). Among these, clones being most closely related to several groups of uncultured Clostridia were the most abundant (52.8% and 67.7% in F-B and M-B). Some of these uncultured Clostridia have been previously detected in thermophilic anaerobic bioreactors (8). The ecophysiological function of these organisms is not certain, but often Clostridium spp. show hydrolytic fermentative (5) and acetate-oxidizing activity (17). Also abundant in both clone libraries were Clostridia most closely related to Halocella cellulosilytica (20.7% and 15.9% in F-B and M-B), a halophilic cellulolytic organism (29). Therefore, halophilic and halotolerant bacteria able to thrive at the high salt concentrations prevailing in the fermenter (20 g liter−1), particularly those of ammonium (2.7 g liter−1), dominated the reactor ecosystem.
It can be concluded that bacterial diversity, based on the number of phylotypes in the clone libraries, increased with the maturation of wastes, but phylogenetically the dominant communities remained mostly unchanged during the process of anaerobic digestion. Presumably, the environmental filter imposed by the saline environment in the biowaste-degrading fermenter selected a phylogenetically stable community dominated by groups physiologically adapted to osmotic stress. Under such conditions, the significant changes detected in other chemical parameters, such as pH or VFAs, did not provoke a major shift in the phylogenetic groups of bacteria. The selective filter imposed by the saline environment would also explain the constant denaturing gradient gel electrophoresis patterns detected in the mature sludge of the same reactor over a whole year of unstable performance (23). Our results contradict previous reports describing bacterial populations as very dynamic under pH-changing conditions (13) as well as in stably performing reactors (9). However, in those studies, lab-scale reactors fed with synthetic wastewater containing glucose as the only carbon and energy source were used, eliminating the possible influence of ammonia on the dynamics.
Finally, a set of singletons appeared in the mature sludge (M-B) that were closely related to syntrophic bacteria, namely, Clostridium ultunense (26), Syntrophomonas sp. (24, 31), and Pelotomaculum sp. (25). No syntrophs were detected in the fresh sludge, whereas these constituted 2.5% of the bacterial community in the mature sludge. Interestingly, no propionate-oxidizing bacteria were detected, and, indeed propionate concentrations did not decrease during the process; thus, propionate was presumably not degraded. It is possible that the anaerobic digestion period was too short to allow the proliferation of propionate degraders (5). Alternatively, high ammonia levels could have inhibited the growth of these syntrophs (3).
Archaeal community structure.
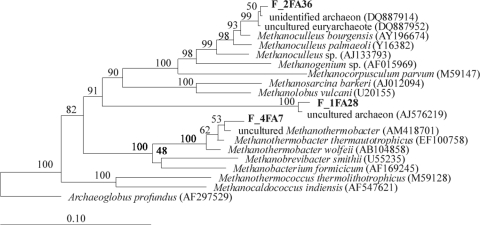
Two archaeal clone libraries were constructed from the fresh and mature sludges (F-A and M-A). After screening 400 clones, four phylotypes were detected in F-A and three in M-A. These corresponded to three OTUs in F-A, of which only one persisted in M-A. This low archaeal diversity has been previously reported in thermophilic reactors (13, 30). Both clone libraries were dominated by organisms related to Methanoculleus bourgensis (Table 4 and Fig. 2), a hydrogenotrophic methanogen (11). Its frequency increased from 90.8% in F-A to 100% in M-A. In F-A, other clones appeared that were most closely related to clone GZK24, detected in the leachate of a municipal solid waste landfill (14), or to the hydrogenotroph Methanothermobacter thermautotrophicus (11). Therefore, there was a decrease in the archaeal clonal diversity following maturation of the biowastes, with a total disappearance of methanogens other than Methanoculleus sp.
TABLE 4.
Distribution of clones in the fresh and mature sludge archaeal clone libraries
| Putative division | No. of clones (no. of phylotypes)
|
Closest cultured species or environmental clone (accession no.)a | % Sequence identity | |
|---|---|---|---|---|
| F-A | M-A | |||
| Methanomicrobiales | 177 (2) | 201 (3) | Methanoculleus bourgensis (AY196674) | 96-98 |
| Methanobacteriales | 1 (1) | 0 | Methanothermobacter thermautotrophicus (EF100758) | 98 |
| Uncertain | 17 (1) | 0 | Clone GZK24 (AJ576219) | 99 |
Environmental clones are given for sequences not closely related to any cultured species.
FIG. 2.
Neighbor-joining tree of partial 16S rRNA gene sequences depicting the phylogenetic relationships of 16S rRNA gene archaeal clones from fresh (F) and mature (M) sludge DNA. Numbers at nodes represent bootstrap values based on 1,000 replications. Archaeoglobus profundus was used as the outgroup of the tree. Sequences from this study are shown in bold.
RT-PCR confirmed the dominance of Methanoculleus (>98%) and the significant increase of its abundance with the maturation of wastes (Table 5). However, the group of uncultured archaeons and Methanothermobacter could be amplified from both sludge types. The lack of detection of these organisms in the clone library constructed from the mature sludge could be due to the low probability of sampling them randomly among all PCR-generated products because of their low abundance.
TABLE 5.
16S rRNA gene copy numbers for the groups of methanogens quantified by RT-PCR in the fresh and mature sludges
| Target | 16S rRNA gene copy no. (copies g−1 sludge) ina:
|
|
|---|---|---|
| Fresh sludge | Mature sludge | |
| Methanoculleus | 3.9 × 108 (1.2 × 108) | 9.0 × 108 (2.2 × 108)* |
| Uncultured group | 5.7 × 106 (1.3 × 106)* | 2.3 × 106 (4.3 × 105) |
| Methanothermobacter | 1.6 × 106 (6.0 × 105) | 1.4 × 106 (7.4 × 105) |
| Methanosaeta | ND | ND |
| Methanosarcina | ND | ND |
Standard deviations are given in parentheses (n = 5). Asterisks indicate significant differences at P < 0.05, after Student's t test with SPSS 15.0. ND, not detected (<600 gene copies g−1 sludge).
RT-PCR also confirmed the absence in the samples of the two genera of known acetotrophic methanogens (Methanosarcina and Methanosaeta). This indicates that the syntrophic oxidation of acetate was the only pathway for methanogenesis from biowastes. Acetate concentrations above 1 mM, as was the case here, should be enough to support an acetoclastic community of methanogens (13). On the other hand, VFAs in the reactor were not above the reported inhibitory concentrations for acetotrophic methanogens (27). Therefore, the most plausible explanation for the inhibition of the acetotrophic pathway is the high concentration of salts, particularly those of ammonium (2.7 g liter−1) (27). Also the prevalence of Methanoculleus over other hydrogenotrophic methanogens might be related to its tolerance to high salt concentrations (27).
In conclusion, the prokaryotic phylogenetic groups in the anaerobic sludge of a thermophilic plug-flow reactor remained relatively unaltered despite changes in the environmental conditions concurrent with the maturation of the biowastes. We demonstrated that biogas production was based on syntrophic acetate oxidation by a community of organisms able to thrive at the high salt levels prevailing in the reactor. This environmental factor presumably determined the minor shift in the phylogenetic groups between the fresh and mature sludges, despite significant changes in pH and VFAs, which have been shown to be influential for anaerobic community dynamics in previous studies. Whether the saline conditions impose a selective filter that determines phylogenetic stability in anaerobic reactors should be further investigated.
Nucleotide sequence accession numbers.
The EMBL Nucleotide Sequence Database accession numbers of the 16S rRNA gene sequences obtained in this study are AM947508 to AM947560.
Acknowledgments
We thank the staff of the biogas plant, Roppen, Austria, particularly G. Gstraunthaler, for providing us with basic data about the reactor. D. Sperl and A. Wagner helped us with the batch incubation experiment and high-pressure liquid chromatography measurements. L. Strömbom made very helpful comments on RT-PCR. Critical comments by three anonymous reviewers are gratefully acknowledged.
M. Goberna was supported by Ministerio de Educación y Ciencia (EX-2006-0094) and Marie Curie Actions (MEIF-CT-2006-041034). I. Franke-Whittle was supported by Fonds zur Förderung der wissenschaftlichen Forschung (FWF-FP200010).
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Aller, J. Y., and P. F. Kemp. 2008. Are Archaea inherently less diverse than Bacteria in the same environments? FEMS Microbiol. Ecol. 65:74-87. [DOI] [PubMed] [Google Scholar]
- 2.Briones, A., and L. Raskin. 2003. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14:270-276. [DOI] [PubMed] [Google Scholar]
- 3.Calli, B., B. Mertoglu, B. Inanc, and O. Yenigun. 2005. Methanogenic diversity in anaerobic reactors under extremely high ammonia levels. Enzyme Microb. Technol. 37:448-455. [Google Scholar]
- 4.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 5.Chynoweth, D. P., and P. Pullammanappallil. 1996. Anaerobic digestion of municipal solid wastes, p. 71-114. In A. C. Palmisano and M. A. Barlaz (ed.), Microbiology of solid wastes. CRC Press, Boca Raton, FL.
- 6.Collins, G., S. Kavanagh, S. McHugh, S. Connaughton, A. Kearney, O. Rice, C. Carrig, C. Scully, N. Bhreathnach, T. Mahony, P. Madden, A. M. Enright, and V. O′Flaherty. 2006. Accessing the black box of microbial diversity and ecophysiology: recent advances through polyphasic experiments. J. Environ. Sci. Health A 41:897-922. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkel, C., D. Kemnitz, M. Kube, P. Ricke, K. J. Chin, S. Dedysh, R. Reinhardt, R. Conrad, and W. Liesack. 2005. Retrieval of first genome data for rice cluster I methanogens by a combination of cultivation and molecular techniques. FEMS Microbiol. Ecol. 53:187-204. [DOI] [PubMed] [Google Scholar]
- 9.Fernández, A., S. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke-Whittle, I. H., M. Goberna, and H. Insam. Design and testing of real-time PCR primers for the quantification of Methanoculleus, Methanosarcina, Methanothermobacter and a group of uncultured methanogens. Can. J. Microbiol., in press. [DOI] [PubMed]
- 11.García, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 12.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hori, T., S. Haruta, Y. Ueno, M. Ishii, and Y. Igarashi. 2006. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl. Environ. Microbiol. 72:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, L. N., Y. Q. Chen, H. Zhou, S. Luo, C. Y. Lan, and L. H. Qu. 2003. Characterization of methanogenic Archaea in the leachate of a closed municipal solid waste landfill. FEMS Microbiol. Ecol. 46:171-177. [DOI] [PubMed] [Google Scholar]
- 15.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 16.Karakashev, D., D. J. Batstone, and I. Angelidaki. 2005. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microbiol. 71:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karakashev, D., D. J. Batstone, E. Trably, and I. Angelidaki. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 72:5138-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaPara, T. M., C. H. Nakatsu, L. M. Pantea, and J. E. Alleman. 2002. Stability of the bacterial communities supported by a seven-stage biological process treating pharmaceutical wastewater as revealed by PCR-DGGE. Water Res. 36:638-646. [DOI] [PubMed] [Google Scholar]
- 19.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 20.Levén, L., A. R. B. Eriksson, and A. Schnürer. 2007. Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol. Ecol. 59:683-693. [DOI] [PubMed] [Google Scholar]
- 21.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malin, C., and P. Illmer. 2008. Ability of DNA content and DGGE analysis to reflect the performance condition of an anaerobic biowaste fermenter. Microbiol. Res. 163:503-511. [DOI] [PubMed] [Google Scholar]
- 24.McInerney, M. J., M. P. Bryant, R. B. Hespell, and J. W. Costerton. 1981. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl. Environ. Microbiol. 41:1029-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu, Y.-L., Y. Sekiguchi, S. Hanada, H. Imachi, I.-C. Tseng, S.-S. Cheng, A. Ohashi, H. Harada, and Y. Kamagata. 2006. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185:172-182. [DOI] [PubMed] [Google Scholar]
- 26.Schnürer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 27.Schnürer, A., G. Zellner, and B. H. Svensson. 1999. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 29:249-261. [Google Scholar]
- 28.Shigematsu, T., Y. Tang, H. Kawaguchi, K. Ninomiya, J. Kijima, T. Kobayashi, S. Morimura, and K. Kida. 2003. Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J. Biosci. Bioeng. 96:547-558. [DOI] [PubMed] [Google Scholar]
- 29.Simankova, M. V., N. A. Chernych, G. A. Osipov, and G. A. Zavarzin. 1993. Halocella cellulolytica gen. nov., sp. nov., a new obligately anaerobic, halophilic, cellulolytic bacterium. Syst. Appl. Microbiol. 16:385-389. [Google Scholar]
- 30.Tang, Y., T. Shigematsu, Ikbal, S. Morimura, and K. Kida. 2004. The effects of micro-aeration on the phylogenic diversity of microorganisms in a thermophilic anaerobic municipal solid-waste digester. Water Res. 38:2537-2550. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, C., X. Liu, and X. Dong. 2005. Syntrophomonas erecta sp. nov., a novel anaerobe that syntrophically degrades short-chain fatty acids. Int. J. Syst. Evol. Microbiol. 55:799-803. [DOI] [PubMed] [Google Scholar]
- 32.Zumstein, E., R. Moletta, and J. J. Godon. 2000. Examination of two years of community dynamics in an anaerobic bioreactor using fluorescence polymerase chain reaction (PCR) single-strand conformation polymorphism analysis. Environ. Microbiol. 2:69-78. [DOI] [PubMed] [Google Scholar]