Abstract
In industrial fermentation processes, the yeast Saccharomyces cerevisiae is commonly used for ethanol production. However, it lacks the ability to ferment pentose sugars like d-xylose and l-arabinose. Heterologous expression of a xylose isomerase (XI) would enable yeast cells to metabolize xylose. However, many attempts to express a prokaryotic XI with high activity in S. cerevisiae have failed so far. We have screened nucleic acid databases for sequences encoding putative XIs and finally were able to clone and successfully express a highly active new kind of XI from the anaerobic bacterium Clostridium phytofermentans in S. cerevisiae. Heterologous expression of this enzyme confers on the yeast cells the ability to metabolize d-xylose and to use it as the sole carbon and energy source. The new enzyme has low sequence similarities to the XIs from Piromyces sp. strain E2 and Thermus thermophilus, which were the only two XIs previously functionally expressed in S. cerevisiae. The activity and kinetic parameters of the new enzyme are comparable to those of the Piromyces XI. Importantly, the new enzyme is far less inhibited by xylitol, which accrues as a side product during xylose fermentation. Furthermore, expression of the gene could be improved by adapting its codon usage to that of the highly expressed glycolytic genes of S. cerevisiae. Expression of the bacterial XI in an industrially employed yeast strain enabled it to grow on xylose and to ferment xylose to ethanol. Thus, our findings provide an excellent starting point for further improvement of xylose fermentation in industrial yeast strains.
It is widely acknowledged that fuels from regenerative resources are becoming increasingly important in times of a dwindling crude oil supply and the growing environmental concern of the public. Plant biomass, particularly when accruing as a waste product, is an attractive feedstock for bioethanol production. An important prerequisite for such an alternative strategy would be the complete conversion of all available sugars in biomass hydrolysates into ethanol. While the hexose sugars are easily fermentable, no suitable microorganism is available for fermenting pentose into ethanol. Calculations have resulted in an estimate that production of lignocellulosic ethanol would reduce the cost of producing ethanol by nearly 20% (3). Therefore, ethanol production from pentose sugars has received considerable attention (4, 9).
Although some anaerobic fungi and bacteria are able to metabolize xylose, they are not suitable for industrial bioethanol production due to low and inefficient production rates and the mixed acid fermentation life-style (28), which generates too many by-products. The baker's yeast Saccharomyces cerevisiae remains the organism of choice for industrial production of ethanol. However, while hexoses are converted rapidly to high yields of ethanol, wild-type S. cerevisiae strains are not able to ferment pentose sugars, such as d-xylose and l-arabinose, efficiently. Several different approaches in genetic engineering have been used to enable d-xylose fermentation in yeast.
Successful xylose fermentation in recombinant S. cerevisiae strains was previously achieved by heterologous expression of the XYL1 and XYL2 genes (encoding xylose reductase [XR] and xylitol dehydrogenase [XDH], respectively) from Pichia stipitis (8, 12, 14, 15) or by expression of a xylA gene (encoding xylose isomerase [XI]) from Piromyces sp. strain E2 (17) or Thermus thermophilus (33). Both approaches resulted in strains growing on xylose and fermenting it into ethanol. Although expression of XR and XDH resulted in rapid fermentation of xylose, NADPH/NAD cofactor imbalance under anaerobic conditions led to considerable accumulation of xylitol (6, 14, 15, 30, 32). However, employing XI instead of XR/XDH avoids cofactor imbalance and xylitol accumulation, as d-xylose is converted directly into d-xylulose without a redox reaction being involved.
Many attempts to express an active prokaryotic XI in S. cerevisiae have failed. None of the efforts to express XI from Escherichia coli (25), Bacillus subtilis (2), Lactobacillus pentosus (10), or Clostridium thermosulfurogenes (23) in S. cerevisiae resulted in active XI, arguing for the inability of yeast either to express xylA or to synthesize active enzyme (25). The first successful attempt was made with the xylA gene from the thermophilic bacterium Thermus thermophilus. XI from T. thermophilus could be expressed in S. cerevisiae in an active form, but the activity of this thermophilic enzyme, with a temperature optimum at 85°C, was very low at 30°C (33). In subsequent rounds of mutagenesis, the enzyme could be considerably improved but, however, still not enough for efficient xylose conversion in yeast (22).
For the first time, Kuyper et al. (17) successfully expressed a xylA gene from the anaerobic fungus Piromyces sp. strain E2 in S. cerevisiae with high enzymatic activity. However, a drawback of this enzyme was its strong inhibition by xylitol. A laboratory haploid yeast strain which exhibited fast anaerobic growth on d-xylose and also high ethanol production rates was constructed (18, 20). Furthermore, mixed sugar utilization of d-glucose and d-xylose could recently be achieved by evolutionary engineering of recombinant yeast strains (19).
In this paper, we report the cloning and successful expression of the first XI of prokaryotic origin with high activity in S. cerevisiae. As an advantage, the new enzyme is far less susceptible to inhibition by xylitol than is the enzyme from the Piromyces strain.
MATERIALS AND METHODS
Strains and media.
Yeast strains and plasmids used in this work are listed in Table 1. S. cerevisiae was grown aerobically in synthetic complete (SC) medium (6.7 g liter−1 Difco yeast nitrogen base without amino acids), supplemented with amino acids as described previously (38), supplemented with 20 g liter−1 d-glucose or 20 g liter−1 d-xylose as a carbon source, and buffered at pH 5.5 with 20 mM KH2PO4. In case the sugar concentration exceeded 20 g liter−1 in the medium, the concentration of yeast nitrogen base was doubled. For maintenance of plasmids, media lacked uracil or contained 200 mg/liter G418 (Calbiochem) for selection in industrial strains. Screening for functional XIs was conducted on the same solid medium with 18 g liter−1 agar but with 20 g liter−1 d-xylose instead of glucose as the sole carbon source. In anaerobic fermentations, defined medium (31) was used, supplemented with amino acids as described previously (38) (30 g liter−1 d-xylose, 200 mg/liter G418, and 150 μl liter−1 of silicone antifoam [Sigma]) as well as with the anaerobic growth factors ergosterol (0.01 g liter−1) and Tween 80 (0.40 g liter−1) dissolved in ethanol (resulting in 2.5 g liter−1 ethanol in the medium).
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/phenotype | Source or reference |
|---|---|---|
| S. cerevisiae strains | ||
| CEN.PK2-1C | MATaleu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC2 | K. D. Entian, Frankfurt, Germany |
| MKY9 | (MATaleu2-3,112 ura3-52 trp1-289 his3-Δ1MAL2-8cSUC2 PromTKL1::loxP-Prom-vkHXT7 PromRPE1::loxP-Prom-vkHXT7 PromRKI1::loxP-Prom-vkHXT7 PromGAL2::loxP-Prom-vkHXT7 PromXKS1::loxP-Prom-vkHXT7) + unknown beneficial mutations for pentose growth | Boles lab stock |
| BarraGrande | Industrial strain for bioethanol production | Brazilian ethanol plant |
| BWY10Xyl | BarraGrande with plasmid YEp-opt.XI-Clos-K evolved on xylose growth | This work |
| Plasmids | ||
| p426H7 | URA3 | 11 |
| YEp-XI-Agro | pHXT7; xylA from A. tumefaciens; tCYC1 URA3 | This work |
| YEp-XI-Arab | pHXT7; xylA from A. tumefaciens; tCYC1 URA3 | This work |
| YEp-XI-BaLi | pHXT7; xylA from B. licheniformis; tCYC1 URA3 | This work |
| YEp-XI-Burk | pHXT7; xylA from B. xenovorans; tCYC1 URA3 | This work |
| YEp-XI-Clos | pHXT7; xylA from C. phytofermentans; tCYC1 URA3 | This work |
| YEp-opt.XI-Clos | pHXT7; codon-optimized xylA from C. phytofermentans; tCYC1 URA3 | This work |
| YEp-XI-Lacto | pHXT7; xylA from L. pentosus; tCYC1 URA3 | This work |
| YEp-opt.XI-Piro | pHXT7; codon-optimized xylA from Piromyces sp. strain E2; tCYC1 URA3 | Boles lab stock |
| YEp-XI-Pseudo | pHXT7; xylA from Pseudomonas syringae; tCYC1 URA3 | This work |
| YEp-XI-Robi | pHXT7; xylA from R. biformata; tCYC1 URA3 | This work |
| YEp-XI-Saccha | pHXT7; xylA from S. degradans; tCYC1 URA3 | This work |
| YEp-XI-Salmo | pHXT7; xylA from S. enterica serovar Typhimurium; tCYC1 URA3 | This work |
| YEp-XI-Staph | pHXT7; xylA from S. xylosus; tCYC1 URA3 | This work |
| YEp-XI-Xantho | pHXT7; xylA from X. campestris; tCYC1 URA3 | This work |
| YEp-opt.XI-Clos-K | pHXT7; codon-optimized xylA from C. phytofermentans; tCYC1 URA3 loxP-kanMX-loxP FAA2 locus | This work |
Bacillus licheniformis (DSM 13), Burkholderia xenovorans (DSM 17367), Clostridium phytofermentans (DSM 18823), Lactobacillus pentosus (DSM 20314), Leifsonia xyli subsp. cynodontis (DSM 46306), Pseudomonas savastanoi pv. phaseolicola (DSM 50282), Robiginitalea biformata (DSM 15991), Saccharophagus degradans (DSM 17024), Staphylococcus xylosus (DSM 20266), Streptomyces diastaticus subsp. diastaticus (DSM 40496) and Xanthomonas campestris pv. campestris (DSM 3586) were cultivated according to the recommendations of the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) (http://www.dsmz.de/) in DSM media 1, 220, 520, 11, 751, 54, 515, 514a, 53, 65, and 1, respectively. Salmonella enterica serovar Typhimurium (71-098L) (obtained from Molecular Toxicology Incorporated) was cultivated in Luria-Bertani (LB) medium.
Plasmids were propagated in Escherichia coli SURE (Stratagene, La Jolla, CA) grown on LB medium with 40 μg ml−1 ampicillin. E. coli was transformed via electroporation according to the methods of Dower et al. (7) and Wirth (36).
Preparation of genomic DNA.
Bacterial DNA was prepared as described in reference 24; alternatively, PCRs were performed using broken cells as templates. Genomic DNA from Agrobacterium tumefaciens and cDNA from Arabidopsis thaliana were kind gifts from C. Weber, Frankfurt, Germany.
Plasmid construction.
The coding regions of the xylA genes encoding XIs from various organisms were amplified by PCR from genomic DNA or cDNA from the strains listed under “Strains and media” and cloned into the EcoRI/BamHI-linearized vector p426H7 (URA3) by recombination cloning employing the methods described by Wieczorke et al. (34) but omitting the six histidine codons. The open reading frames were amplified by using the specific primer pairs x-for and x-rev (where x is specific for the organism [Table 2]). Vector YEp-opt.XI-Clos-K is based on vector p426H7 and was generated by cloning the dominant selection marker gene kanMX, amplified by PCR using primer pair FAA2-kanMX-f/FAA2-kanMX-r, into SacI/KpnI-linearized vector p426H7. The kanMX gene was flanked by sequences homologous to sequences in the genome of yeast near the FAA2 gene obtained by PCR using primer pair FAA2-1-f/FAA2-1-r and FAA2-2-f/FAA2-2-r. Homologous regions should enable later genomic integration of the construct. 5′ of the kanMX gene, the codon-optimized gene version of xylA from C. phytofermentans, obtained by PCR with primers FAA2-optXI-Clos-f and FAA2-optXI-Clos-r, was inserted into PmeI-linearized vector, resulting in YEp-opt.XI-Clos-K.
TABLE 2.
Primers used in this work
| Primer | Sequence |
|---|---|
| XI-Agro-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAAATTTTCAAACACCACCC |
| XI-Agro-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAAACGTAACGATTGACCACG |
| XI-Arab-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAAGAAAGTTGAGTTTTTTATGC |
| XI-Arab-2rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTACATTGCAGATTGGAAAATC |
| XI-BaLi-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGTTTTTTAGAAATATCGGAATG |
| XI-BaLi-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGCTAGTGTGTCTCCTTTCCTGCCG |
| XI-Burk-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGTCCTATTTCGAACACATTCCCG |
| XI-Burk-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAGCGTCCGCTGTAAATAGCC |
| XI-Clos-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAAAAATTACTTTCCAAATG |
| XI-Clos-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTATCTAAATAAAATATTATTTACG |
| XI-Lacto-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGACAAACGAATATTGGCAAGG |
| XI-Lacto-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTATTTACTTAACGTCTCGATAAT |
| XI-Leif-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGCTGGCTCGAGCGCAGGCTCC |
| XI-Leif-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAGCGCGCGCCGAGCAGATGC |
| XI-Pseudo-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGCCGTACTTCCCCGCCGTCG |
| XI-Pseudo-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAGAAGTAGATAAAGCGGTTGACC |
| XI-Robi-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGATTACCACCGGAGACAAAG |
| XI-Robi-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAGATAAATTGGTTGATAATG |
| XI-Saccha-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAGCGTAGTACTTGGCGATAAAG |
| XI-Saccha-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTAACGAATATATTGGTTAATAATG |
| XI-Salmo-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGCAGGCTTATTTTGACCAACTCG |
| XI-Salmo-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTATTTATCAAACAGATAACGGTTAAC |
| XI-Staph-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGTCTTATTTTGATATCAATAAAG |
| XI-Staph-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAATCCTTATTATTAATATTTAAG |
| XI-Strep-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAGCTACCAGCCCACCCCCG |
| XI-Strep-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGCTAGCCCCGCGCGCCCAGCAGG |
| XI-Xantho-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAGCAACACCGTTTTCATCGGC |
| XI-Xantho-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTCAACGCGTCAGGTACTGATTGATC |
| opt.XI-Clos-for | CAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGAAGAACTACTTCCCAAACG |
| opt.XI-Clos-rev | GTAAGCGTGACATAACTAATTACATGACTCGAGTTATCTGAACAAAATGTTGTTAAC |
| opt.XI-Piro-for | AACACAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAATGGCTAAGGAATACTTCCCA |
| opt.XI-Piro-rev | GAATGTAAGCGTGACATAACTAATTACATGACTCGAGTTATTGGTACATAGCAACAAT |
| FAA2-1-f | CAAGCGCGCAATTAACCCTCACTAAAGGGAACAAAAGCTGTACTTATGACGATTTGGAAC |
| FAA2-1-r | ATATCAGGGCTCACTACATG |
| FAA2-2-f | CGGGGCGTAATCACCTAACTC |
| FAA2-2-r | CAGTCACGACGTTGTAAAACGACGGCCAGTGAGCGCGCGTCTAATGGCGATTTAGCATATG |
| FAA2-kanMX-f | GATGGAGGTGCAGTGTTTTCCATGTAGTGAGCCCTGATATGTTTAAACTTCGTACGCTGCAGGTCGAC |
| FAA2-kanMX-r | GAGAAAGGTTTACCTTTGCTGAGTTAGGTGATTACGCCCCGGCATAGGCCACTAGTGGATCTG |
| FAA2-optXI-Clos-f | CAGATGGAGGTGCAGTGTTTTCCATGTAGTGAGCCCTGATATGTTTAAACGAGCTCGTAGGAACAATTTC |
| FAA2-optXI-Clos-r | ATTAAGGGTTGTCGACCTGCAGCGTACGAAGCTTCAGCGGCGAATTGGGTACCGGCCG |
Codon-optimized gene versions were obtained from Geneart AG (Regensburg, Germany) after changing the original codons of the respective genes to those used in the genes encoding glycolytic enzymes in S. cerevisiae as described in the work of Wiedemann and Boles (35) and cloned into the vector p426H7 by recombination cloning as described above. Using the coding region of C. phytofermentans xylA and Piromyces sp. xylA resulted in YEp-opt.XI-Clos and YEp-opt.XI-Piro, respectively.
Phylogenetic analysis.
Amino acid sequences of the XIs were obtained from GenBank and compared using the BLAST algorithm (National Center for Biotechnology Information). Sequences were aligned to plot the phylogenetic tree, using MEGA version 4 (27).
Growth assays (shake flasks).
Cultures of laboratory strains (50 ml) were grown in 500-ml shake flasks (Erlenmeyer flasks) at 30°C in a shaker. Precultures were grown into the stationary phase in SC medium lacking uracil and containing 20 g liter−1 d-xylose as the sole carbon and energy source. Cells were washed with sterile water and inoculated to an optical density at 600 nm (OD600) of 0.5 in the same medium. Growth experiments were performed in triplicate with the given standard deviations, but cultures were started from the same precultures. Cultures of industrial strain BWY10Xyl (20 ml) were grown in 200-ml Erlenmeyer flasks at 30°C in a shaker. Precultures were grown to stationary phase in SC medium containing 20 g liter−1 d-xylose as the sole carbon and energy source. For maintenance of plasmids, media were made with 200 mg/liter G418 (21). Cells were washed with sterile water and used to inoculate the same medium to an OD600 of 0.2. Growth experiments were performed in triplicate with the given standard deviations.
Selection of industrial strain growing on d-xylose.
Mutants of BWY10Xyl able to grow on d-xylose were selected by serial transfer in shake flasks. For serial transfer experiments, a 200-ml shake flask containing 20 ml of SC medium supplemented with 20 g liter−1 d-xylose, 1 g liter−1 yeast extract, 2 g liter−1 peptone, and 200 mg/liter G418 was inoculated with strain BarraGrande containing plasmid YEp-opt.XI-Clos-K. This transfer procedure was repeated four times, covering a period of 28 days. Another two transfers in medium containing only 20 g liter−1 d-xylose as the sole carbon and energy source resulted in strain BWY10Xyl. From the final culture, a sample was streaked out on mineral medium with xylose. Single colonies were picked and restreaked on identical medium. From these plates, single colonies were taken and used for further growth and fermentation experiments.
Metabolite analysis.
The concentrations of glucose, d-xylose, xylitol, glycerol, acetic acid, and ethanol were determined by high-performance liquid chromatography (Dionex) using a Nugleogel Sugar 810 H exchange column (Macherey-Nagel GmbH & Co, Germany). The column was eluted with 5 mM H2SO4 as mobile phase and a flow rate of 0.6 ml min−1 at the temperature of 65°C. Detection was by means of a Shodex RI-101 refractive index detector. For data evaluation, Chromeleon software (version 6.50) was used. Rates of d-xylose consumption were determined in the phase of d-xylose growth.
Determination of culture dry weight.
Dry weight was determined (in duplicate) by filtering 10 ml of the culture through a preweighed nitrocellulose filter (0.45-μm pore size; Roth, Germany). The filters were washed with demineralized water, dried in a microwave oven for 20 min at 140 W, and weighed again.
Anaerobic batch fermentations.
Anaerobic batch fermentations were performed in Minifors bioreactors with a working volume of 2 liters (Infors AG, Bottmingen, Switzerland). Shake-flask precultures were grown until late exponential phase in SC medium supplemented with 200 mg/liter G418 and with 20 g liter−1 d-xylose. Cells were washed with sterile water. Cultures were inoculated at an OD600 of about 0.6 and incubated at 30°C with 250-rpm stirring and at pH 5.5, maintained by addition of 4 M KOH. The synthetic medium contained 30 g liter−1 d-xylose. Cells were grown under aerobic conditions until about 5 g liter−1 d-xylose was consumed and then shifted to anaerobic conditions by sparging with nitrogen gas (containing less than 5 ppm of O2; Air Liquide, Düsseldorf, Germany) for 30 min with a flow rate of 1 liter min−1. Evaporation of ethanol was minimized by using a reflux condenser at 4°C and was not calculated. The experiment was performed in duplicate.
Enzyme assays.
Yeast transformants expressing xylA from C. phytofermentans and codon-optimized xylA from Piromyces sp. strain E2 and C. phytofermentans (carried on multicopy vectors) were cultivated until early exponential growth phase in selective medium. Cells were harvested and disrupted with glass beads (diameter, 0.45 mm) using a Vibrax cell disrupter (Janke & Kunkel, Staufen, Germany). Protein concentration was determined with the method of Bradford (5) by using bovine serum albumin as a standard. Enzyme assays were performed immediately after preparation of crude extracts.
XI activity in cell extracts of recombinant yeast strains was determined at 30°C. Assays were carried out in reaction mixtures containing 0.23 mM NADH, 10 mM MgCl2, 2 U sorbitol dehydrogenase in 100 mM Tris-HCl (pH 7.5), and crude cell extracts, as described previously (17). The reaction was started by addition of d-xylose to a final concentration of 500 mM and monitored by measuring oxidation of NADH (during conversion of d-xylulose to xylitol by sorbitol dehydrogenase) spectrophotometrically at 340 nm. For determination of the kinetic parameters, 6.25 to 500 mM d-xylose was used.
Xylitol inhibition of the XI was measured by adding various concentrations of xylitol (10 to 50 mM) in the presence of 6.25 to 500 mM d-xylose (37). The inhibition constant Ki was calculated from Km′ = Km × (1 + i/Ki) with i as the xylitol concentration used and Km′ as the apparent Km for d-xylose at the respective xylitol concentration. All enzyme assays were carried out at least in triplicate.
RESULTS
Screen for novel functionally expressed XIs in S. cerevisiae.
The yeast S. cerevisiae is able to metabolize xylose only after heterologous expression of an XI or an XR/XDH enzyme pair. However, all attempts to express nonfungal, nonthermophilic XIs with high activity in S. cerevisiae have failed so far.
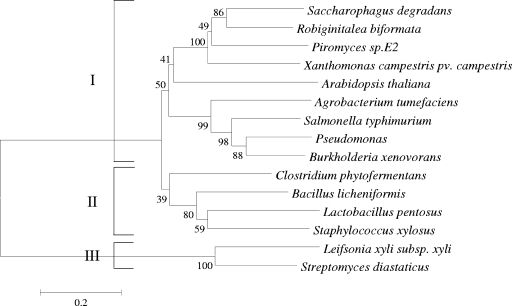
Therefore, our aim was to screen for a new kind of heterologous XI with high activity in S. cerevisiae cells and not subject to, or less subject to, xylitol inhibition. To this end, we selected XIs from 14 organisms of different phylogenetic affiliations which exhibited identities from 17% to 60% to the XI from the Piromyces strain (Fig. 1). The coding sequences of the selected genes were amplified by PCR and cloned via homologous recombination into the high-copy-number yeast expression vector p426H7 (11), placing the respective genes under the control of a strong and constitutive HXT7 promoter fragment and the CYC1 terminator. The coding sequences from L. xyli subsp. cynodontis and S. diastaticus subsp. diasticus could not be amplified and were thus not analyzed in the screen. A codon-optimized xylA gene version from Piromyces sp. strain E2 (YEp-opt.XI-Piro) was used as a positive control in the screening system. The activity of the recombinant XIs was assessed by conferring growth on the yeast strain MKY9 on a synthetic medium with xylose as the only carbon source. In strain MKY9 all the enzymes of the nonoxidative part of the pentose phosphate pathway, the xylulokinase, and the GAL2 permease are overexpressed due to the replacement of their native promoters by the strong HXT71-392 promoter fragment. A similar approach was shown previously to improve growth of the yeast cells on a xylose medium (18).
FIG. 1.
Phylogenetic tree of the amino acid sequences of the tested XIs reported in the GenBank database.
The 12 different expression plasmids, the empty vector control (p426H7), and the Piromyces xylA positive control (p426H7-opt.XI-Piro) were transformed into strain MKY9, first selecting for growth on a medium with 20 g liter−1 glucose but without uracil as the plasmid selection marker. Transformants were replica plated on a synthetic medium without uracil and with 20 g liter−1 xylose as the sole carbon source. The test was scored as positive if yeast colonies could be detected after 4 to 5 days of incubation at 30°C. Nonfunctional XIs did not confer the ability to grow on d-xylose even after 2 weeks of incubation and were thus scored as negative. Yeast transformants expressing the XI from C. phytofermentans and the positive control expressing XI from the Piromyces strain could grow on the xylose medium; all other XIs tested in the screen did not enable yeast transformants to grow on d-xlyose.
Analysis of the codon usage of the xylA gene from C. phytofermentans using CODONW (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form_codonw) revealed that, compared to S. cerevisiae, its codon adaptation index is very low (0.136), which may result in rather inefficient gene expression in S. cerevisiae. Therefore, the codon usage of xylA was adapted to that of the genes encoding glycolytic enzymes in S. cerevisiae (35) to further improve xylose conversion in yeast. This approach has previously been reported to improve l-arabinose conversion via heterologously expressed genes (35). The codon-optimized gene exhibited a codon adaptation index of 0.991 and, when provided on plasmid YEp-opt.XI-Clos, enabled growth of S. cerevisiae with d-xylose, as expected. The codon-optimized gene version of xylA from C. phytofermentans was further examined and compared to the codon-optimized xylA variant from Piromyces sp. strain E2.
Characterization of the kinetic properties of the C. phytofermentans XI.
The kinetic properties of XI of C. phytofermentans were determined in a crude extract of yeast cells containing plasmids YEp-opt.XI-Clos and YEp-XI-Clos. Yeast cells were grown in minimal medium with 20 g liter−1 glucose into the exponential growth phase and harvested, and crude extracts were prepared. As the XI amino acid sequences encoded by the two plasmids are the same, the XI reactions exhibited comparable apparent Km values for xylose (66.01 ± 1.00 mM for codon-optimized XI and 61.85 ± 3.41 mM for native XI). Cells expressing XI from Piromyces sp. strain E2 showed an apparent Km for d-xylose of 49.85 ± 2.82 mM.
Next, in order to compare the performances of the two isomerases within the yeast cells, the reaction velocities (Vmax; μmol min−1 mg protein−1) of the XI from Piromyces sp. strain E2 and from C. phytofermentans, respectively, were determined in crude extracts. Extracts from cells containing the native clostridial XI gene from C. phytofermentans catalyzed conversion of d-xylose to xylulose at a maximal rate of 0.0076 μmol min−1 mg protein−1 whereas the reaction in extracts derived from cells containing the codon-optimized gene version proceeded at a rate of 0.0344 μmol min−1 mg protein−1. Thus, codon adaptation resulted in a Vmax increased by 450% on average with a deviation of no more than 10% for every measured value. For the codon-optimized xylA variant from Piromyces sp. strain E2, a Vmax of 0.0538 μmol min−1 mg protein−1 was determined.
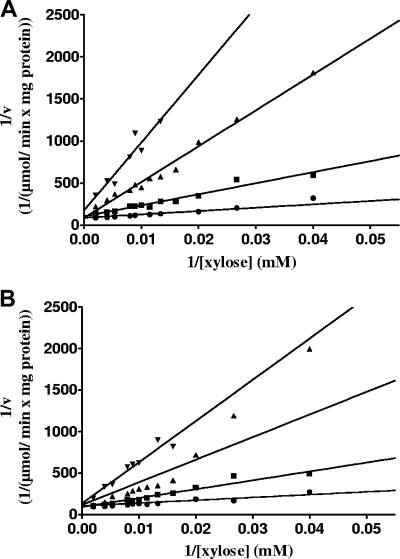
An important feature of XIs is their inhibition by xylitol, a side product during xylose fermentations which negatively affects the efficiency of the overall xylose fermentation process (13). To characterize the influence of xylitol inhibition on XIs from C. phytofermentans and Piromyces sp. strain E2, their apparent Ki values for xylitol were determined. Inhibition kinetics turned out to follow a competitive mechanism, as also previously reported for XI from Lactobacillus brevis (37). However, the apparent inhibition constant Ki of the XI from C. phytofermentans was only 14.51 ± 1.08 mM, whereas that of the enzyme from Piromyces sp. strain E2 was 4.6 ± 1.777 mM (Fig. 2). Thus, the enzyme from C. phytofermentans is three times less inhibited by xylitol than is that from Piromyces.
FIG. 2.
Inhibition of XI from C. phytofermentans (A) and Piromyces sp. strain E2 (B) by xylitol. Strains carrying the gene for XI from C. phytofermentans or Piromyces sp. strain E2, respectively, on a multicopy vector were grown as shake-flask cultures at 30°C into the exponential growth phase in synthetic medium with 20 g liter−1 glucose and without uracil. Crude extracts were prepared, and quantitative enzyme activity tests were performed. Symbols: •, 0 mM xylitol; ▪, 10 mM xylitol; ▴, 30 mM xylitol; ▾, 50 mM xylitol.
Growth performance of yeast transformants expressing the XI of C. phytofermentans.
To test the performance of yeast cells expressing the native and the codon-optimized versions of the XI gene from C. phytofermentans, growth on synthetic medium with 20 g liter−1 xylose of strain MKY9 carrying plasmids YEp-XI-Clos and YEp-opt.XI-Clos, respectively, was analyzed under aerobic conditions in shake-flask cultures. As controls, growth of strains expressing the codon-optimized XI gene from Piromyces from plasmid YEp-opt.XI-Piro and that of a strain containing the empty vector p426H7 were also examined. Precultures of the strains were grown aerobically in the same medium, harvested in the stationary phase, and used to inoculate fresh medium to an OD600 of 0.5.
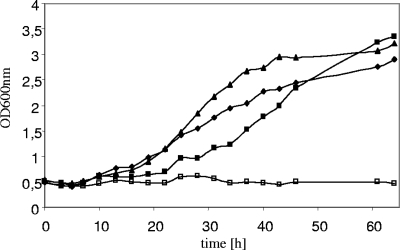
S. cerevisiae cells expressing the native XI gene from C. phytofermentans grew slowly with d-xylose at a maximal rate of 0.039 ± 0.0017 h−1. Recombinant strains expressing the codon-optimized gene version from C. phytofermentans grew slightly faster (maximal growth rate, 0.057 ± 0.0029 h−1) but exhibited a somewhat longer lag phase. Their growth rate was nearly the same as that of the control strain expressing the codon-optimized xylA gene from Piromyces (maximal growth rate, 0.056 ± 0.0030 h−1). The differing lag phases probably depend on subtle differences in the time of harvest of the precultures. The strain carrying the empty vector could not grow at all on d-xylose (Fig. 3).
FIG. 3.
Growth of recombinant S. cerevisiae strains expressing different XIs. SC medium (without uracil) contained 20 g liter−1 d-xylose as the sole carbon source. Yeast strains were grown aerobically as shake-flask cultures at 30°C. Yeast strain MKY9 contained the different XI genes. Symbols: ▪, opt-XI-Clos; ▴, opt-XI-Piro; ⧫, XI-Clos; □, empty vector. Shown are results of a typical experiment.
Growth on xylose of an industrial S. cerevisiae strain.
Industrial S. cerevisiae strain BarraGrande expressing the xylA gene from plasmid YEp-opt.XI-Clos-K could initially not grow on d-xylose as the sole carbon and energy source. To select for spontaneous mutants growing on xylose, the strain was subjected to serial transfer in shake flasks containing a synthetic medium with 20 g liter−1 xylose, 1 g liter−1 yeast extract, and 2 g liter−1 peptone. After only four transfers, covering a period of 28 days, the strain showed growth on xylose. Another two transfers on xylose medium resulted in a strain growing well on xylose medium.
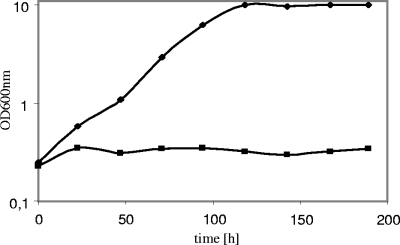
First, growth on d-xylose medium was tested under aerobic conditions using shake-flask cultures (Fig. 4). It turned out that strain BWY10Xyl, containing the codon-optimized xylA gene, could grow on xylose medium with a maximal specific growth rate of 0.04 ± 0.004 h−1. The strain reached a final OD of 10 in less than 120 h. The wild-type strain could not grow at all on d-xylose.
FIG. 4.
Aerobic growth of industrial S. cerevisiae strain expressing the codon-optimized XI from C. phytofermentans. SC medium contained 20 g liter−1 d-xylose as the sole carbon source. Yeast strains were grown aerobically as shake-flask cultures at 30°C. Symbols: ⧫, BWY10Xyl; ▪, BarraGrande (wild type).
Fermentation characteristics of an industrial yeast strain expressing XI.
To analyze d-xylose consumption and ethanol production of yeast transformants containing the codon-optimized XI from C. phytofermentans, two anaerobic batch fermentor cultivations were performed in synthetic medium with 30 g liter−1 of d-xylose. Precultures of strain BWY10Xyl containing the codon-optimized gene were pregrown aerobically in shaking flasks containing 100 ml of SC medium with 20 g liter−1 d-xylose. Cells were harvested, washed, and inoculated into batch fermentors. To generate enough biomass, cells were first grown under aerobic conditions until 5 g liter−1 of d-xylose was consumed (Fig. 5). Anaerobic conditions were maintained by sparging with nitrogen gas until no oxygen was left in the medium (see Materials and Methods).
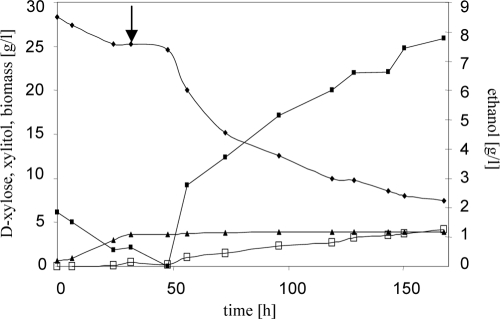
FIG. 5.
Anaerobic batch fermentation by recombinant S. cerevisiae. Shown is a graph of anaerobic batch fermentation of strain BWY10Xyl. The strain was grown in mineral medium supplemented with amino acids and with 30 g liter−1 d-xylose as the sole carbon source. The strain was pregrown in the fermentor under aerobic conditions until about 5 g liter−1 d-xylose was consumed and then shifted to anaerobic conditions (indicated by the arrow). Symbols: ⧫, d-xylose; ▪, ethanol; □, xylitol; ▴, biomass. Shown are results of a typical experiment.
d-Xylose consumption and ethanol production rates were determined for both cultures in the anaerobic phase of the fermentation, i.e., starting with approximately 25 g liter−1 d-xylose in the medium. d-Xylose was consumed at a rate of 0.07 and 0.06 g d-xylose h−1 g (dry weight)−1, respectively. A residual of ca. 7 g liter−1 d-xylose was still left in the medium after 170 h. The ethanol production rate was 0.03 (0.03) g ethanol h−1 g (dry weight)−1, and the ethanol yield was 0.43 (0.42) g ethanol g d-xylose consumed−1. As by-products, xylitol, glycerol, and acetate were produced (0.03 g glycerol g d-xylose consumed−1, 0.02 g acetate g d-xylose consumed−1, and 0.18 g xylitol g d-xylose consumed−1).
DISCUSSION
The yeast S. cerevisiae is not able to metabolize the pentose sugar d-xylose as it lacks the enzyme activities to convert xylose into xylulose. One possible approach for efficient d-xylose fermentation in S. cerevisiae would be the heterologous expression of an XI. However, most of the XIs expressed in S. cerevisiae before now were not active. Also, in our screening only expression of 1 out of 12 tested XI genes conferred on the yeast cells the ability to grow on d-xylose medium as the sole carbon and energy source, indicating that the others were not functional. The reasons for the absence of XI activity in S. cerevisiae were discussed previously (25). The isomerases tested in this study fall into three different clusters (Fig. 1). The functional XI from the bacterium C. phytofermentans shows only 52% identity to the corresponding enzyme from Piromyces sp. strain E2, which is so far the only enzyme exhibiting significant activity in S. cerevisiae at its growth temperature but is of eukaryotic origin.
In yeast crude extracts, the XI from C. phytofermentans exhibited kinetics similar to those of the enzyme from Piromyces. By adapting the codon usage of the corresponding gene to that of glycolytic genes in S. cerevisiae, the enzyme's apparent Vmax increased more than 450% to 0.0344 μmol min−1 mg protein−1, possibly due to its higher abundance. Compared to previous work (reference 9 and references therein), growth rates of our recombinant strains are rather low, independently of whether they express the C. phytofermentans or the Piromyces XI. For instance, in the work of Kuyper et al. (20) a recombinant yeast strain containing the xylA gene from Piromyces sp. strain E2 grows aerobically on xylose at a rate of 0.18 h−1. Also, Sonderegger and Sauer (26) reported a yeast strain containing the XK and XDH genes from P. stipitis and exhibiting xylose-dependent growth with a rate of 0.12 h−1. However, in both cases efficient metabolization of xylose could be achieved only after extensive metabolic and evolutionary engineering (18, 20, 26). Remarkably, we could, for the first time, manipulate yeast strains currently used in industrial fermentation processes for xylose fermentation. Already, after a very short period of optimization the industrial strain BWY10Xyl exhibited significant growth on xylose and also xylose fermentation. Further efforts to improve our XI-containing yeast strains are ongoing and will be reported elsewhere.
The most important finding of our work is that the XI from C. phytofermentans is significantly less sensitive to inhibition by xylitol (Ki, 14.51 ± 1.08 mM) than is the XI from Piromyces (Ki, 4.67 ± 1.77 mM). While xylitol formation under anaerobic conditions is strongly reduced in yeast cells expressing an XI compared to XR/XDH (20), other reactions in the cell still produce xylitol from xylose, such as the unspecific aldose reductase encoded by the GRE3 gene (16). While deletion of the GRE3 gene is possible in laboratory strains and results in a significantly decreased xylitol formation (29), gene deletion of GRE3 in industrial strains has failed so far (our unpublished data), probably due to the difficulties in genetically manipulating these polyploid/aneuploid strains. Moreover, GRE3 deletion should also not be desired for industrial applications, because the aldose reductase is known to be involved in detoxification of hydrolysates (1). Therefore, an XI with less xylitol inhibition is highly desired, and the XI from C. phytofermentans will be advantageous during d-xylose conversion with industrial strains. Our work provides a promising starting point for further improving xylose fermentation for industrial ethanol production.
Acknowledgments
We thank Marco Keller for strain MKY9 and plasmid YEp-opt.XI-Piro. In addition, we thank Christian Weber for preparing genomic DNA from A. tumefaciens and cDNA from A. thaliana.
Part of this work was financed by the European Commission through contract no. 019882 (New Improvements for Lignocellulosic Ethanol).
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Aguilera, J., and J. A. Prieto. 2001. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 39:273-283. [DOI] [PubMed] [Google Scholar]
- 2.Amore, R., M. Wilhelm, and C. P. Hollenberg. 1989. The fermentation of xylose—an analysis of the expression of Bacillus and Actinoplanes xylose isomerase genes in yeast. Appl. Microbiol. Biotechnol. 30:351-357. [Google Scholar]
- 3.Ångspanneföreningen. 1994. Report: P23332-1. NUTEK, Stockholm, Sweden.
- 4.Aristidou, A., and M. Penttila. 2000. Metabolic engineering applications to renewable resource utilization. Curr. Opin. Biotechnol. 11:187-198. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bruinenberg, P. M., J. P. van Dijken, and W. A. Scheffers. 1983. An enzymic analysis of NADPH production and consumption in Candida utilis. J. Gen. Microbiol. 129:965-971. [DOI] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hagerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn-Hagerdal, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jonsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 10.Hallborn, J. 1995. Metabolic engineering of Saccharomyces cerevisiae: expression of genes involved in pentose metabolism. Lund University, Lund, Sweden.
- 11.Hamacher, T., J. Becker, M. Gardonyi, B. Hahn-Hagerdal, and E. Boles. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783-2788. [DOI] [PubMed] [Google Scholar]
- 12.Ho, N. W., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karhumaa, K., B. Hahn-Hagerdal, and M. F. Gorwa-Grauslund. 2005. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359-368. [DOI] [PubMed] [Google Scholar]
- 14.Kotter, P., R. Amore, C. P. Hollenberg, and M. Ciriacy. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18:493-500. [DOI] [PubMed] [Google Scholar]
- 15.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 16.Kuhn, A., C. van Zyl, A. van Tonder, and B. A. Prior. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuyper, M., H. R. Harhangi, A. K. Stave, A. A. Winkler, M. S. Jetten, W. T. de Laat, J. J. den Ridder, H. J. Op den Camp, J. P. van Dijken, and J. T. Pronk. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 4:69-78. [DOI] [PubMed] [Google Scholar]
- 18.Kuyper, M., M. M. Hartog, M. J. Toirkens, M. J. Almering, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399-409. [DOI] [PubMed] [Google Scholar]
- 19.Kuyper, M., M. J. Toirkens, J. A. Diderich, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925-934. [DOI] [PubMed] [Google Scholar]
- 20.Kuyper, M., A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 4:655-664. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 22.Lonn, A., M. Gardonyi, W. van Zyl, B. Hahn-Hagerdal, and R. C. Otero. 2002. Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and protein characterization. Eur. J. Biochem. 269:157-163. [DOI] [PubMed] [Google Scholar]
- 23.Moes, C. J., I. S. Pretorius, and W. H. van Zyl. 1996. Cloning and expressing of the Clostridium thermosulfurogenes d-xylose isomerase gene (xylA) in Saccharomyces cerevisiae. Biotechnol. Lett. 18:269-274. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. M. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sarthy, A. V., B. L. McConaughy, Z. Lobo, J. A. Sundstrom, C. E. Furlong, and B. D. Hall. 1987. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 53:1996-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonderegger, M., and U. Sauer. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 69:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 28.Teunissen, M. J., H. J. Op den Camp, C. G. Orpin, J. H. Huis in 't Veld, and G. D. Vogels. 1991. Comparison of growth characteristics of anaerobic fungi isolated from ruminant and non-ruminant herbivores during cultivation in a defined medium. J. Gen. Microbiol. 137:1401-1408. [DOI] [PubMed] [Google Scholar]
- 29.Traff, K. L., R. R. Otero Cordero, W. H. van Zyl, and B. Hahn-Hagerdal. 2001. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl. Environ. Microbiol. 67:5668-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dijken, J. P., E. van den Bosch, J. J. Hermans, L. R. de Miranda, and W. A. Scheffers. 1986. Alcoholic fermentation by ‘non-fermentative’ yeasts. Yeast 2:123-127. [DOI] [PubMed] [Google Scholar]
- 31.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. Van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 32.Wahlbom, C. F., A. Eliasson, and B. Hahn-Hagerdal. 2001. Intracellular fluxes in a recombinant xylose-utilizing Saccharomyces cerevisiae cultivated anaerobically at different dilution rates and feed concentrations. Biotechnol. Bioeng. 72:289-296. [DOI] [PubMed] [Google Scholar]
- 33.Walfridsson, M., X. Bao, M. Anderlund, G. Lilius, L. Bulow, and B. Hahn-Hagerdal. 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 35.Wiedemann, B., and E. Boles. 2008. Codon-optimized bacterial genes improve l-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:2043-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth, R. 1989. Elektroporation: eine alternative Methode zur Transformation von Bakterien mit Plasmid-DNA. Formum Mikrobiol. 11:507-515. [Google Scholar]
- 37.Yamanaka, K. 1969. Inhibition of d-xylose isomerase by pentitols and d-lyxose. Arch. Biochem. Biophys. 131:502-506. [DOI] [PubMed] [Google Scholar]
- 38.Zimmermann, F. K. 1975. Procedures used in the induction of mitotic recombination and mutation in the yeast Saccharomyces cerevisiae. Mutat. Res. 31:71-86. [DOI] [PubMed] [Google Scholar]