Abstract
The application of a selected Acetobacter pasteurianus strain for traditional balsamic vinegar production was assessed. Genomic DNA was extracted from biofilms after enrichment cultures on GYC medium (10% glucose, 1.0% yeast extract, 2.0% calcium carbonate) and used for PCR/denaturing gradient gel electrophoresis, 16S rRNA gene sequencing, and enterobacterial repetitive intergenic consensus/PCR sequencing. Results suggested that double-culture fermentation is suitable for traditional balsamic vinegar acetification.
The use of selected starter cultures (SSC) in fermented food production is widely applied throughout the food industry, in particular for wine, dairy products, sausages, and a variety of vegetables (3, 11). The advantages of their use are related to the improvement of the process control, hygiene, and quality with respect to fermented foods obtained through indigenous fermentation. Vinegar is one of the fermented beverages produced without SSC inoculation, in both small- and large-scale production, mainly for the following reasons: (i) the majority of vinegars have low commercial value, and often technological innovation is not considered profitable, and (ii) there is limited knowledge of the ecophysiology of acetic acid bacteria (AAB) due to the difficulty in accessing, sampling, isolating, and preserving strains (2, 12, 15, 16, 17). Among vinegars, traditional balsamic vinegar (TBV) is an Italian aged condiment produced by “seed vinegar,” the so-called “mother of vinegar” that is an indigenous starter culture withdrawn from acetifying vinegar through back-slopping procedures. The raw material is a fermented and cooked grape must (here indicated as must) at a soluble solids content ranging from 20 to 60°Bx (10). TBV production is regulated by denomination of protected origin guidelines that specify procedures and final product features. In particular, the raw material characteristics, the production process (e.g., must cooking, alcoholic fermentation, acetic oxidation, and ageing), features of the production area (no environmental condition management is permitted), and analytical and sensorial parameters are stated as follows: acidity (not less than 4.5% [wt/wt], expressed as grams of acetic acid per 100 g of product), density at 20°C (not less than 1.240 g per liter), color, aroma, and taste. The production is performed in wood barrels, and the process is carried out by sequential refilling to acetify the must and replace the volume lost by evaporation. AAB grow on the surface of liquid by biofilm formation. No addition of any substance can be made except for the acetifying must as a starter (7). Microbial studies of TBV reported culture-dependent and -independent approaches to evaluating AAB occurrence in TBV musts (5, 10). These studies highlighted the occurrence of Gluconacetobacter europaeus as a widespread indigenous species, as well as Acetobacter pasteurianus, Acetobacter aceti, and Acetobacter malorum. However, no comprehensive studies of AAB diversity and the correlation between species occurrence and technological steps of TBV production have been published, due mainly to the difficulty of easy access to AAB microflora in vinegar matrix by both culture-dependent and -independent approaches.
Regarding production technology, at least one drawback of current production procedures has been acknowledged. It concerns the difficulty of start-up acetification, which affects the minimum acidity value required for the final product. In fact, some studies showed that many variables regulate AAB growth and activity. Above all is the sugar concentration among substrates and the temperature among physical parameters. To efficiently control the acetification start-up, it is necessary to understand the function of AAB responsible for the initial colonization of musts and to investigate the microbial succession suitable to complete the acetification. Our previous researches on TBV showed that AAB strains exhibit different growing abilities. In particular, strains of Acetobacter pasteurianus grow quickly on laboratory synthetic media, wine, and cooked must. In contrast, strains belonging to G. europaeus do not grow or grow very slowly on cooked and fermented must (9, 10).
The goal of this study was to implement a laboratory SSC to test it on a factory scale for TBV production purposes. In particular, we focused our attention on the effect of A. pasteurianus strain AB0220 on the acetification and dynamics of species at the end of the process. The SSC effectiveness was assessed by monitoring analytical parameters (acetic acid, ethanol, and pH), species succession, and strain persistence during three stages by the following molecular analyses: PCR/denaturing gradient gel electrophoresis (DGGE), 16S rRNA gene sequencing, and enterobacterial repetitive intergenic consensus (ERIC)/PCR sequencing using genomic DNA extracted from biofilms recovered on GYC (10% glucose, 1.0% yeast extract, 2.0% calcium carbonate) plates.
Strains and growth conditions.
Strain AB0220 was grown on GYC medium (for solid medium, 1.5% agar was added) and incubated at 30°C, and type strains DSMZ 3509 and DSMZ 6160 were grown according to the manufacturer's procedures (Deutsche Sammlung von Mikroorganismen und Zellkulturen, GmbH, Germany).
Technological performance of Acetobacter pasteurianus.
The technological performance of AB0220 was tested as follows: using wine, Passmore and Carr medium, and three cooked musts (must A, must A plus yeast extract [YE], and must B) at different levels of ethanol, acetic acid, pH, and soluble solids (Table 1); and monitoring percent ethanol (vol/vol; gravimetric analysis), percent acetic acid (wt/wt; neutralizing samples at pH 7.2 with 0.1 N NaOH; it was assumed that all medium acidities were due to acetic acid), pH (MicropH 2002; Crison), and soluble solids as degrees Brix (2WAJ refractometer; Abbe) acquired over a period of 24 days at 30°C. Five milliliters of AB0220 culture was inoculated in a 100-ml flask containing 50 ml of broth. AB0220 showed the best trend in terms of increase in acetic acid content for must A plus YE (5.07%) and wine (4.29%), whereas the lowest increase in acetic acid content was observed for must B (0.78%) and Passmore and Carr medium (0.53%). Although the increase in acetic acid content was not the highest for must A (3.59%), a constant production was observed over 24 days. The less-productive performance observed for must B and Passmore and Carr medium could be explained by the combined effect of ethanol and pH levels and concentration of soluble solids, resulting in stressful physiological conditions.
TABLE 1.
Composition of grape musts, wine, and defined media to test performance of Acetobacter pasteurianus AB0220
| Medium | pH | % Acetic acid (wt/wt) | °Bx | % Ethanol (vol/vol) |
|---|---|---|---|---|
| Must A | 3.58 | 0.23 | 7 | 7.5 |
| Must B | 3.5 | 0.65 | 13 | 10 |
| Must A + YE (2%) | 3.58 | 0.23 | 7 | 7.5 |
| Wine | 3.7 | 0.22 | 6 | 5.5 |
| Passmore and Carr | 4.26 | 0.39 | 14 | 0 |
SSC design, implementation, and analytical monitoring.
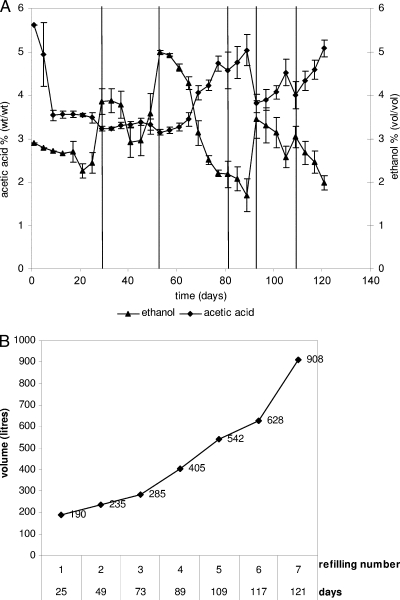
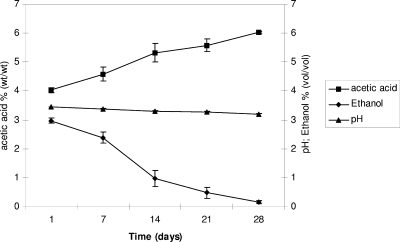
To obtain about 900 liters of culture for TBV production, the implementation of the SSC was performed for the following three stages: stage 1, the start-up under laboratory conditions; stage 2, the factory scale-up performed in a tank system; and stage 3, acetification of factory barrels. The refilling procedure was performed for stage 1 and stage 2 by fixing 1 and 3% as the upper and lower limits for ethanol content and 3% as the lower limit for acetic acid content, and then the must volume was added accordingly. Three trial aliquots of uninoculated must A were used as the acetification control starting from stage 1. The acetification trend was monitored by measuring the ethanol consumption pH and levels of acetic acid and soluble solids. Stage 1 was started up using 5 ml of GYC liquid AB0220 culture. Must A was used as the first refilling of stage 1, and then further refilling steps were performed by adding pasteurized must at increasing soluble solids contents (from 7 to 20°Bx). Stage 1 was stopped at 70 liters of SSC (pH 3.37, 5.62% acetic acid, 1.22% ethanol, 20.7°Bx) and required 30 days. The SSC volume of stage 1 was used to inoculate the must for the scale-up operation, which was performed in a tank system (stage 2). Stage 2 was carried out for 121 days, during which seven refilling steps were done (Fig. 1B), resulting in a final volume of 628 liters (pH 3.4, 5.0% acetic acid, 2.0% ethanol, 20°Bx) (Fig. 1A). Stage 3 was performed in wood barrels, mixing the SSC volume previously obtained (628 liters) and 280 liters of must for refilling purposes. A total of 21 barrels plus the control trial were filled, and no further refilling was done. After 28 days, a constant increase in acetic acid concentration was obtained, starting from 4% to 6% together with ethanol consumption (Fig. 2). Degrees Brix increased from 20% (at the end of stage 2) to 25% (at the end of stage 3); this was due mainly to evaporation, resulting in an increase of soluble solids.
FIG. 1.
(A) Acetification trend during scale-up (stage 2) monitored by acetic acid and ethanol measures. Vertical lanes correspond to the refilling step. The data are averages based on three trials, and error bars indicate the standard determination. (B) Volume increase through seven refilling steps of scale-up (stage 2).
FIG. 2.
Acetification trend during barrel acetification (stage 3) monitored by measuring acetic acid and ethanol levels and pH. The data are averages based on three trials, and error bars indicate the standard determination.
All acetification controls for each stage consisted of the initial must A without refilling, since no increase in acetic acid was observed. This demonstrated that uncultivable AAB were not involved in the acetification of this process.
Acetobacter pasteurianus persistence and species succession evaluation.
To perform molecular analyses, samples were collected in triplicate for stage 1 (at 30 days), stage 2 (at 0, 60, and 120 days), and stage 3 (at 7, 14, 21, and 28 days). Aliquots (0.1 mg) of superficial biofilm were recovered from three different points on the surfaces of tanks and barrels, respectively, streaked on GYC plates, and incubated at 30°C for 24 h. The biofilm was totally recovered from plates and used for genomic DNA extraction. The extraction was performed by sodium dodecyl sulfate proteinase-cetyl trimethyl ammonium bromide treatment as previously reported (10), and DNA was quantified by spectrophotometric measure (NanoDrop ND-1000). 16S rRNA PCR/DGGE and ERIC/PCR were applied as previously described (5, 8, 13, 19). Individual DGGE bands for stage 2 at day 60 and stage 3 at day 28 were excised, reamplified, and subjected to sequence analysis (5). Direct sequencing of the 16S rRNA gene was performed on 16S rRNA amplicons as a template using the following primers: 616F (5′-AGAGTTTGATYMTGGCTCA-3′; positions 8 to 26 on 16S rRNA gene; Escherichia coli numbering), 16S F343 (5′-TACGGGAGGCAGCAG-3′; positions 342 to 356 on the 16S rRNA gene; Escherichia coli numbering), and 16S 1492 (5′-GGCTACCTTGTTACGACTT-3′; positions 1490 to 1510 on the rRNA gene; Escherichia coli numbering). PCR analysis was performed on template DNAs using Ex Taq DNA polymerase (Takara Bio, Inc., Japan) according to the manufacturer's conditions and procedures. PCR products from the 16S rRNA gene were purified using the Montage PCR cleanup kit and automatically sequenced (Eurofins MWG Operon service, Germany). Sequence contigs were assembled using ChromasPro (v1.41), and similarities were searched using the BLAST program (1). Molecular analyses were performed on triplicates of GYC plates at each of the three stages.
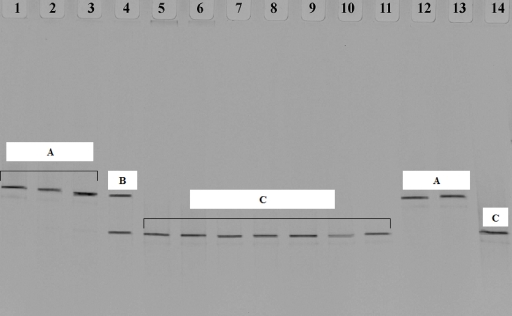
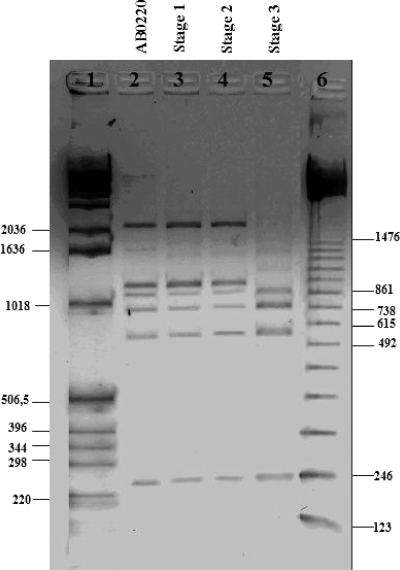
PCR/DGGE analysis allowed distinguishing between two different species groups along the three stages (Fig. 3). In particular, a single band was observed corresponding to the end of the laboratory phase (stage 1) as well as at the beginning and after 60 days of stage 2, with the same migration distance as the bands corresponding to AB0220 and the type strain A. pasteurianus DSM 3509, respectively (Fig. 3, lanes 1, 2, 3, 12, and 13). At the end of stage 2, the migration pattern showed two bands (Fig. 3, lane 4), one with the same migration distance as that of strains AB0220 and DSM 3509 and the other with the same distance as that of G. europaeus DSM 6160 (Fig. 3, lane 14). Instead, for each time point of barrel acetification (stage 3), a single dominant band with the same migration distance as that of G. europaeus DSM 6160 was observed (Fig. 3, lanes 5 to 11). Results from replicates were consistent. Species occurrence and succession were also confirmed by sequencing DGGE bands of stage 2 (60 days) and stage 3 (28 days). These data highlighted the development of a stable set of bacterial population deducible by a simple and clear electrophoretic pattern compared to DGGE patterns of samples derived from a high level of biodiversity that show the typical smear (6). Moreover, results indicated that the population during stage 1 and stage 2 was distinctly different from that occurring in stage 3. 16S rRNA gene sequencing analysis confirmed DGGE results; in particular, a large percentage of sequence homology was found with A. pasteurianus at stages 1 and 2 at 60 days (100%) and with G. europaeus at stage 3 (99 and 100%) (Table 2). At the end of stage 2 (120 days), the 16S rRNA gene sequence exhibited the presence of more than one bacterial species as indicated by a mixed sequence chromatogram and confirmed by two bands in DGGE analysis (Fig. 3, lane 4). Finally, an ERIC/PCR fingerprinting assay has been performed at the end of each acetification stage to evaluate strain occurrence. A similar electrophoretic profile was obtained for stages 1 and 2, comparable to that of strain AB0220, whereas a different pattern was obtained for stage 3 (Fig. 4). This result confirmed a change in the population at stage 3.
FIG. 3.
Acetic acid bacteria species monitoring from the laboratory stage to scale-up detected by PCR/DGGE. Lane 1, stage 1 (at 30 days); lane 2, stage 2 (at 0 days); lane 3, stage 2 (at 60 days); lane 4, stage 2 (at 120 days); lanes 5 and 6, stage 3 (at 7 days); lanes 7 and 8, stage 3 (at 14 days); lanes 9 and 10, stage 3 (at 21 days); lane 11, stage 3 (at 28 days); lane 12, AB0220; lane 13, A. pasteurianus DSM 3509; and lane 14, G. europaeus DSM 6160. A, B, and C (white rectangles) indicate profile groups.
TABLE 2.
AAB species monitoring from laboratory scale (stage 1) to factory scale (stages 2 and 3) as detected by PCR/DGGE and 16S rRNA gene sequencing
| Acetification stage | Days | Speciesa | 16S sequence accession no.b | Size (bp) | Species name, accession no.c | % Sequence similarity |
|---|---|---|---|---|---|---|
| 1 | 30 | A. pasteurianus | FM177227 | 1,410 | A. pasteurianus, DQ523493.1 | 100 |
| 2 | 60 | A. pasteurianus | FM177228 | 1,410 | A. pasteurianus, DQ523493.1 | 100 |
| 120 | A. pasteurianus/G. europaeusd | |||||
| 3 | 7 | G. europaeus | FM177229 | 1,451 | G. europaeus, EU096233.1 | 99 |
| 14 | G. europaeus | FM177230 | 1,450 | G. europaeus, EU096233.1 | 100 | |
| 21 | G. europaeus | FM177231 | 1,450 | G. europaeus, EU096233.1 | 100 | |
| 28 | G. europaeus | FM177232 | 1,450 | G. europaeus, EU096233.1 | 100 |
PCR/DGGE species designation obtained by clustering AAB species according to 16S rRNA/DGGE analysis (4). Bands of stage 2 (at 60 days) and stage 3 (at 28 days) were excised and sequenced.
Accession number obtained in this study.
Accession number of sequence of closest relative.
A mixed sequence chromatogram was obtained.
FIG. 4.
AAB strain occurrence from SSC implementation (stage 1) to stages 2 and 3 detected by ERIC/PCR. Lane 1, 1-kb DNA ladder (Invitrogen); lane 2, strain AB0220; lane 3, stage 1; lane 4, stage 2; lane 5, stage 3; and lane 6, 123-bp DNA ladder (Invitrogen).
Molecular analyses suggested that the effect of AB0220 SSC implementation favors a microbial succession (A. pasteurianus toward G. europaeus) corresponding to specific technological steps.
On the basis of all of this evidence, our hypothesis is that A. pasteurianus is a pioneer species for the growth of G. europaeus. In particular, the persistence of the inoculated A. pasteurianus strain during stages 1 and 2 (corresponding to day 150 of a total of 178 days) was ensured by the scale-up procedure, during which new must was periodically added to increase the SSC volume, and the strain was not exposed to the negative effect of acetic acid. Moreover, the concentration of ethanol, which is the preferred carbon source for Acetobacter spp., was maintained in a range between 1.7 and 5%, positively affecting the AB0220 growth. Due to these favorable conditions, the contribution of AB0220 along step 1 and step 2 was to start up the acetification, to drive the process through the refilling steps, and to promote the growth of indigenous G. europaeus cells suitable for stage 3. G. europaeus is a species of AAB that requires acetic acid for growth (14, 18), and for this reason it was not chosen as the SSC for starting acetification. The microbial succession of a different species with respect to the inoculated one derives from the acetic acid-driven selection of the indigenous strain. According to the production procedure of TBV, the end of the refilling corresponded to a decrease in ethanol and a remarkable increase in acetic acid, promoting the growth of indigenous G. europaeus. Therefore, our A. pasteurianus SSC had the following two positive effects: (i) to start the acetification process from cooked must without acetic acid and (ii) to promote the growth of naturally occurring G. europaeus strains that are able to quickly finish the acetification. The lack of growth of AAB in the acetification control of the three stages, in which the refilling was done using uninoculated must, confirmed our observations.
Current TBV production is strongly conditioned by the cyclic changing environment of the medium; the inoculation of pioneer AAB has the main impact on starting acetification and on the establishment of a suitable growth environment for subsequent species. The best way to carry out the acetification could be double-culture fermentation by two selected strains with different phenotypic traits suitable to establish a microbial succession. The exploitation of the double-culture fermentation using A. pasteurianus strains as pioneer bacteria and G. europaeus as a subsequent species could allow a multistep transformation that would be impossible for a single microorganism in the TBV environment.
In this study, for the first time an SSC was carried out at a laboratory scale and applied to a TBV production scale. Indirect monitoring of acetification by analytical and molecular tools allowed an understanding of the effectiveness of the designed procedure. Results suggested that double-SSC of A. pasteurianus and G. europaeus could be the main input in view of a more reliable and stable production system.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene were deposited into EMBL databases under the following accession numbers: FM177227, FM177228, FM177229, FM177230, FM177231, and FM177232.
Acknowledgments
This work was supported by funding from the European Communities (Project WINEGAR, Cooperative Research under the Sixth Framework Programme).
We thank the Cavalli Cav. Ferdinando factory involved in the project for providing samples and factory technical assistance.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camu, N., T. De Winter, K. Verbrugghe, I. Cleenwerck, P. Vandamme, J. S. Takrama, M. Vancanneyt, and L. De Vuyst. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 73:1809-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocolin, L., R. Urso, K. Rantsiou, C. Cantoni, and G. Comi. 2006. Multiphasic approach to study the bacterial ecology of fermented sausages inoculated with a commercial starter culture. Appl. Environ. Microbiol. 72:942-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vero, L., and P. Giudici. 2008. Genus-specific profile of acetic acid bacteria by 16S rDNA PCR-DGGE. Int. J. Food Microbiol. 125:96-101. [DOI] [PubMed] [Google Scholar]
- 5.De Vero, L., E. Gala, M. Gullo, L. Solieri, S. Landi, and P. Giudici. 2006. Application of denaturing gradient gel electrophoresis (DGGE) analysis to evaluate acetic acid bacteria in traditional balsamic vinegar. Food Microbiol. 23:809-813. [DOI] [PubMed] [Google Scholar]
- 6.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. 2000. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl. Environ. Microbiol. 66:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazzetta Ufficiale della Repubblica Italiana. 30 May 2000. Aceto balsamico tradizionale di Modena. Production disciplinary of the denomination of protected origin. Gazzetta Ufficiale della Repubblica Italiana, p. 124. Istituto Poligrafico e Zecca dello Stato, Rome, Italy.
- 8.González, A., N. Hierro, M. Poblet, N. Rozes, A. Mas, and J. M. Guillamon. 2005. Application of molecular methods to demonstrate species and strain evolution of acetic acid bacteria population during a wine production. Int. J. Food Microbiol. 102:295-304. [DOI] [PubMed] [Google Scholar]
- 9.Gullo, M., and P. Giudici. 2008. Acetic acid bacteria in traditional balsamic vinegar: phenotypic traits relevant for starter cultures selection. Int. J. Food Microbiol. 125:46-53. [DOI] [PubMed] [Google Scholar]
- 10.Gullo, M., C. Caggia, L. De Vero, and P. Giudici. 2006. Characterization of acetic acid bacteria in “traditional balsamic vinegar.” Int. J. Food Microbiol. 106:209-212. [DOI] [PubMed] [Google Scholar]
- 11.Holzapfel, W. H. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75:197-212. [DOI] [PubMed] [Google Scholar]
- 12.Kittelmann, M., W. W. Stamm, H. Follmann, and H. G. Truper. 1989. Isolation and classification of acetic acid bacteria from high percentage vinegar fermentations. Appl. Microbiol. Biotechnol. 30:47-52. [Google Scholar]
- 13.Lopez, I., F. Ruiz-Larrea, L. Cocolin, E. Orr, T. Phister, M. Marshall, J. VanderGheynst, and D. A. Mills. 2003. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:6801-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sievers, M., S. Sellmer, and M. Teuber. 1992. Acetobacter europaeus sp. nov., a main component of industrial vinegar fermenters in central Europe. Syst. Appl. Microbiol. 15:386-392. [Google Scholar]
- 15.Sokollek, S. J., and W. P. Hammes. 1997. Description of a starter culture preparation for vinegar fermentation. Syst. Appl. Microbiol. 20:481-491. [Google Scholar]
- 16.Sokollek, S. J., C. Hertel, and W. P. Hammes. 1998. Cultivation and preservation of vinegar bacteria. J. Biotechnol. 60:195-206. [Google Scholar]
- 17.Trcek, J. 2005. Quick identification of acetic acid bacteria based on nucleotide sequences of the 16S-23S rDNA internal transcribed spacer region and of the PQQ-dependent alcohol dehydrogenase gene. Syst. Appl. Microbiol. 28:735-745. [DOI] [PubMed] [Google Scholar]
- 18.Trcek, J., K. Jernejc, and K. Matsushita. 2007. The highly tolerant acetic acid bacterium Gluconacetobacter europaeus adapts to the presence of acetic acid by changes in lipid composition, morphological properties and PQQ-dependent ADH expression. Extremophiles 11:627-635. [DOI] [PubMed] [Google Scholar]
- 19.Versalovic, J., T. Koeuth, and R. J. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]