Abstract
Direct enumeration and genetic analyses indicate that aquatic sediments harbor abundant and diverse viral communities. Thus far, synecological analysis of estuarine sediment viral diversity over an annual cycle has not been reported. This oversight is due in large part to a lack of molecular genetic approaches for assessing viral diversity within a large collection of environmental samples. Here, randomly amplified polymorphic DNA PCR (RAPD-PCR) was used to examine viral genotypic diversity within Chesapeake Bay sediments. Using a single 10-mer oligonucleotide primer for all samples, RAPD-PCR analysis of sediment viral assemblages yielded unique banding patterns across spatial and temporal scales, with the occurrence of specific bands varying among the sample set. Cluster analysis of RAPD-PCR amplicon banding patterns indicated that sediment viral assemblages changed with season and to a lesser extent with geographic location. Sequence analysis of RAPD-PCR amplicons revealed that 76% of sediment viral sequences were not homologous to any sequence in the GenBank nonredundant protein database. Of the GenBank sequence homologs, the majority belonged to viruses within the Podoviridae (24%) and Myoviridae (22%) viral families, which agrees with the previously observed frequencies of these morphological families in Chesapeake Bay sediments. Furthermore, the majority of the sediment viral sequences homologous to GenBank nonredundant protein sequences were phages or prophages (57%). Hence, RAPD-PCR proved to be a reliable and useful approach for characterization of viral assemblages and the genetic diversity of viruses within aquatic sediments.
Large numbers of viruses, an estimated abundance greater than 1031 viruses worldwide (11, 26), have been found in a variety of environments, including seawater (38), freshwater (19), sediments (25, 28), and soils (34). Viruses are not only abundant but also likely to significantly influence the population dynamics and genotypic composition of their bacterial host populations (29, 33). Process-level investigations of viral activity in sediments have shown that viruses are an active component of sediment microbial communities (23). Glud and Middelboe (23) found that bacterial growth rates and viral production increased in parallel with respiration, suggesting that viruses are active members of benthic microbial communities. Previous studies have shown that sediment viral abundance exceeds coexisting bacterial abundance by 10- to 1,000-fold (15, 17, 25), creating the potential for viral processes to influence the microbial ecology of aquatic sediments. However, with the exception of small-scale metagenomic investigations (4, 8), there exists little information on the genetic content of viriobenthos assemblages or how the composition of these assemblages changes over ecological gradients.
Despite the high abundances of viruses in nature, the lack of a shared genetic marker creates a difficult problem when attempting to examine viral genetic diversity in environmental samples (31). Gene g20 encodes a multifunctional protein within the collar between the capsid and tail in T4-like bacteriophages and has been of significant importance in examining the genetic diversity of cyanomyoviruses (22, 24, 32). As well, others have been able to evaluate the diversity of unidentified aquatic picornavirus-like viruses using the RNA-dependent RNA polymerase gene (13). Other studies have attempted to examine phage genetic diversity based on the DNA polymerase gene (6, 21). Unfortunately, not all known phages contain these specific genes; hence, their use as universal markers is markedly inadequate. Thus, molecular methods that do not rely on polymorphism analysis of a single gene product must be used to circumvent these limitations.
Recently, metagenomic approaches (i.e., sequencing of random genomic DNA fragments from whole microbial assemblages) have been used to examine genetic diversity within viral (18) and prokaryotic (10) assemblages. For sediment environments, metagenomic analysis has revealed that the viriobenthos is perhaps the most diverse of all viral assemblages, having been estimated to contain more than 10,000 genotypes per kg of sediment (4). Viral assemblages within a wide range of environments including marine (2, 8) and estuarine (3) waters, soils (20), stromatolites (16), and equine (9) and human feces (5, 40) have been examined. Overall, these studies have shown that a relatively low proportion (∼30%) of viral metagenome sequences are similar to sequences found in the nonredundant GenBank database (nr database), but the probability of detecting significant BLAST homologs increases twofold when queries against other viral metagenome sequence libraries are included (3). Thus, the function of most viral genes is currently unknown; however, these genes are broadly distributed among viruses.
While large-scale metagenomics offers unprecedented resolution of the diversity and composition of a viral assemblage, the significant costs and computational requirements preclude routine application in a large collection of environmental samples. Recently, Winget and Wommack (36) introduced a new, low-cost, high-throughput means for genetic analysis of viral diversity utilizing random amplified polymorphic DNA PCR (RAPD-PCR). In this approach, a single 10-bp oligonucleotide serves as both the forward and reverse primers in a single thermocycler reaction. Target sequences in the template DNA are randomly selected; thus, development of a RAPD-PCR assay requires no prior information on the DNA coding content within the sample or organism—a significant advantage considering the largely unknown nature of most viral genes.
In this study, we assess the potential of RAPD-PCR as a tool to examine genotype-scale compositional changes in the Chesapeake Bay viriobenthos and to explore the genetic diversity of viruses within Chesapeake Bay sediments. To our knowledge, this is the first study to use RAPD-PCR for evaluating sediment viral diversity and documenting compositional changes in viriobenthos assemblages over time and geographic location.
MATERIALS AND METHODS
Sampling sites and collection.
Sediment samples were collected from the Chesapeake Bay during nine cruises from April 2003 to October 2005 using a four-tube multicorer device (MC-400 Hedrick/Marrs; Ocean Instruments). Sediment samples were collected from surface depths of 15 m, 25 m, and 9 m for station 724 (37°24′N, 76°5′W), station 804 (38°4′N, 76°13′W), and station 908 (39°8′N, 76°20′W), respectively. Each 10-cm-diameter core was subsampled once with a sterile 60-ml-cutoff syringe, and the top 2 cm was processed immediately onboard.
Viral extractions.
Sediments were processed for removal of viruses as described by Helton et al. (25). Briefly, 2 ml of surface sediment was placed inside sterile 50-ml centrifuge tubes to which 8 ml of 10 mM disodium pyrophosphate and 5 mM EDTA were added. Samples were vortexed at high speed horizontally for 20 min. Larger particles were removed by centrifugation at 2,000 × g for 25 min postagitation. The supernatant was filtered through a 0.45-μm filter (Sterivex; Millipore Corp.). The filtrate was then passed through a 0.22-μm filter (Sterivex) to remove any remaining particles and bacteria. Viral particles in the 0.22-μm filtrate (ca. 8 ml) were concentrated using Centricon-YM30 filters (30,000-molecular-weight cutoff; Millipore), thrice rinsed with sterile TE (100 mM Tris-HCl, 10 mM EDTA), and filtered again with a 0.22-μm filter (Sterivex) prior to storage at −20°C. Several viral concentrate samples were treated with DNase alone, as well as samples treated with heat plus DNase as described by Helton et al. (25) to test for the presence or effects of any free DNA remaining in the concentrated filtrates. Treated samples from both DNase and heat plus DNase treatments were used as templates for testing RAPD-PCR amplification of viral DNA.
RAPD-PCR.
Primer OPA-9 (5′-GGGTAACGCC-3′) was used in all PCRs. PCR mixtures (25 μl) contained 2.0 mM MgCl2 (included in 10× buffer), 0.8 mM each deoxyribonucleoside triphosphate (TaKaRa Bio Inc.), 4 μM primer, and 2.5 U of TaKaRa ExTaq HotStart Version (TaKaRa Bio Inc.). Template concentrations were standardized by adding one microliter of a sediment viral extract containing ca. 7 × 107 virus particles to each RAPD-PCR assay mixture. Reactions were carried out in an MJ Research PTC-200 thermocycler using the following parameters: initial denaturation for 10 min at 94°C; 33 cycles of 3 min at 35°C, 1 min at 72°C, and 30 s at 94°C; and a final extension of 10 min at 72°C. All RAPD-PCR products were visualized by gel electrophoresis on a 1.8% Metaphor (FMC BioProducts) agarose gel in 0.5× Tris-borate-EDTA buffer. Gels were stained in a 1× SYBR gold (Invitrogen) bath for 30 min and imaged with a Typhoon 8600 variable-mode imager (Molecular Dynamics, Amersham Pharmacia Biotech) under 560 BP 30/green 532 nm, focal plane plus 3 mm. Resulting band patterns were analyzed using GelCompar II (version 4.50; Applied Maths). The similarity of RAPD-PCR banding patterns was determined using Jaccard's coefficient, and a dendrogram depicting banding pattern similarity was generated using the unweighted pair group method of averages algorithm.
Cloning and sequencing.
Resulting RAPD-PCR amplicons from all three locations (908, 804, and 724) and from three seasons (autumn 2003, spring 2004, and autumn 2004) were used for genetic analyses. In addition, a recurring band of ∼625 bp (in 12 of 20 samples) was selected from several samples, excised from the agarose gel, and purified with the QIAquick gel purification kit (Qiagen). Entire collections of RAPD-PCR amplicons were purified with the QIAquick PCR purification kit (Qiagen). Both purified products (gel excised bands and whole collections) were cloned using the pCR8/GW/TOPO TA cloning kit (Invitrogen) in accordance with the manufacturer's protocol. This vector was chosen as it includes terminator sequences on either side of the multiple cloning site to prevent the transcription of insert DNA. Cloned products were chemically transformed into One Shot Mach1-T1 (Invitrogen) chemically competent Escherichia coli cells in accordance with the manufacturer's instructions. Clones with plasmid DNA containing RAPD-PCR inserts were purified using the DirectPrep 96 MiniPrep kit (Qiagen).
A total of 518 clones were sequenced using an ABI Prism 3130XL genetic analyzer (Applied Biosystems). From that, 448 sequences were obtained and edited for contaminating vector and primer sequence using Sequencher 4.6 (Gene Codes Corp.) software. Sequence results for the ∼625-bp band were aligned and compared using Sequencher 4.6 (Gene Codes Corp.) software. All sediment viral sequences were subjected to several BLAST analyses (1). A translated query versus protein (BLASTx) was used for homology searches against the GenBank nr and environmental nonredundant (env-nr) databases. A translated query versus the translated database (tBLASTx) was used for homology searches against the GenBank nucleotide (nt) and environmental nucleotide (env-nt) databases as well as databases for three viral metagenomes: (i) Chesapeake Bay virioplankton (CBV) (3), (ii) Delaware soil viruses (DSV) (K. E. Wommack, S. R. Bench, K. E. Williamson, and M. Radosevich, presented at the International Symposium on Microbial Ecology, ISME-10, Cancun, Mexico, 22 to 27 August 2004), and (iii) other viral (OV) databases (4, 5, 8). Only BLAST results showing expectation values (E) of <0.001 were considered for phylogenetic and putative functional identification.
Nucleotide sequence accession numbers.
All sediment viral sequences were deposited in the GenBank database with accession numbers FJ640107 to FJ640554.
RESULTS
Banding patterns and sequences.
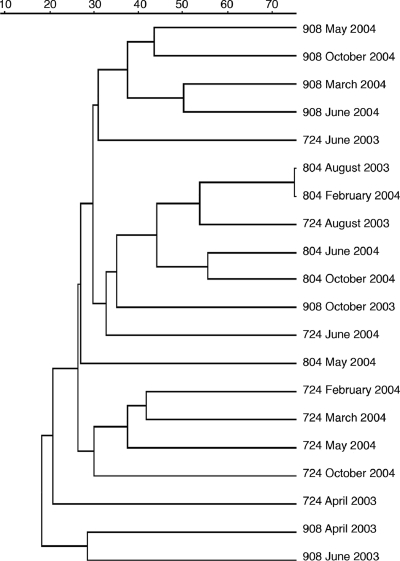
Nine RAPD-PCR 10-mer primers were tested on sediment viral extract concentrates (see Table S1 in the supplemental material), and only primer OPA-9 yielded banding patterns for each location and sampling date (see Fig. S1 in the supplemental material). To determine the accuracy and fidelity of viral-community RAPD-PCR, replicate samples from three stations were amplified in separate thermocycler reactions and the resulting banding patterns were compared. The level of similarity between replicate banding patterns ranged between 80% and 100% (see Fig. S2 in the supplemental material). In subsequent analyses of RAPD-PCR banding patterns, 80% or greater similarity was considered identical. Banding patterns from viral concentrates collected across the time series were compared by cluster analysis, which showed that sample location was the strongest factor determining the clustering of RAPD-PCR fingerprints (Fig. 1). The maximum similarity among nonreplicate sample banding patterns was 75% for station 804 between August 2003 and February 2004 samples. The average similarity across all banding patterns was 30%, indicating that many of the sediment samples shared more than one-half of their total amplicons. Viral assemblages from midbay station 804 showed the least change across the time series, with the majority of samples having ∼45% similarity in RAPD banding patterns. In contrast, sediments from upper bay station 908 had the greatest variability in viral assemblages (i.e., least overall similarity), with all samples showing slightly less than 20% similarity. Viriobenthos assemblages within sediments from station 724 also had a high degree of variability, with samples from 2003 scattered across several clades and separate from 2004 samples. Within each station, sediment viral assemblages from 2004 samples were typically more similar to one another than they were to 2003 samples, indicating a possible interannual change in the Chesapeake Bay viriobenthos. Samples that did not amplify or showed less than four bands per lane were excluded from analyses due to probable degradation of DNA or incomplete amplifications (n = 5 of 25). DNase-treated viral concentrates showed no loss of RAPD-PCR bands compared to untreated controls (data not shown) (36). Samples treated with heat plus DNase prior to RAPD-PCR also showed no loss of bands. However, a noticeable decrease in band intensity was observed.
FIG. 1.
Unweighted pair group method of averages tree of viriobenthos RAPD-PCR banding patterns. Jaccard's similarity scale is listed along the top.
BLAST homology analysis of RAPD-PCR amplicon sequences.
Although the bay stations are quite different according to chemical and physical composition (25), one recurring band of ∼625 bp was observed within 12 RAPD-PCR banding patterns from the total population of 20 samples (see Fig. S1 in the supplemental material). DNA within this band was cloned from the RAPD-PCR patterns of two samples (one each from stations 908 and 804). Subsequently, two clones from each of these bands were sequenced. Also included in this analysis was a single clone containing a sequence homologous to the ∼625-bp band that occurred within a clone library of RAPD-PCR amplicons from another station 804 sample. Alignment of these five amplicon sequences showed a 1.6% overall divergence at the nucleotide level, and each sequence gave the same top BLAST homolog when assessed against environmental sequence databases (CBV, E < 10−20; env-nt, E < 10−9; DSV, E < 10−4). No significant BLAST homology was found in searches against the GenBank nt/nr databases.
All RAPD-PCR sequences were compared to sequences in the GenBank nr and env-nr databases as well as three databases of viral metagenomic sequences. In all cases only those homologs showing a BLAST E value of <0.001 were considered significant. Median expectation values for all BLAST sequence homologs within each database and according to taxonomic classification (e.g., virus, bacteria, or environmental) are listed in Table 1. Of the 448 analyzed sediment viral sequences, 54% showed no significant homology to sequences within any of the subject BLAST databases (e.g., nr, nt, env-nr, env-nt, and viral metagenome databases). The frequencies of significant homology to sequences in the GenBank nr/nt and environmental databases were similar, 24 and 22%, respectively.
TABLE 1.
Log transformed median BLAST E values for RAPD-PCR sequences with significant BLAST homology (E < 0.001)
| Database or domain | Median E | No. of sequencesa | % of totala |
|---|---|---|---|
| GenBank nt/nr | −9.48 | 107 | 24 |
| Bacteria | −6.78 | (75) | (17) |
| Virus | −7.66 | (31) | (7) |
| Environmentalb | −8.00 | 97 | 22 |
| env-nt/env-nrc | −8.01 | (147) | (32) |
| CBV | −9.36 | (122) | (27) |
| DSV | −6.40 | (33) | (7) |
| OV | −7.54 | (86) | (19) |
| No homology | 244 | 54 | |
| Total | NAd | 448 | 100 |
Values in parentheses were not included in the total.
Each sequence was similar to at least one environmental database sequence (env-nt, env-nr, or viral metagenome) but showed no similarity to any sequence in the nt or nr database.
For each environmental database the number of sequences includes all sequences that had a significant BLAST homolog to that database regardless of hits to other databases.
NA, not applicable.
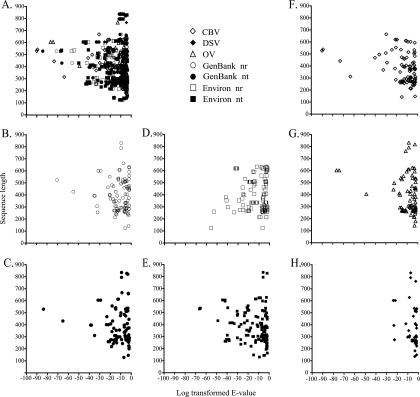
To determine whether sequence length significantly covaried with BLAST expectation value, the lengths of RAPD-PCR amplicons were plotted against E values of their best BLAST alignments. Overall, the mean read length for RAPD-PCR amplicon sequences showing significant BLAST homology was 359 bp, with an average 45% G+C content. However, when examined according to the subject database, RAPD-PCR average sequence length varied slightly: 409 bp for homologs to the nr/nt database, 404 bp for homologs to the env-nr/env-nt database, 411 bp for homologs to the OV metagenome, 390 bp for homologs to the CBV metagenome, and 388 bp for homologs to the DSV metagenome (Fig. 2). Longer sequence lengths did not appear to result in lower E values, as many of the longest RAPD-PCR sequences showed relatively high BLAST expectation values. Indeed, the lowest E values were seen among sequences near the average sequence length for the entire collection of RAPD-PCR amplicons.
FIG. 2.
Sequence length distribution of RAPD-PCR amplicons versus their log transformed E values (E less than 10−3). Overall average sequence length was 359 bp. (A) All databases combined; (B) GenBank nr database; (C) GenBank nt database; (D) env-nr database; (E) env-nt database; (F) CBV metagenome; (G) OV metagenome; (H) DSV metagenome.
The median E value for the collection of BLAST homologs to RAPD-PCR amplicons within each subject database or taxonomic domain provides a global indication of the genetic similarity between viriobenthos assemblages and a given collection of sequences. By these criteria, sequences within the CBV subject database showed better overall similarity (lowest log median E value) to viriobenthos RAPD-PCR sequences than those in any other environmental database (Table 1). Indeed, this median E value (log −9.36) was nearly as low as that for the GenBank nr/nt database (log −9.48), a database containing nearly 1,000-fold more sequences. In contrast to results for the CBV database, the median E value of BLAST homologs to metagenome sequences from the DSV database was the highest of all those from subject databases (log −6.40), broadly indicating the larger genetic distance between viriobenthos assemblages and soil viral assemblages.
Taxonomic and functional characterization of RAPD-PCR amplicon sequences.
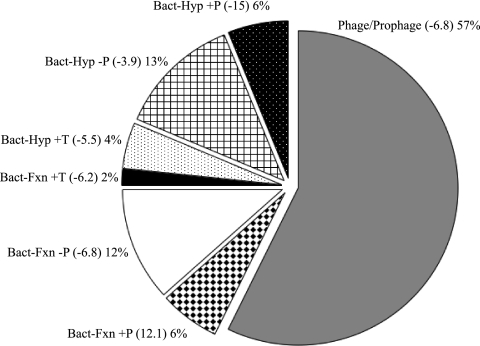
For each amplicon sequence, the top five hits were analyzed and the best hit was selected. Best hits were selected based on E value and most appropriate taxonomic identification. Analysis of significant homologs (E < 0.001) to sequences within GenBank (nr/nt database) showed that 29% of the GenBank (nr/nt database) sequence homologs were classified as viral and 70% bacterial. Among the RAPD-PCR amplicon sequences with a viral best hit, 46% belonged to bacteriophages within the order Caudovirales (i.e., the tailed phages) and another 11% were of algal virus origin (i.e., Phycodnaviridae). Analysis of viral homologous sequences showed that a large proportion of these belonged to prophages (34%), as well as the Podoviridae (24%) or Myoviridae (22%) viral families (see Fig. S3 in the supplemental material). Proteobacteria comprised the majority of bacterial best hits to RAPD-PCR amplicon sequences, of which most were from the class Alphaproteobacteria. Among the bacterial homologs, 57% were listed as prophage/phage related and 23% were classified as hypothetical or conserved hypothetical proteins. Of the hypothetical proteins, 6% were encoded by genes located within 60 kb of phage-related genes (e.g., terminase and integrase genes) and 4% near transposase genes within their host genomes (Fig. 3).
FIG. 3.
Comparison of protein homologs to sediment viral concentrate RAPD-PCR sequences (112 of a total of 448 sequenced). Phage, phage-related, or near-phage sequences comprised 69% of the total. Values in parentheses are median log transformed E values. Results are based on tBLASTx E values (E < 0.001). Abbreviations are as follows: Bact, bacteria; Hyp, hypothetical gene; Fxn, gene with defined function; +P, within 60 kb of phage-related gene(s); −P, no close proximity to phage-related gene(s); +T, within 60 kb of transposon-related gene(s).
Gene homologs of RAPD viral sequences are listed in Table 2 and in Table S2 in the supplemental material. The majority of protein homologs were to hypothetical proteins of unknown function in each domain, 51% of the total from viruses, prophages, and bacteria (Table 2). Replication, recombination and repair, and structural protein functional groups accounted for 19% of the homologs within viruses and prophages. Of the bacterial homologs, cell wall and membrane genes were the most dominant functional class. For putative gene identifications, lowest E values for virus sequences were for terminase (E = −31.5) and putative carbohydrate kinase (E = −35.4) (see Table S2 in the supplemental material). Among prophage sequences, a late control gene and a hypothetical protein had the lowest E values, −16.6 and −16.2, respectively. With an E value of −70.7, an acyl-coenzyme A synthetase sequence homolog was the most significant among the bacterial sequences; however, the gene for this homolog also happened to be near (±60 kb) a phage integrase gene.
TABLE 2.
Distribution of homologs to RAPD-PCR amplicon sequences according to protein functional groups and taxonomic domain
| Domain | Protein function | No. of homologs (% of domain) |
|---|---|---|
| Virus | Nucleotide transport and metabolism | 5 (16) |
| Replication, recombination, and repair | 6 (19) | |
| Structural | 6 (19) | |
| Unknown function | 13 (45) | |
| Prophage | Replication, recombination, and repair | 4 (19) |
| Structural | 6 (29) | |
| Unknown function | 10 (52) | |
| Bacteria | Energy production and conversion | 1 (2) |
| Cell wall/membrane/envelope biogenesis | 7 (13) | |
| Posttranslational modification, protein turnover, chaperones | 1 (2) | |
| Amino acid transport and metabolism | 2 (4) | |
| Cell motility | 1 (2) | |
| Carbohydrate transport and metabolism | 1 (2) | |
| Cell envelope biogenesis, outer membrane | 1 (2) | |
| Replication, recombination, and repair | 2 (4) | |
| Inorganic ion transport and metabolism | 5 (9) | |
| Intracellular trafficking and secretion/extracellular structures | 4 (7) | |
| Translation, ribosomal structure, and biogenesis | 1 (2) | |
| Unknown function | 28 (52) |
DISCUSSION
The objective of this study was to determine if RAPD-PCR could be an effective and useful method to evaluate the seasonal dynamics and preliminary metagenomic characterization of viruses in environmental samples, specifically in the eutrophic sediments of the Chesapeake Bay. The resulting banding patterns produced were robust and, most importantly, reproducible. In effect, RAPD-PCR provided a two-pronged approach for assessment of viral diversity in estuarine sediments: (i) at the community level, comparison of RAPD-PCR banding patterns allowed for inferences concerning the genetic similarity between sediment viral assemblages; (ii) at the level of individual viral genotypes, sequence analysis of RAPD-PCR amplicons provided a detailed view of the genetic composition of sediment viral assemblages.
RAPD-PCR profiling of viral communities in sediments.
Overall, it is apparent that RAPD banding patterns do not remain absolutely consistent over time at any given location, indicating that viriobenthic assemblages are temporally and geographically dynamic. The high viral production rates and short turnover times reported for many sediment environments (14, 27) lend further support to the notion that the composition of viriobenthos assemblages can change quickly. In contrast to water column environments where pulsed-field gel electrophoresis and RAPD-PCR analysis indicated seasonally linked changes in Chesapeake Bay virioplankton assemblages (36, 39), viriobenthos assemblages in the bay tended to change with station location. The most likely explanation for the variability in sediments from different stations is the spatial heterogeneity of the bay sediments. Unlike the water column, where a given volume of water is in constant motion, sediments are relatively stable and have limited movement compared to the water column. Even though sediments shift due to movements by benthic organisms (e.g., worms, bivalves) as well as resuspension and sedimentation of particulates, these changes are less obtrusive in the sediment environment than in the water column.
RAPD-PCR amplicon sequence analysis.
BLAST sequence homology searches for this study showed that over one-half of the sediment viral RAPD-PCR amplicon sequences were completely novel. This is a high proportion compared to the ca. 30% novel sequences typically reported for environmental double-stranded DNA viral metagenome sequence libraries (3, 4). However, the 24% frequency of BLAST-positive RAPD-PCR amplicon sequences against sequences within the GenBank nt/nr database is similar to that seen in other viral metagenomic studies that have used traditional Sanger sequencing. In many cases, RAPD-PCR amplicons from sediment viruses displayed significant BLAST homology to sequences within more than one database. In those instances, it is clear that frequently the best homologs to sediment amplicon sequences occur in the environmental and metagenomic databases and not in the known organismal databases. Homologs to bacterial functional genes were also found among RAPD-PCR amplicons, a finding echoed in several other metagenomic studies (see the review by Comeau et al. [12]). A large metagenomic study of virioplankton assemblages found a far higher proportion (91%) of unknown sequences (2), a result which was largely attributable to the short read length of sequences (∼100 bp) within those shotgun libraries. Recent in silico metagenome simulation experiments clearly demonstrated that short read sequencing technology performs especially poorly at characterizing the functional genetic diversity within virioplankton assemblages (37). Moreover, the low homolog detection frequency of short viral metagenome reads is not alleviated by the increases in sequence coverage newer technologies can provide.
It is likely that the high proportion of unknown sequences seen for this collection of RAPD-PCR amplicons can be explained, in part, by the relatively short average read lengths of these sequences. Nevertheless examination of the distribution of the sequence length versus significant BLAST expectation values indicated that longer sequence lengths did not necessarily yield lower expectation values (Fig. 2). Thus, even short reads can produce good quality sequence matches. Other studies which have examined the functional and taxonomic diversity of RAPD-PCR amplicons from viral assemblages though BLAST homology searches reported BLAST-positive and novel sequence frequencies similar to those seen here (35, 36). In the case of viral assemblages at deep-sea hydrothermal vents (35), the frequencies of BLAST-positive sequences with respect to novel sequences were similar for RAPD-PCR amplicon libraries and a small random shotgun sequence library which did not involve a selective PCR step. Overall, the similar behaviors of sediment RAPD-PCR sequences in BLAST analysis indicate that these amplicons were derived from viral assemblages and are a reasonable means to superficially survey the functional and taxonomic diversity within sediment viral assemblages. These findings also reinforce the idea that the majority of extant viral diversity in the biosphere is poorly characterized.
A preponderance of sequences with homology to genes belonging to members of the Podoviridae and Myoviridae families was seen within a metagenome library of Chesapeake Bay virioplankton (3) as well as in this study. This finding contrasts with previous viral metagenome studies from marine waters (8), coastal sediments (4), and equine feces (9) which found that the majority of viral BLAST hits belonged to phages within the Siphoviridae family (18). Phages within the Podoviridae and Myoviridae families are often considered to be virulent, while phages within the Siphoviridae family have been considered to be more often temperate (4). Thus, the observation of frequent homologs to sequences belonging to members of the Podoviridae and a low frequency of hits to Siphoviridae sequences indicate that the sediments of the Chesapeake Bay do not contain many phage families known to be temperate.
Persistence of identical RAPD-PCR bands indicates that the same viral genes can be found and are maintained within different sediment environments over time. Conserved genes exist among viral groups, and extensive gene transfer can occur among dissimilar viruses in the environment (26, 30). Whether this ∼625-bp sequence occurred within only one type of virus or within several different viruses cannot be unambiguously ascertained from this study. However, the possibility of finding a common viral gene within a large collection of viral assemblages is not unexpected, as previously reported data have shown that nearly identical T7-like DNA polymerase sequences can be detected in widely divergent environments (6). Moreover, diversity analyses based on metagenomic sequence data have reported that local-scale viral diversity is high but that diversity is more limited on a global scale (7). By extension, this result predicts that specific sequences can be found across a broad range of environments. Thus, there is a good likelihood that this ∼625-bp sequence is distributed across several different viruses and not limited to a single commonly occurring virus.
In the present study, RAPD-PCR has been successfully used to examine the composition of viriobenthos assemblages in sediment samples from the Chesapeake Bay. Diversity measurements based solely on morphological classification of viral particles or analysis of viral metagenome sequences lack the sensitivity, resolution, and/or sample throughput necessary for determining short-term changes in the composition and diversity of viriobenthos assemblages. As the data show, RAPD-PCR banding patterns can address these morphological shortcomings and thus provide the data needed to assess fine-scale synecological effects of viruses within sediment microbial environments.
Supplementary Material
Acknowledgments
This research was supported in part by an NSF Microbial Observatories grant (MCB-0132070) awarded to K.E.W.
We express our sincerest gratitude to S. W. Polson and J. Bhavsar for computational assistance, D. M. Winget and K. E. Williamson for helpful comments, the captain and crew of the R/V Cape Henlopen, Old Dominion University, for use of the multicorer device, and the staff of the DNA Sequencing & Genotyping Center at the Charles C. Allen, Jr., Biotechnology Laboratory at the University of Delaware.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bench, S. R., T. E. Hanson, K. E. Williamson, D. Gosh, M. Radosovich, K. Wang, and K. E. Wommack. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. London Ser. B Biol. Sci. 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart, M., J. H. Miyake, and F. Rohwer. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236:249-256. [DOI] [PubMed] [Google Scholar]
- 7.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cann, A. J., S. E. Fandrich, and S. Heaphy. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151-156. [DOI] [PubMed] [Google Scholar]
- 10.Chen, K., and L. Pachter. 2005. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Computat. Biol. 1:106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brussow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comeau, A. M., G. F. Hatfull, H. M. Krisch, D. Lindell, N. H. Mann, and D. Prangishvili. 2008. Exploring the prokaryotic virosphere. Res. Microbiol. 159:306-313. [DOI] [PubMed] [Google Scholar]
- 13.Culley, A. I., A. S. Lang, and C. A. Suttle. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054-1057. [DOI] [PubMed] [Google Scholar]
- 14.Danovaro, R., A. Dell'Anno, C. Corinaldesi, M. Magagnini, R. Noble, C. Tamburini, and M. Weinbauer. 2008. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454:1084-1087. [DOI] [PubMed] [Google Scholar]
- 15.Danovaro, R., A. Dell'Anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desnues, C., B. Rodriguez-Brito, S. Rayhawk, S. Kelley, T. Tran, M. Haynes, H. Liu, M. Furlan, L. Wegley, B. Chau, Y. Ruan, D. Hall, F. E. Angly, R. A. Edwards, L. Li, R. V. Thurber, R. P. Reid, J. Siefert, V. Souza, D. L. Valentine, B. K. Swan, M. Breitbart, and F. Rohwer. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340-343. [DOI] [PubMed] [Google Scholar]
- 17.Duhamel, S., and S. Jacquet. 2006. Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J. Microbiol. Methods 64:316-332. [DOI] [PubMed] [Google Scholar]
- 18.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 19.Farnell-Jackson, E. A., and A. K. Ward. 2003. Seasonal patterns of viruses, bacteria and dissolved organic carbon in a riverine wetland. Freshwater Biol. 48:841-851. [Google Scholar]
- 20.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filee, J., P. Forterre, T. Sen-Lin, and J. Laurent. 2002. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54:763-773. [DOI] [PubMed] [Google Scholar]
- 22.Fuller, N. J., W. H. Wilson, I. R. Joint, and N. H. Mann. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glud, R. N., and M. Middelboe. 2004. Virus and bacteria dynamics of a coastal sediment: implication for benthic carbon cycling. Limnol. Oceanogr. 49:2073-2081. [Google Scholar]
- 24.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helton, R. R., L. Liu, and K. E. Wommack. 2006. Assessment of factors influencing direct enumeration of viruses within estuarine sediments. Appl. Environ. Microbiol. 72:4767-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 27.Hewson, I., and J. A. Fuhrman. 2003. Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic waters. Microb. Ecol. 46:337-347. [DOI] [PubMed] [Google Scholar]
- 28.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 29.Middelboe, M., A. Hagstrom, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, J. Pinhassi, K. Simu, and M. G. Lorenz. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 30.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 31.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 33.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 34.Williamson, K. E., M. Radosevich, and K. E. Wommack. 2005. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71:3119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson, S. J., S. C. Cary, K. E. Williamson, R. R. Helton, S. R. Bench, D. Winget, and K. E. Wommack. 2008. Lysogenic virus-host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2:1112-1121. [DOI] [PubMed] [Google Scholar]
- 36.Winget, D. M., and K. E. Wommack. 2008. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 74:2612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wommack, K. E., J. Bhavsar, and J. Ravel. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 74:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wommack, K. E., R. T. Hill, M. Kessel, E. Russek-Cohen, and R. R. Colwell. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. L. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. J. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.