Abstract
A segregationally stable expression and secretion vector for Saccharomyces cerevisiae, named pYABD01, was constructed by cloning the yeast gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s) into the S. cerevisiae high-copy-number expression vector pYES2. The structural genes of the two leaderless peptides of enterocin L50 (EntL50A and EntL50B) from Enterococcus faecium L50 were cloned, separately (entL50A or entL50B) and together (entL50AB), into pYABD01 under the control of the galactose-inducible promoter PGAL1. The generation of recombinant S. cerevisiae strains heterologously expressing and secreting biologically active EntL50A and EntL50B demonstrates the suitability of the MFα1s-containing vector pYABD01 to direct processing and secretion of these antimicrobial peptides through the S. cerevisiae Sec system.
Lactic acid bacteria (LAB) are widely known for their ability to produce a variety of ribosomally synthesized proteins or peptides, referred to as bacteriocins, displaying antimicrobial activity against a broad range of gram-positive bacteria and, to a lesser extent, gram-negative bacteria, including spoilage and food-borne pathogenic microorganisms (11, 19, 33, 34, 36, 37). These antimicrobials may be classified into three main classes: (i) the lantibiotics, or posttranslationally modified peptides; (ii) the nonmodified, small, heat-stable peptides; and (iii) the large, heat-labile protein bacteriocins. Class II bacteriocins are further grouped into five subclasses: the subclass IIa (pediocin-like bacteriocins containing the N-terminal conserved motif YGNGVxC), the subclass IIb (two-peptide bacteriocins), the subclass IIc (leaderless bacteriocins), the subclass IId (circular bacteriocins), and the subclass IIe (other peptide bacteriocins) (17, 19, 21, 37). All lantibiotics and most class II bacteriocins are synthesized as biologically inactive precursors containing an N-terminal extension (the so-called double-glycine-type leader sequence or the Sec-dependent signal peptide), which is cleaved off concomitantly with externalization of biologically active bacteriocins by a dedicated ATP-binding cassette transporter and its accessory protein or by the Sec system and the signal peptidases, respectively (11, 17). Interestingly, only a few bacteriocins described to date are synthesized without an N-terminal extension, including enterocin L50 (L50A and L50B) (8), enterocin Q (EntQ) (10), enterocin EJ97 (41), and the bacteriocin LsbB (20).
In recent years, there has been an increasing interest in the application of bacteriocinogenic microorganisms and/or their bacteriocins as biopreservatives to guarantee the safety and quality of foods and beverages, such as fermented vegetables and meats, dairy and fish products, and wine and beer (12, 15, 16, 39, 47). Three main strategies for the use of bacteriocins as food biopreservatives have been proposed: (i) addition of a purified/semipurified bacteriocin preparation as a food additive; (ii) use of a substrate previously fermented by a bacteriocin-producing strain as a food ingredient; and/or (iii) inoculation of a culture to produce the bacteriocin in situ in fermented foods (13, 15). The lantibiotic nisin A is the most widely characterized bacteriocin and the only one that has been legally approved in more than 48 countries as a food additive for use in certain types of cheeses (13, 16). Likewise, nisin A has been approved as a beer additive in Australia and New Zealand (16). However, the difficulties encountered in addressing the regulatory approval of new bacteriocins as food additives have spurred the development of the other bacteriocin-based food biopreservation strategies (13, 17).
Beer is a beverage with a remarkable microbiological stability and is considered as a food substrate difficult to spoil. However, some LAB, such as Lactobacillus brevis, Lactobacillus lindneri, and Pediococcus damnosus, are able to spoil beer and are recognized as the most hazardous bacteria for breweries, being responsible for approximately 70% of microbial beer spoilage incidents (40, 47). The ever-growing consumer demand for less-processed and less chemically preserved foods and beverages is promoting the development of alternative biocontrol strategies, such as those based on the use of bacteriocins as biopreservatives (12, 15, 39, 47). However, beyond the strict requirements to fulfill legal regulations, the commercial application of bacteriocins as beer additives is hindered mainly by low bacteriocin production yields and increases in production costs (44). Considering that Saccharomyces cerevisiae is commonly used as starter culture for brewing (24, 28, 35), a novel beer biopreservation strategy based on the development of bactericidal S. cerevisiae brewing strains has been proposed to overcome the aforementioned challenges (44, 46, 47). In this respect, the heterologous production of LAB bacteriocins, namely, pediocin PA-1 (PedPA-1) from Pediococcus acidilactici PAC1.0 and plantaricin 423 from Lactobacillus plantarum 423, by laboratory strains of S. cerevisiae has been reported (44, 46).
Enterocin L50 (EntL50) is a commonly found bacteriocin composed of two highly related leaderless antimicrobial peptides, enterocin L50A (EntL50A) and enterocin L50B (EntL50B), which possesses a broad antimicrobial spectrum against LAB, food-borne pathogenic bacteria, and human and animal clinical pathogens (8, 9, 10, 11). Previous work by our group showed that EntL50 (EntL50A and EntL50B) may be used as a beer biopreservative to inhibit the growth of beer spoilage bacteria (1). Therefore, genetically engineered strains of S. cerevisiae heterologously expressing and secreting EntL50A and EntL50B have been developed in this work. For this purpose, we constructed the segregationally stable expression and secretion vector pYABD01, which allowed the secretion of biologically active EntL50A and EntL50B directed by MFα1s through the S. cerevisiae Sec system.
MATERIALS AND METHODS
Microorganisms, plasmids, media, and culture conditions.
The sources and relevant genotypes of the microorganisms and plasmids used in this work are listed in Table 1. The EntL50 (EntL50A and EntL50B)-producing strain Enterococcus faecium L50 (8) and the indicator strain P. damnosus CECT4797 (EntL50 sensitive; EntL50s) (1) were grown aerobically in MRS broth (pH 6.2; Oxoid Ltd., Basingstoke, United Kingdom) at 30°C. Escherichia coli cells (Invitrogen Life Technologies, Carlsbad, CA) were propagated in Luria-Bertani (LB) broth (Sigma-Aldrich Inc., St. Louis, MO) at 37°C with shaking (200 to 250 rpm). Kanamycin (50 μg/ml), ampicillin (Amp) (50 to 100 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (80 μg/ml) (Sigma-Aldrich) were added to LB medium for selection of E. coli transformants. S. cerevisiae INVSc1 (Invitrogen) was cultured in YPD medium (10 g/liter yeast extract [Oxoid], 20 g/liter peptone [Oxoid], and 20 g/liter glucose [Panreac Química S. A., Barcelona, Spain]) (44) at 30°C with shaking (200 to 250 rpm). Yeast transformants were grown in YPD medium supplemented with Amp (100 or 50 μg/ml in liquid or solid medium, respectively) and/or in synthetic complete minimal medium (SC) containing 0.67% (wt/vol) yeast nitrogen base (Invitrogen) without amino acids, 2% (wt/vol) glucose, and all the required growth factors except uracil (SC−Ura) (44). Solid media contained 1.5 or 2% (wt/vol) agar (Oxoid) for LAB and E. coli or S. cerevisiae, respectively. Soft MRS agar used in the spot-on-agar test (SPAT) described below contained 0.8% (wt/vol) agar.
TABLE 1.
Microorganisms and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or referenceb |
|---|---|---|
| Bacterial strains | ||
| E. faecium L50 | EntL50 (EntL50A and EntL50B), EntP, and EntQ producer | DNBTA |
| P. damnosus 4797 | Indicator microorganism, EntL50s-EntPs-EntQr | CECT |
| E. coli TOP10 | Host strain, F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| E. coli MAX Efficiency DH5α | Host strain, supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi−1 relA1 | Invitrogen |
| Yeast (S. cerevisiae) strains | ||
| INVSc1 | Host strain, MATα his3Δ1 leu2 trp1-289 ura3-52; His− Leu− Trp− Ura− | Invitrogen |
| INVSc1-α | S. cerevisiae INVSc1 derivative carrying pYABD01, Ampr | This work |
| L50A-20 | S. cerevisiae INVSc1 derivative carrying pYABD02, EntL50A producer, Ampr | This work |
| L50B-4 | S. cerevisiae INVSc1 derivative carrying pYABD03, EntL50B producer, Ampr | This work |
| L50AB-2 | S. cerevisiae INVSc1 derivative carrying pYABD04, EntL50A producer, Ampr | This work |
| Plasmids | ||
| PCR2.1-TOPO | 3.9-kb cloning vector, Ampr Kanr | Invitrogen |
| pPICZαA | P. pastoris 3.6-kb protein expression and secretion vector carrying a methanol-inducible promoter (PAOX1), 5′AOX1 region, MFα1s, Zeor | Invitrogen |
| pYES2 | S. cerevisiae 5.9-kb protein expression vector carrying galactose-inducible promoter PGAL1, Ura3, Ampr | Invitrogen |
| pRSETB-entL50A | pRSETB derivative carrying entL50A | 8 |
| pRSETB-entL50B | pRSETB derivative carrying entL50B | 8 |
| pTBS01 | PCR2.1-TOPO derivative carrying MFα1s | This work |
| pTBS02 | PCR2.1-TOPO derivative carrying entL50A | This work |
| pTBS03 | PCR2.1-TOPO derivative carrying entL50B | This work |
| pYABD01 | pYES2-derived expression and secretion vector carrying MFα1s | This work |
| pYABD02 | pYABD01 derivative carrying entL50A | This work |
| pYABD03 | pYABD01 derivative carrying entL50B | This work |
| pYABD04 | pYABD02 derivative carrying MFα1s fused in frame to entL50B | This work |
MFα1s, yeast gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s).
Abbreviations: CECT, Colección Española de Cultivos Tipo (Valencia, Spain); DNBTA, Departamento de Nutrición, Bromatología y Tecnología de los Alimentos, Facultad de Veterinaria, Universidad Complutense de Madrid (Madrid, Spain).
DNA isolation and manipulation.
Small- and large-scale plasmid DNA isolation from E. coli recombinants was carried out using the High Pure plasmid isolation kit (Roche Diagnostics SL, Madrid, Spain) and the QIAgen Plasmid Midi kit (Qiagen GmbH, Hilden, Germany), respectively. Total genomic DNA from recombinant yeasts was isolated using the Wizard DNA Purification kit (Promega Corporation, Madison, WI). Oligonucleotide primers (Table 2) were obtained from Sigma-Genosys Ltd. (Cambridge, United Kingdom). For PCR amplifications, samples were subjected to an initial cycle of denaturation (97°C for 2 min), followed by 35 cycles of denaturation (94°C for 45 s), annealing (50 to 56°C for 30 s), and elongation (72°C for 30 to 60 s), ending with a final extension step at 72°C for 7 min. The PCR-generated fragments were extracted from 0.8% (wt/vol) agarose gels using the Real Clean Matrix kit (Durviz SLU, Madrid, Spain) and purified using the QIAquick PCR purification kit (Qiagen). Nucleotide sequencing of both strands of purified PCR products was done at the DNA Sequencing Service of Sistemas Genómicos (Valencia, Spain). Platinum Taq polymerase (Invitrogen), DNA restriction enzymes (New England Biolabs Inc., Beverly, MA), and T4 DNA ligase (Promega) were used as suggested by the manufacturers. E. coli TOP10 and E. coli MAX Efficiency DH5α competent cells, as well as S. cerevisiae INVSc1 competent cells from the S.c. EasyCom transformation kit, were transformed according to the supplier's instructions (Invitrogen).
TABLE 2.
Primers and PCR products used in this study
| Primer or PCR product | Nucleotide sequence (5′-3′)a or PCR product descriptionb | Fragment(s) amplified |
|---|---|---|
| Primers | ||
| Alfa1-HindIII | AAGCTTACGATGAGATTTCCTTCAATTTTTACTGC | α1 |
| Alfa3-XbaI | TCTAGAAAGCTGGCGGCCGCCGCGGCTC | α1 |
| Alfa4-SmaI | CCCGGGACGATGAGATTTCCTTCAATTTTTACTG | α-L50B |
| L50A1-XhoI | CTCGAGAAAAGAATGGGAGCAATCGCAAAATTAGTAGCAAAG | L50A |
| L50A2-XbaI | TCTAGAGTATAACCCCCGGGATTTTAAATATGTTTTTTAATCCACTCAATG | L50A |
| L50B1-XhoI | CTCGAGAAAAGAATGGGAGCAATCGCAAAACTAGTGAC | L50B |
| L50B2-XbaI | TCTAGAAACATTAATGTCTTTTTAGCCATTTTTCAATTTGATC | L50B, α-L50B |
| pYES2-F | AACCCCGGATCGGACTACTAG | α2 |
| pYES2-R | GGCGTGAATGTAAGCGTGAC | α2 |
| PCR products | ||
| Fragment α1 | 345-bp HindIII-XbaI fragment containing the Kozak sequence fused to MFα1s | |
| Fragment α2 | 442-bp fragment containing the Kozak sequence fused to MFα1s | |
| Fragment L50A | 170-bp XhoI-XbaI fragment containing MFα1s fused to entL50A | |
| Fragment L50B | 154-bp XhoI-XbaI fragment containing MFα1s fused to entL50B | |
| Fragment α-L50A | 422-bp HindIII-XbaI fragment containing MFα1s fused to entL50A | |
| Fragment α-L50B | 406-bp SmaI-XbaI fragment containing MFα1s fused to entL50B |
Cleavage sites for restriction enzymes are underlined; Kozak sequence (ACGATGA), and the nucleotides encoding the Kex2 signal cleavage site (AAAAGA) of the mating pheromone α-factor 1 secretion signal (MFα1s) are shown in bold.
MFα1s refers to the yeast gene region encoding MFα1s including the Kex2 cleavage site.
Construction of protein expression and secretion vector pYABD01 for S. cerevisiae.
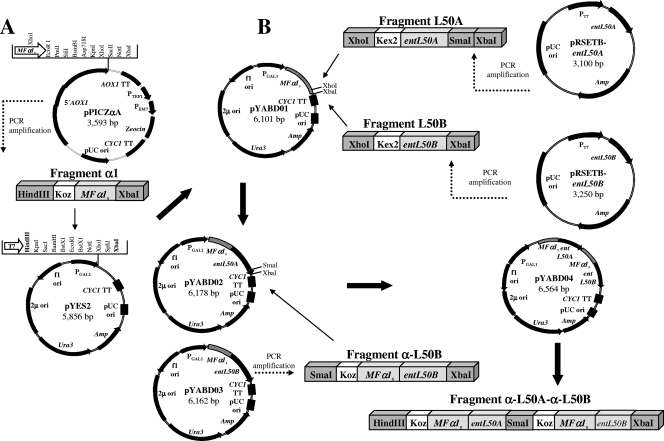
The strategy employed for construction of pYABD01 is depicted in Fig. 1. The Pichia pastoris expression and secretion vector pPICZαA was used as the template for PCR amplification of fragment α1 containing a Kozak translation initiation sequence (ACGATGA) in addition to the gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s), which includes the Kex2 signal cleavage site (Glu-Lys-Arg) required for processing of the fusion protein. The purified fragment α1 was cloned into PCR2.1-TOPO (Invitrogen), generating the plasmid pTBS01, which was chemically transformed into E. coli TOP10 cells. Plasmid pTBS01 was digested with HindIII and XbaI and the resulting 339-bp fragment purified from the agarose gel and ligated with T4 DNA ligase into the HindIII and XbaI sites of the S. cerevisiae protein expression vector pYES2 to give plasmid pYABD01, in which fragment α1 is under the control of the PGAL1 promoter (induced by galactose and repressed by glucose) and the corresponding enhancer sequences. The recombinant plasmid was chemically transformed into E. coli MAX Efficiency DH5α cells and transformants selected on LB plates with Amp (50 μg/ml) at 37°C for 24 h. The plasmid pYES2 was also chemically transformed into this host, and the resulting strain was used as a control. The presence of the plasmid pYABD01 in transformed cells was confirmed by PCR and plasmid isolation and restriction analysis, and the correct nucleotide sequence of pYABD01 was confirmed by DNA sequencing as described above. Primers used for PCR amplification and DNA sequencing were designed from the nucleotide sequence of plasmids pPICZαA and pYES2 (Invitrogen website) (Table 2).
FIG. 1.
(A) Construction of the S. cerevisiae protein expression and secretion vector pYABD01, derived from pPICZαA and pYES2, containing the yeast gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s), including the nucleotides encoding the Kex2 signal cleavage site under the control of the galactose-inducible promoter PGAL1. (B) Construction of the recombinant plasmids pYABD02, pYABD03, and pYABD04, derived from pYABD01, containing MFα1s fused in frame to entL50A and/or entL50B. Plasmid sizes are given in base pairs. Only relevant restriction enzyme sites are shown. 5′AOX1, promoter region; AOX1 TT, transcription termination; PTEF1, transcription elongation factor 1, driving expression of the Sh ble gene in Pichia; PEM7, constitutive promoter driving expression of the Sh ble gene in E. coli; Zeocin gene, zeocin resistance (Sh ble gene); CYC1 TT, transcription terminator; pUC ori, maintenance and high-copy-number replication in E. coli; Amp, ampicillin resistance gene; Ura3, gene for selection of yeast transformants in uracil-deficient medium; 2μ ori, maintenance and high-copy-number replication in yeast; f1 ori, rescue of single-stranded DNA; entL50A, structural gene of EntL50A; entL50B, structural gene of EntL50B; PT7, T7 promoter driving expression of heterologous gene expression; Koz, Kozak translation initiation sequence of yeasts.
In order to assess the segregational stability of pYABD01 (carrying Ura3), this plasmid was transformed into S. cerevisiae INVSc1 (auxotrophic for Ura) competent cells, and the resulting S. cerevisiae INVSc1-α(pYABD01) transformant (see below) was grown in SC−Ura at 30°C overnight (optical density at 600 nm [OD600] of 2.0). Subsequently, the culture was transferred to YPD broth (initial inoculum of approximately 1 × 105 CFU/ml) and further incubated for 50 generations (seven serial culture transfers using 0.5% inocula every 24 h) to determine the plasmid stability without selective pressure during continuous exponential growth phase. At 24-h intervals, samples were withdrawn from the culture for bacterial counting on SC−Ura and YPD plates.
Cloning of EntL50A (entL50A) and EntL50B (entL50B) structural genes, separately and together, in pYABD01.
The strategy employed for cloning of entL50A and entL50B in pYABD01 is summarized in Fig. 1. The recombinant plasmids pRSETB-entL50A and pRSETB-entL50B, previously constructed (8), were used as DNA templates for PCR amplification of fragments L50A and L50B carrying the nucleotide sequence encoding the Kex2 signal cleavage site (AAAAGA) of MFα1s fused to entL50A and entL50B, respectively. The purified fragments L50A and L50B were cloned into the plasmid pYABD01, essentially as described above, generating the recombinant plasmids pYABD02 and pYABD03, respectively. In these plasmids, entL50A and entL50B are fused in frame to MFα1 and gene expression is under the control of PGAL1. The recombinant plasmids were chemically transformed into E. coli MAX Efficiency DH5α cells and S. cerevisiae INVSc1 cells, and the resulting S. cerevisiae L50A and S. cerevisiae L50B transformants were selected on SC−Ura plates at 30°C for 2 to 4 days. In order to express and secrete EntL50A and EntL50B simultaneously, the recombinant plasmid pYABD04 was constructed and analyzed essentially as described for pYABD02 and pYABD03 (Fig. 1). Briefly, the fragment α-L50B, which contains a Kozak sequence and MFα1 fused to entL50B, was cloned into pYABD02 downstream of the fragment α-L50A, generating the plasmid pYABD04. This plasmid was transformed into S. cerevisiae INVSc1 competent cells, generating S. cerevisiae L50AB transformants. The plasmid pYABD01, lacking the bacteriocin structural genes, was also transformed into S. cerevisiae INVSc1, and the resulting S. cerevisiae INVSc1-α strain was used as a control.
The presence of pYABD01 in S. cerevisiae INVSc1-α, pYABD02 in S. cerevisiae L50A, pYABD03 in S. cerevisiae L50B, and pYABD04 in S. cerevisiae L50AB transformants was confirmed by PCR and plasmid isolation and restriction analysis, and the correct nucleotide sequences of the recombinant plasmids were confirmed by DNA sequencing as described above. Primers used for PCR amplification and DNA sequencing were designed from the published nucleotide sequence of the EntL50 (EntL50A and EntL50B) operon (8) and plasmid pPICZαA (Table 2).
Detection and quantification of EntL50A and EntL50B heterologous production by antimicrobial and immunochemical assays.
The direct antimicrobial activities of cultures from several S. cerevisiae L50A(pYABD02) and S. cerevisiae L50B(pYABD03) transformants were screened by using a SPAT essentially as described previously (7, 44). Briefly, transformants were precultured in both YPD and SC−Ura broth overnight (OD600 of 2.0). Subsequently, equal amounts (20 μl) of yeast precultures were spotted onto plates of both media supplemented with 20 g/liter galactose (Merck Farma y Química S. A., Barcelona, Spain) and 10 g/liter raffinose (Sigma-Aldrich) (YPGR and SCGR, respectively), and incubated at 30°C for 30 days. Periodically, one of the plates was overlaid with MRS soft agar previously seeded with approximately 1 × 105 CFU/ml of the indicator microorganism P. damnosus CECT4797 and further incubated for 16 h. After incubation, the direct antimicrobial activity was detected by the presence of growth inhibition zones of the indicator microorganism around the spotted producer strain. Likewise, the direct antimicrobial activities of cultures from several S. cerevisiae L50AB(pYABD04) transformants were screened as described above but only on SCGR plates.
In order to determine bacteriocin production kinetics, selected S. cerevisiae L50A(pYABD02) and S. cerevisiae L50B(pYABD03) transformants, as well as S. cerevisiae L50AB(pYABD04) transformants, were precultured in YPD and SC−Ura broth or in SC−Ura broth, respectively. Cells were harvested by centrifugation (5,000 × g at 4°C for 10 min), washed with YPD or SC−Ura (both without glucose), and resuspended to an OD600 of 2.0 in YPGR and SCGR broth. These recombinant yeasts were grown in YPGR and SCGR broth or in SCGR broth at 30°C for 8 days. Yeast growth (OD600), cell dry weight (CDW), and the bacteriocin activity and concentration were determined periodically in duplicate. Supernatants were obtained by centrifugation (12,000 × g at 4°C for 10 min), further adjusted to pH 6.2, and filter sterilized through 0.22-μm-pore-size filters (Millipore Corp., Bedford, MA). Subsequently, aliquots of the supernatants were lyophilized and further dissolved to 1/20 of their original volume in 20 mM phosphate buffer (pH 6.0). For determination and quantification of bacteriocin activity, supernatants and 20-fold-concentrated supernatants (CS) were analyzed by an agar well diffusion test (ADT) (7) and a microtiter plate assay (MPA) (10, 25), respectively, using P. damnosus CECT4797 as the indicator microorganism. One bacteriocin unit (BU) was defined as the highest dilution of the supernatant causing 50% growth inhibition of the indicator microorganism. Cultures and supernatants from the S. cerevisiae INVSc1-α (pYABD01) transformants, lacking the bacteriocin structural genes, were used as negative controls in the antimicrobial assays.
For quantification of EntL50A and EntL50B heterologous production in the supernatants of the recombinant yeasts, a noncompetitive indirect enzyme-linked immunosorbent assay (NCI-ELISA) using rabbit polyclonal antibodies with specificity for EntL50A (anti-LR1-KLH) and EntL50B (anti-LR2-KLH) (14) was performed essentially as described previously (14, 22). The ELISA plates included control wells coated with (i) 0.1 M sodium carbonate-bicarbonate buffer (pH 9.6) (coating buffer [CB])-MRS, CB-YPGR, and CB-SCGR broth to set the plate background level and (ii) six twofold dilutions of samples containing known concentrations of pure EntL50A and EntL50B (determined from the A280 using the molar extinction coefficient) in CB-MRS, CB-YPGR, and CB-SCGR broth to set plate standard curves. Supernatants from S. cerevisiae INVSc1-α (pYABD01) were used as negative controls in the immunochemical assays.
To measure the synergistic activities of heterologously produced EntL50A and EntL50B, supernatants from the S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) strains were challenged against P. damnosus CECT4797, separately and in combination, to achieve a 1:1 bacteriocin peptide ratio, by a SPAT and an MPA.
Purification and mass spectrometry analysis of recombinant EntL50A and EntL50B.
EntL50A and EntL50B, heterologously produced by S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03), respectively, were purified essentially as described previously (7). Briefly, yeast precultures grown in SC−Ura broth at 30°C were used to inoculate 1-liter cultures that were grown in SCGR broth at 30°C with shaking until maximum EntL50A and EntL50B concentrations were achieved (ca. 144 and 120 h, respectively). After the cells were removed by centrifugation (12,000 × g at 4°C for 30 min), the supernatants were subjected to precipitation with ammonium sulfate (50% [wt/vol]; Merck) and subsequently desalted by gel filtration (PD-10 columns; GE Healthcare Life Sciences, Barcelona, Spain). The obtained fractions were further subjected to cation exchange (SP Sepharose Fast Flow) and hydrophobic interaction (Octyl Sepharose CL-4B) chromatographies, followed by reversed-phase chromatography (PepRPC HR 5/5) in a fast-protein liquid chromatography (FPLC) system (GE Healthcare Life Sciences). The antimicrobial activities of the fractions obtained during the purification procedure were determined by an MPA using P. damnosus CECT4797 as the indicator microorganism. Fractions displaying a high and specific bacteriocin activity were pulled together and rechromatographed on the same reversed-phase column until chromatographically pure bacteriocin peptides were obtained. The concentrations of EntL50A and EntL50B in the supernatants and the fractions obtained at the end of the purification procedure were determined by an NCI-ELISA.
The purified recombinant bacteriocin peptides were subjected to mass spectrometry analysis by using a matrix-assisted laser desorption ionization-time-of-flight Voyager-DE STR mass spectrometer (MALDI-TOF MS) (PerSeptive Biosystems, Foster City, CA) at the mass spectrum service of the Centro de Genómica y Proteómica, UCM (Madrid, Spain).
Tricine-SDS-PAGE, Western blotting, and gel overlay assay.
FPLC-purified EntL50A and EntL50B were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 16% (wt/vol) Tricine gels (Tricine-SDS-PAGE) (42) after silver staining. Western blotting using anti-LR1-KLH (specific for EntL50A) and anti-LR2-KLH (specific for EntL50B) antibodies was performed essentially as described previously (22). To determine the antimicrobial activities of purified recombinant EntL50A and EntL50B, a gel overlay assay (3) was performed using P. damnosus CECT4797 as the indicator microorganism.
RESULTS
Cloning of entL50A and entL50B, separately and together, into the S. cerevisiae protein expression and secretion vector pYABD01.
The MFα1s-containing pYABD01 vector (6,101 bp) constructed in this work (Fig. 1) was maintained by approximately 100% of the S. cerevisiae INVSc1-α(pYABD01) cells grown for at least 50 generations in the absence of selective pressure (results not shown). The bacteriocin structural genes entL50A and entL50B were cloned, separately and together, into pYABD01, resulting in pYABD02, pYABD03, and pYABD04, respectively (Fig. 1). Specific PCR amplifications, as well as restriction analysis and DNA sequencing, confirmed the correct construction of the plasmids obtained in this work (results not shown). In the case of the recombinant plasmid pYABD04, several attempts to sequence the fragment α-L50A-α-L50B were made, but the sequencing signal was lost immediately after the last codon of MFα1s fused to entL50A. In order to overcome this problem, pYABD04 was digested with HindIII-SmaI and SmaI-XbaI, generating the fragments α-L50A and α-L50B, respectively, which were sequenced and shown to contain the correct sequences.
Heterologous expression and secretion of biologically active EntL50A and EntL50B by S. cerevisiae.
S. cerevisiae L50A(pYABD02) and S. cerevisiae L50B(pYABD03) cultures spotted onto YPGR and SCGR plates and grown at 30°C for 30 days showed no direct antimicrobial activity. However, the supernatants and/or CS from cultures of these recombinant strains grown at 30°C in YPGR and SCGR broth for 8 days displayed antimicrobial activity (results not shown). It is pertinent to note that no antimicrobial activity was found in the supernatants and CS from control cultures of S. cerevisiae INVSc1-α(pYABD01) (results not shown), which rules out the possibility that the extracellular antimicrobial activity exerted by S. cerevisiae L50A(pYABD02) and S. cerevisiae L50B (pYABD03) transformants was due to metabolites other than bacteriocins. The S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) transformants were selected for further microbiological and immunochemical quantification of recombinant EntL50A and EntL50B production, respectively (Table 3). The antimicrobial activities of the supernatants and CS from cultures of S. cerevisiae L50A-20(pYABD02) grown in the minimal medium SCGR were first detected 72 and 24 h after induction of gene expression (at time zero), respectively, and the maximum antimicrobial activity was found after incubation for 144 h (3.7 and 255 BU/mg CDW, respectively) (results not shown and Table 3). Although no antimicrobial activity was detected in the supernatants from S. cerevisiae L50B-4(pYABD03) grown in the same medium up to 192 h, the CS of this recombinant strain displayed antimicrobial activity, which was first detected at 24 h of incubation and maximal after 120 h (165 BU/mg CDW). The presence of the recombinant bacteriocin peptides in the supernatants from these transformants and their absence in those from S. cerevisiae INVSc1-α(pYABD01) were demonstrated by an NCI-ELISA. The production of EntL50A and EntL50B by S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03), respectively, started immediately after induction of gene expression and was parallel to cell growth, and the bacteriocin peptides reached their maximum concentration at the beginning of the stationary phase or at the late stage of the exponential growth phase, respectively (Table 3). In this respect, the maximum concentrations of EntL50A (19.3 ng/ml or 8.4 ng/mg CDW) and EntL50B (52.5 ng/ml or 24.0 ng/mg CDW) in the supernatants from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) were detected at 144 and 120 h of incubation, respectively. The maximum specific activity of EntL50A in the CS from S. cerevisiae L50A-20(pYABD02) (30 BU/ng EntL50A) was about fourfold higher than that of EntL50B in the CS from S. cerevisiae L50B-4(pYABD03) (7.1 BU/ng EntL50B). On the other hand, these transformants grew much faster and to a higher cell density in the complex medium YPGR (OD600 of 8.5) than in SCGR (OD600 of 4.8 to 5.1). However, the extracellular antimicrobial activity of S. cerevisiae L50A-20(pYABD02) was lower than that in SCGR and was detected only in the CS, and no antimicrobial activity was found either in the supernatants or in the CS from S. cerevisiae L50B-4(pYABD03) (results not shown). The antimicrobial activity and production of EntL50A by S. cerevisiae L50A-20(pYABD02) started 48 h after gene expression induction, corresponding to the early stationary phase of growth in YPGR. The maximum antimicrobial activity (diameter of inhibition zone of 9.3 mm) and EntL50A concentration (15.0 ng/ml or 5.2 ng/mg CDW) were detected after incubation for 96 h. Surprisingly, notwithstanding the fact that S. cerevisiae L50B-4(pYABD03) did not show antimicrobial activity in YPGR, EntL50B was detected in the corresponding supernatants and reached its maximum concentration (56.3 ng/ml, corresponding to 19.1 ng/mg CDW) after 6 days of incubation. Moreover, it is interesting to note that the antimicrobial activity of these EntL50A and EntL50B concentrations was lower than that exerted by similar peptide concentrations (3.6 to 6.3 and 17.1 to 20.0 ng/mg CDW, respectively) produced in SCGR (diameters of 16.7 to 18.1 and 13.4 to 13.7 mm, respectively) (Table 3).
TABLE 3.
Production and antimicrobial activity of recombinant EntL50A and EntL50B from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) cultures, respectivelya
| Time (h) |
S. cerevisiae L50A-20(pYABD02)
|
S. cerevisiae L50B-4(pYABD03)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD600b | EntL50A concn inc:
|
Antimicrobial activity as assessed by:
|
SAAf (BU/ng EntL50A) | OD600 | EntL50B concn inc:
|
Antimicrobial activity as assessed by:
|
SAAf (BU/ng EntL50B) | |||||||||
| ADT (mm)d
|
MPA ine:
|
ADT (mm)d
|
MPA ine:
|
|||||||||||||
| ng/ml | ng/mg CDW | S | CS | BU/ml | BU/mg CDW | ng/ml | ng/mg CDW | S | CS | BU/ml | BU/mg CDW | |||||
| 0 | 2.0 | NDd | ND | ND | ND | ND | ND | NE | 2.0 | ND | ND | ND | ND | ND | ND | NE |
| 2 | 2.0 | 0.7 | 0.4 | ND | ND | ND | ND | NE | 2.0 | 0.3 | 0.2 | ND | ND | ND | ND | NE |
| 8 | 2.1 | 1.5 | 0.9 | ND | ND | ND | ND | NE | 2.0 | 7.2 | 4.2 | ND | ND | ND | ND | NE |
| 12 | 2.5 | 1.6 | 0.9 | ND | ND | ND | ND | NE | 2.1 | 9.8 | 5.7 | ND | ND | ND | ND | NE |
| 18 | 3.4 | 1.7 | 0.9 | ND | ND | ND | ND | NE | 2.5 | 13.4 | 7.4 | ND | ND | ND | ND | NE |
| 24 | 4.4 | 4.6 | 2.1 | ND | 12.4 s2 | 650 | 15 | 7 | 3.9 | 17.8 | 8.4 | ND | 11.1 s2 | 600 | 15 | 1.8 |
| 36 | 4.5 | 4.8 | 2.2 | ND | 16.3 s2 | 900 | 20 | 9 | 4.3 | 24.4 | 11.1 | ND | 13.2 s2 | 1,400 | 30 | 2.7 |
| 48 | 4.6 | 5.0 | 2.3 | ND | 16.5 s2 | 1,900 | 45 | 20 | 4.3 | 37.5 | 17.1 | ND | 13.4 s2 | 1,900 | 45 | 2.6 |
| 72 | 4.6 | 8.0 | 3.3 | 8.2 s | 16.7 s2 | 2,100 | 50 | 14 | 4.4 | 44.5 | 20.0 | ND | 13.7 s2 | 3,200 | 70 | 3.5 |
| 96 | 4.9 | 7.6 | 3.6 | 8.2 s | 16.7 s2 | 3,500 | 75 | 23 | 4.4 | 52.2 | 23.4 | ND | 13.9 s2 | 4,800 | 110 | 4.6 |
| 120 | 4.9 | 14.4 | 6.3 | 9.5 s | 18.1 s2 | 4,900 | 110 | 17 | 4.5 | 52.5 | 24.0 | ND | 16.1 s2 | 7,500 | 165 | 7.1 |
| 144 | 5.0 | 19.3 | 8.4 | 10.4 s | 18.9 s2 | 11,600 | 255 | 30 | 4.7 | 51.0 | 22.4 | ND | 15.2 s2 | 3,500 | 75 | 3.3 |
| 168 | 5.0 | 14.4 | 6.3 | 10.3 s | 18.6 s2 | 8,000 | 175 | 28 | 4.8 | 42.3 | 18.4 | ND | 14.7 s2 | 3,700 | 80 | 4.3 |
| 192 | 5.1 | 10.9 | 4.7 | 9.0 s | 17.8 s2 | 6,000 | 130 | 28 | 4.8 | 40.5 | 17.6 | ND | 14.1 s2 | 2,700 | 60 | 3.4 |
Cultures were grown in SCGR at 30°C.
Gene expression was induced at time zero.
Calculated by using an NCI-ELISA using specific polyclonal antibodies. ND, no bacteriocin detected.
Antimicrobial activity against P. damnosus CECT4797 as determined by an ADT. Inhibition zones are differentiated as follows: s, sharp; s2, extremely sharp. ND, no inhibition zone detected using 50 μl of supernatant (S) or 20-fold-concentrated supernatant (CS).
Antimicrobial activity of CS against P. damnosus CECT4797 as determined by an MPA. ND, no inhibition detected using 100 μl of CS.
Specific antimicrobial activity, i.e., the antimicrobial activity of CS (BU/mg CDW), calculated by an MPA, divided by the EntL50A or EntL50B concentration (ng/mg CDW). NE, not evaluable.
The simultaneous production of EntL50A and EntL50B by a single yeast strain was investigated using S. cerevisiae L50AB (pYABD04) transformants. As in the case of S. cerevisiae L50A (pYABD02) and S. cerevisiae L50B(pYABD03) transformants, no direct antimicrobial activity was detected in cultures spotted onto SCGR plates and grown for up to 30 days (results not shown). However, bacteriocin production was detected and quantified by antimicrobial and immunochemical assays in the supernatants and CS from cultures of the selected transformant S. cerevisiae L50AB-2(pYABD04) grown in SCGR broth for 8 days. The antimicrobial activities of the supernatants and CS were first detected 24 and 8 h after induction of gene expression (at time zero), respectively, and the maximum antimicrobial activities were found after incubation for 144 h (4.4 and 303 BU/mg CDW, respectively). The maximum EntL50A concentration and specific activity in the supernatants and CS of S. cerevisiae L50AB-2(pYABD04) were 24 ng/ml (10.4 ng/mg CDW) and 29 BU/ng EntL50A, respectively, but strikingly, EntL50B was not produced.
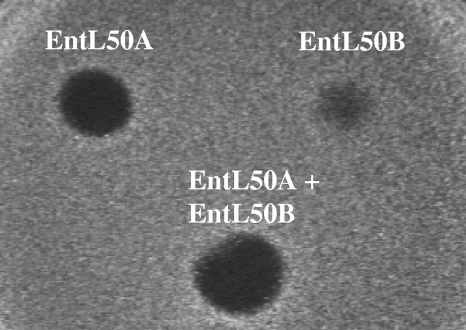
In order to determine whether EntL50A and EntL50B could be produced in a mixed culture, S. cerevisiae L50A-20 (pYABD02) and S. cerevisiae L50B-4(pYABD03) were cocultured in SCGR broth at a 1:1 ratio at 30°C for 8 days. Although growth was similar to that obtained when yeasts were grown independently, the antimicrobial activity of the nonconcentrated coculture supernatants was detected earlier and its maximum value was higher (Table 4). In this respect, the maximum antimicrobial activity of the supernatants, found after 96 h of incubation, was 170 BU/mg CDW, corresponding to 10.3 ng/ml (4.7 ng/mg CDW) of EntL50A and 46.4 ng/ml (21.7 ng/mg CDW) of EntL50B. Interestingly, the independent antimicrobial activities of similar amounts of EntL50A (3.6 to 6.3 ng/mg CDW) and EntL50B (20 to 24 ng/mg CDW) in the CS from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4 (pYABD03), respectively, were 70 to 110 BU/mg CDW. These results demonstrate the production of both EntL50A and EntL50B by the coculture and suggest that the recombinant bacteriocin peptides act synergistically. To further confirm and quantify this synergistic effect, supernatants from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) were challenged against P. damnosus CECT4797 independently and mixed in a 1:1 bacteriocin peptide ratio. As expected, the equimolar mixture of EntL50A and EntL50B exerted a greater antimicrobial effect (133 BU/mg CDW) than the additive effect of the peptides acting independently (53 and 23 BU/mg CDW, respectively), demonstrating a degree of synergy of approximately 1.8-fold (Fig. 2).
TABLE 4.
Production and antimicrobial activity of recombinant EntL50A and EntL50B from an S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) coculturea
| Time (h) | OD600b | Bacteriocin peptide concn inc:
|
Antimicrobial activity as assessed by:
|
|||||
|---|---|---|---|---|---|---|---|---|
| ng/ml
|
ng/mg CDW
|
ADT (mm)d | MPA ine:
|
|||||
| EntL50A | EntL50B | EntL50A | EntL50B | BU/ml | BU/mg CDW | |||
| 0 | 1.9 | ND | ND | ND | ND | ND | ND | ND |
| 24 | 4.0 | 3.3 | 20.5 | 1.6 | 9.7 | 7.9 s | 40 | 20 |
| 48 | 4.2 | 6.8 | 42.0 | 3.1 | 19.1 | 8.1 s | 70 | 30 |
| 72 | 4.3 | 8.8 | 44.3 | 4.0 | 20.0 | 10.2 s | 310 | 140 |
| 96 | 4.4 | 10.3 | 46.4 | 4.7 | 21.7 | 10.7 s | 360 | 170 |
| 120 | 4.4 | 9.4 | 55.3 | 4.2 | 24.8 | 10.3 s | 320 | 140 |
| 144 | 4.5 | 8.4 | 45.7 | 3.7 | 20.4 | 9.8 s | 260 | 120 |
| 168 | 4.5 | 7.1 | 36.0 | 3.2 | 16.1 | 9.3 s | 210 | 90 |
S. cerevisiae L50A-20(pYABD02) and L50B-4(pYABD03) were grown at a 1:1 ratio in SCGR broth at 30°C.
Gene expression was induced at time zero.
EntL50A and EntL50B concentration calculated by an NCI-ELISA using specific polyclonal antibodies for EntL50A or EntL50B, and expressed as ng/ml and ng/mg CDW. ND, no bacteriocin detected.
Antimicrobial activity against P. damnosus CECT4797 as determined by an ADT. Inhibition zones are differentiated as in Table 3. ND, no inhibition zone detected using 50 μl of supernatant.
Antimicrobial activity of supernatant against P. damnosus CECT4797 as determined by an MPA. ND, no inhibition detected using 100 μl of supernatant.
FIG. 2.
Antimicrobial activity of supernatants from S. cerevisiae L50A-20(pYABD02) (EntL50A) and S. cerevisiae L50B-4(pYABD03) (EntL50B) cultures grown in SCGR broth at 30°C, independently and combined to achieve a 1:1 bacteriocin peptide ratio (EntL50A+EntL50B), against P. damnosus CECT4797 as determined by a SPAT.
Purification and characterization of EntL50A and EntL50B heterologously produced by S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03), respectively.
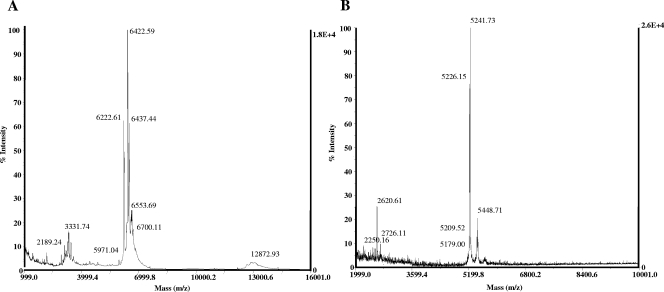
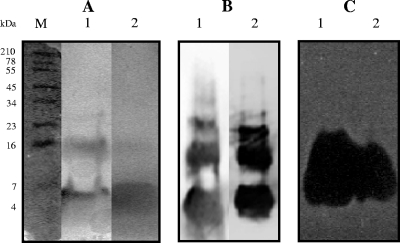
The specific antimicrobial activities of purified EntL50A and EntL50B were 452- and 2,000-fold greater than those of culture supernatants from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) grown in SCGR broth at 30°C for 144 and 120 h, respectively. Interestingly, although the antimicrobial activity in these fractions represented a recovery of only 1.3 and 2.6% of the EntL50A and EntL50B original activity, respectively, the corresponding amounts of recombinant EntL50A and EntL50B were 8.6 and 23.5 μg, respectively, which represent 46.5 and 45.8% of the original bacteriocin peptide amounts, respectively (Table 5). The purity and molecular mass of recombinant EntL50A and EntL50B were analyzed by MALDI-TOF MS. The results obtained for recombinant EntL50A revealed three major peptides with molecular masses ranging from 6.2 to 6.4 kDa (Fig. 3A), while those for recombinant EntL50B showed two major peptides with molecular masses (ca., 5,242 and 5,226 Da) closely similar to that of natural EntL50B (5,178 Da), as well as a minor peptide with the expected molecular mass (5,179 Da) (Fig. 3B). Recombinant EntL50A and EntL50B were also analyzed on silver-stained Tricine-SDS-PAGE gels (Fig. 4A), revealing a major band of the expected size, as well as two and one upper band, in the samples containing EntL50A and EntL50B, respectively, suggesting that both recombinant peptides display a strong tendency to form aggregates. The peptides and peptide aggregates were also detected by Western blotting using antibodies specific for EntL50A and EntL50B (Fig. 4B), and they were shown to be biologically active by a gel overlay assay (Fig. 4C).
TABLE 5.
Purification of recombinant EntL50A and EntL50B produced by S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03), respectivelya
| Purification stage | Vol (ml) | Total A254b | Total activity (BU)c | Sp act (BU/A254)d | Increase in sp act (fold)e | Total activity (%) | Enterocin yield (ng)f | Enterocin yield (%) | Enterocin sp act (BU/ng)g |
|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae L50A-20(pYABD02) | |||||||||
| Culture supernatant | 1000 | 24,580 | 142,100 | 6 | 1 | 100 | 18,500 | 100 | 7.7 |
| Ammonium sulfate precipitation | 100 | 221 | 15,700 | 71 | 12 | 11 | ND | ND | ND |
| Gel filtration chromatography | 200 | 37.4 | 12,800 | 342 | 57 | 9 | ND | ND | ND |
| Cation exchange chromatography | 50 | 3.9 | 1,100 | 282 | 47 | 0.8 | ND | ND | ND |
| Hydrophobic-interaction chromatography | 10 | 0.8 | 14,200 | 17,750 | 2,958 | 10 | ND | ND | ND |
| Reversed-phase chromatography | 2.6 | 0.7 | 1,900 | 2,714 | 452 | 1.3 | 8,600 | 46.5 | 0.2 |
| S. cerevisiae L50B-4(pYBAD03) | |||||||||
| Culture supernatant | 1000 | 14,400 | 46,000 | 3 | 1 | 100 | 51,300 | 100 | 0.9 |
| Ammonium sulfate precipitation | 100 | 157 | 10,000 | 64 | 21 | 22 | ND | ND | ND |
| Gel filtration chromatography | 200 | 29.6 | 6,400 | 216 | 72 | 14 | ND | ND | ND |
| Cation exchange chromatography | 50 | 0.5 | 0 | 0 | 0 | 0 | ND | ND | ND |
| Hydrophobic-interaction chromatography | 10 | 3.0 | 2,700 | 900 | 300 | 5.9 | ND | ND | ND |
| Reversed-phase chromatography | 1.9 | 0.2 | 1,200 | 6,000 | 2,000 | 2.6 | 23,500 | 45.8 | 0.05 |
Cultures were grown in SCGR broth at 30°C.
Absorbance at 254 nm multiplied by the volume in milliliters.
Antimicrobial activity in bacteriocin units per milliliter (BU/ml) as determined by an MPA multiplied by the total volume.
Specific activity expressed as the total activity (BU) divided by the total A254.
The specific activity of a fraction (BU/A254) divided by the specific activity of the culture supernatant (BU/A254).
EntL50A and EntL50B concentration as determined by an NCI-ELISA using specific polyclonal antibodies for EntL50A or EntL50B. ND, not determined.
Specific activity expressed as the total activity (BU) divided by the enterocin yield (ng).
FIG. 3.
Mass spectrometry analysis of recombinant EntL50A (A) and EntL50B (B) purified from S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) cultures, respectively, grown in SCGR broth at 30°C.
FIG. 4.
(A) Tricine-SDS-PAGE of purified recombinant EntL50A and EntL50B after silver staining. (B) Western blotting using rabbit polyclonal antibodies with specificity for EntL50A (anti-LR1-KLH) and EntL50B (anti-LR2-KLH). (C) Antimicrobial activity after gel overlay with the indicator strain P. damnosus CECT4797. Lane 1, purified EntL50A; lane 2, purified EntL50B. SeeBlue Pre-Stained Standard molecular mass markers band sizes (M) (Invitrogen) are indicated on the left.
DISCUSSION
This study reports for the first time the heterologous expression and secretion of a non-pediocin-like bacteriocin, EntL50 (EntL50A and EntL50B), by a yeast. This was achieved using pYABD01, a segregationally stable expression and secretion vector derived from the S. cerevisiae 2μm episomal high-copy-number plasmid pYES2. The recombinant plasmid pYABD01 generated in this work contains the PGAL1 promoter for high-level inducible protein expression by galactose and the yeast 13-residue signal peptide MFα1s, including the Kex2 cleavage site required for processing of fusion proteins during Sec-dependent secretion. The generated recombinant S. cerevisiae strains exerted bacteriocinogenic activity in liquid media (SCGR and/or YPGR broth). Interestingly, although yeast growth was higher in YPGR than in SCGR broth, the extracellular antimicrobial activity was much lower in the former medium, with recombinant EntL50A and EntL50B production representing 62 and 80%, respectively, of that found in SCGR broth. Given the high segregational stability of pYABD01 grown in YPD broth without selective pressure, these differences in bacteriocin extracellular concentrations are unlikely to be due to plasmid instability in the cells grown under nonselective conditions (YPGR broth). Strikingly, EntL50A and EntL50B produced in SCGR broth were more active than similar peptide amounts produced in YPGR broth. The lower antimicrobial activity found in the complex medium YPGR broth may be ascribed to one or more of the following factors: (i) the presence of compounds (e.g., salt, proteins, and/or collagen-like materials) interfering with bacteriocin activity (2, 23, 26, 44, 45), (ii) (higher) aggregation of bacteriocin peptide monomers, rendering less active oligomers and/or complexes with medium constituents (14), and/or (iii) higher proteolytic bacteriocin peptide degradation due to a higher cell density and lysis, yielding a higher concentration of vacuolar proteases (2, 14, 23).
The maximum amounts of EntL50A produced by S. cerevisiae L50A-20(pYABD02) in SCGR and YPGR broth were approximately three- to fourfold lower, respectively, than those of EntL50B produced by S. cerevisiae L50B-4(pYABD03) in the same media. At first glance, these differences may be ascribed to a higher C-terminal proteolytic degradation of EntL50A and/or to a higher secretion of EntL50B. With respect to the latter possibility, it has been demonstrated that the efficiency of secretion partially depends on the nature of the heterologous protein (6, 45); however, we consider it unlikely that this phenomenon is responsible for the differences in EntL50A and EntL50B concentrations found in the respective supernatants, since these peptides show similar physicochemical characteristics and are highly related (72% identity, most pronounced at the N terminus), sharing 31 out of 44 and 43 amino acid residues, including the first 8 amino acid residues (8, 10). Moreover, hydrophobic proteins or peptides, such as most bacteriocins, are known to interact with the producer cell membranes (43, 44). In this respect, EntL50A is more hydrophobic than EntL50B (GRAVY [grand average of hydropathy] indexes of 0.202 and −0.144, respectively) (10), which may lead to a higher retention within the yeast cytoplasmic membrane after an initial pH-dependent cell surface adsorption and thus to lower EntL50A concentrations in the respective supernatants. In a previous study, we showed that EntL50A and EntL50B are produced in equimolar amounts by the wild-type strain E. faecium L50 (14), and therefore, we have evaluated in this work the recombinant paired production of EntL50A and EntL50B by a single S. cerevisiae strain. The extracellular production of EntL50A by S. cerevisiae L50AB-2(pYABD04) was 24% higher than that by S. cerevisiae L50A-20(pYABD02), but strikingly, EntL50B was not produced. Considering the signal loss found during sequencing of the fragment α-L50A-α-L50B and the high similarity between entL50A and entL50B, it is likely that DNA secondary structures are established in this fragment and probably in the corresponding mRNA, leading to the repression of entL50B expression at the transcriptional and/or translational level and to the higher expression of entL50A. In this respect, it is known that stem-loop structures may block polymerase progress (32) and inhibit translational initiation in S. cerevisiae (38, 48). Although the paired production of EntL50A and EntL50B by a single S. cerevisiae strain was not achieved, we succeeded in the coproduction of both peptides by coculturing S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03) and showed that recombinant EntL50A and EntL50B act synergistically at a 1:1 ratio, as reported in previous studies for in vitro-synthesized EntL50A and EntL50B (1, 8). To date, bacteriocin secretion by S. cerevisiae has been described only for PedPA-1 (44) and pediocin-like plantaricin 423 (46). Contrary to our results, recombinant strains of S. cerevisiae heterologously expressing and secreting PedPA-1 or plantaricin 423, under the control of the alcohol dehydrogenase I gene promoter (PADH1), showed antimicrobial activity on solid medium. However, only low extracellular bacteriocin activity was obtained in the CS from the recombinant yeasts, likely due to bacteriocin retention within the cytoplasmic membrane and/or adsorption to the producer cells (44, 46).
Notwithstanding the fact that the maximum amounts of EntL50A and EntL50B heterologously produced by S. cerevisiae L50A-20(pYABD02) and S. cerevisiae L50B-4(pYABD03), respectively, represent only approximately 4 and 11% of the maximum production by E. faecium L50 (217 and 210 ng/mg CDW, respectively) (14), the relevance of the strategy developed in this work is highlighted by the fact that it has allowed for the first time the independent purification of these highly related peptides, resulting in a high recovery of purified EntL50A and EntL50B, as determined by using specific antibodies. The MALDI-TOF MS analysis of the last reversed-phase FPLC fraction containing recombinant EntL50B showed a minor peptide with the same molecular mass as natural EntL50B (5,179 Da), demonstrating that the EntL50B precursor is correctly processed by the Kex2 enzyme, as well as two major peptides with molecular masses of 5,242 and 5,226 Da. These discrepancies may be ascribed to the spontaneous modification of the two methionine residues of EntL50B (i.e., Met1 and Met24). Regarding this, it is known that Met is the amino acid residue most sensitive to reactive oxygen, resulting in a wide range of peptide and proteins with a reduced biological activity (4, 49). By assuming that both Met residues in recombinant EntL50B have become oxidized to methionine sulfone (MetSO2), an addition of 64 Da to the theoretical mass, leading to a molecular mass of 5,242 Da, would be obtained. Likewise, the peptide with a molecular mass of 5,226 Da would be the result of the conversion of Met residues to MetSO2 (addition of 32 Da) and methionine sulfoxide (MetSO) (addition of 16 Da). On the other hand, MALDI-TOF MS analysis of the fraction containing recombinant EntL50A revealed three major peptides with molecular masses of 6.2 to 6.4 kDa. At first glance, this result could indicate that the EntL50A precursor is not correctly processed by the Kex2 enzyme as a consequence of a reduced recognition of the cleavage site (Glu-Lys-Arg) due to a conformational interference exerted by its N terminus; however, the high similarity between EntL50A and EntL50B (most pronounced at the N terminus) does not favor this possibility. Alternatively, recombinant EntL50A may be associated to a hitherto-unknown compound, as has been reported for PedPA-1 heterologously produced by P. pastoris, which is tightly associated with some “collagen-like” material, rendering a biologically inactive bacteriocin (2). Although the last reversed-phase FPLC fractions contained approximately 45% of the EntL50A and EntL50B found in the respective culture supernatants, their antimicrobial activities represented only approximately 1 and 3%, respectively, which also favors the bacteriocin oxidation events cited above. In this respect, it has been reported that Met in bacteriocins become spontaneously oxidized, especially during their purification to homogeneity, leading to a loss or reduction of their antimicrobial activities (5, 10, 29). Moreover, a study carried out simultaneously with the one described herein reported the spontaneous chemical modification of EntL50A and EntL50B during their purification due to Met1 formylation and Met24 oxidation to MetSO, rendering active and inactive peptides, respectively (27). Considering that beer production includes a wort aeration step to promote brewing yeast growth and enhance the start-up of fermentation (47), the oxidative partial inactivation of EntL50A and EntL50B may hamper their application as beer biopreservatives at this stage of the brewing process. Therefore, it would be convenient to develop engineered variants of EntL50A and EntL50B with an increased stability against oxidation by replacing one or both Met residues with other amino acid residues, provided that this replacement is not deleterious for bacteriocin activity, similar to work described for PedPA-1 (29). In this context, replacement of Met at the C-terminal half of bacteriocins (e.g., Met24 in EntL50A and EntL50B) should be carefully addressed, since this region penetrates the hydrophobic part of the target cell membrane, mediating membrane leaking and subsequent cellular death (18, 30, 31).
The results obtained in this work demonstrate the suitability of the generated MFα1s-containing recombinant vector pYABD01 to direct the heterologous secretion of biologically active EntL50A and EntL50B through the S. cerevisiae Sec system. Moreover, these results highlight the feasibility of beer biopreservation by the use of genetically engineered bacteriocinogenic S. cerevisiae brewing yeasts as part of a hurdle preservation technology to obtain safer and more stable beers (44, 46, 47). However, despite our promising results, further research is needed to optimize bacteriocin production, increase bacteriocin stability under oxidative stress, and achieve paired production of EntL50A and EntL50B by a single yeast strain. Finally, the development of bacteriocinogenic laboratory and brewing yeast strains overexpressing EntL50 (EntL50A and EntL50B) may facilitate not only the future applications of this broad-spectrum two-peptide bacteriocin as a biopreservative in breweries and other food industries but also the obtention of larger amounts of recombinant bacteriocin peptides to conduct studies such as those on structure-function relationships and molecular mode of action, which are of great applied and scientific interest and remain to be unraveled.
Acknowledgments
This work was partially supported by projects PR248/02-11688 from the Fundación Danone/Complutense (Madrid, Spain), PR41/06-15051 from the Grupo Santander Central Hispano/Universidad Complutense de Madrid (Madrid, Spain), AGL2003-01508 and AGL2006-01042 from the Ministerio de Educación, Cultura y Deporte (MECD), Spain, and S-0505/AGR/0265 from the Comunidad de Madrid (CAM), Spain. A. Basanta holds a fellowship from CAM, Spain. J. Gutiérrez was a recipient of a fellowship from Ministerio de Ciencia y Tecnología, Spain. R. Criado held a fellowship from MECD, Spain.
We thank the anonymous reviewers for their constructive comments and suggestions for improvement of the manuscript. The help of B. Gómez-Sala, J. Sánchez, and M. Martín is also recognized.
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Basanta, A., J. Sánchez, B. Gómez-Sala, C. Herranz, P. E. Hernández, and L. M. Cintas. 2008. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 125:293-307. [DOI] [PubMed] [Google Scholar]
- 2.Beaulieu, L., D. Groleau, C. B. Miguez, J. F. Jetté, H. Aomari, and M. Subirade. 2005. Production of pediocin PA-1 in the methylotrophic yeast P. pastoris reveals unexpected inhibition of its biological activity due to the presence of collagen-like material. Protein Expr. Purif. 43:111-125. [DOI] [PubMed] [Google Scholar]
- 3.Bhunia, A. K., M. G. Johnson, and B. Ray. 1987. Direct detection of an antimicrobial peptide of Pediococcus acidilactici in sodium dodecyl sulphate-polyacrylamide gel electrophoresis. J. Ind. Microbiol. 2:319-322. [Google Scholar]
- 4.Brot, N., and H. Weissbach. 1991. Biochemistry of methionine sulfoxide residues in proteins. Biofactors 3:91-96. [PubMed] [Google Scholar]
- 5.Casaus, M. P., T. Nilsen, L. M. Cintas, I. F. Nes, and P. E. Hernández. 1997. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143:2287-2294. [DOI] [PubMed] [Google Scholar]
- 6.Cereghino, J. L., and J. M. Cregg. 2000. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 24:45-66. [DOI] [PubMed] [Google Scholar]
- 7.Cintas, L. M., J. M. Rodríguez, M. F. Fernández, K. Sletten, I. F. Nes, P. E. Hernández, and H. Holo. 1995. Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas, L. M., P. Casaus, H. Holo, P. E. Hernández, I. F. Nes, and L. S. Håvarstein. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cintas, L. M., P. Casaus, M. F. Fernández, and P. E. Hernández. 1998. Comparative antimicrobial activity of enterocin L50, pediocin PA-1, nisin A and lactocin S against spoilage and foodborne pathogenic bacteria. Food Microbiol. 15:289-298. [Google Scholar]
- 10.Cintas, L. M., P. Casaus, C. Herranz, L. S. Håvarstein, H. Holo, P. E. Hernández, and I. F. Nes. 2000. Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182:6806-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cintas, L. M., P. Casaus, C. Herranz, I. F. Nes, and P. E. Hernández. 2001. Bacteriocins of lactic acid bacteria. Food Sci. Technol. Int. 7:281-305. [Google Scholar]
- 12.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 13.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 14.Criado, R., J. Gutiérrez, M. Martín, C. Herranz, P. E. Hernández, and L. M. Cintas. 2006. Immunochemical characterization of temperature-regulated production of enterocin L50 (EntL50A and EntL50B), enterocin P, and enterocin Q by Enterococcus faecium L50. Appl. Environ. Microbiol. 72:7634-7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deegan, L. H., P. D. Cotter, C. Hill, and P. Ross. 2006. Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 16:1058-1071. [Google Scholar]
- 16.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholtz. 1996. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 17.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fimland, G., V. G. H. Eijsink, and J. Nissen-Meyer. 2002. Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology 148:3661-3670. [DOI] [PubMed] [Google Scholar]
- 19.Franz, C. M. A. P., M. J. van Belkum, W. H. Holzapfel, H. Abriouel, and A. Gálvez. 2007. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31:293-310. [DOI] [PubMed] [Google Scholar]
- 20.Gajic, O., G. Buist, M. Kojic, L. Topisirovic, O. P. Kuipers, and J. Kok. 2003. Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278:34291-34298. [DOI] [PubMed] [Google Scholar]
- 21.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577-592. [DOI] [PubMed] [Google Scholar]
- 22.Gutiérrez, J., R. Criado, R. Citti, M. Martín, C. Herranz, M. F. Fernández, L. M. Cintas, and P. E. Hernández. 2004. Performance and applications of polyclonal antipeptide antibodies specific for the enterococcal bacteriocin enterocin P. J. Agric. Food Chem. 52:2247-2255. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez, J., D. Bourque, R. Criado, Y. J. Choi, L. M. Cintas, P. E. Hernández, and C. B. Míguez. 2005. Heterologous extracellular production of enterocin P from Enterococcus faecium P13 in the methylotrophic bacterium Methylobacterium extorquens. FEMS Microbiol. Lett. 248:125-131. [DOI] [PubMed] [Google Scholar]
- 24.Hammond, J. R. 1995. Genetically-modified brewing yeasts for the 21st century. Progress to date. Yeast 11:1613-1627. [DOI] [PubMed] [Google Scholar]
- 25.Holo, H., Ø. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilgen, C., J. L. Cereghino, and J. M. Cregg. 2005. Pichia pastoris, p. 143-162. In G. Gellissen (ed.), Production of recombinant proteins: novel microbial and eucaryotic expression systems. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
- 27.Izquierdo, E., A. Bednarczyk, C. Schaeffer, Y. Cai, E. Marchioni, A. van Dorsselaer, and S. Ennahar. 2008. Production of enterocins L50A, L50B, and IT, a new enterocin, by Enterococcus faecium IT62, a strain isolated from Italian ryegrass in Japan. Antimicrob. Agents Chemother. 52:1917-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jespersen, L. 2003. Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages. FEMS Yeast Res. 3:191-200. [DOI] [PubMed] [Google Scholar]
- 29.Johnsen, L., G. Fimland, V. Eijsink, and J. Nissen-Meyer. 2000. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl. Environ. Microbiol. 66:4798-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnsen, L., G. Fimland, and J. Nissen-Meyer. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J. Biol. Chem. 280:9243-9250. [DOI] [PubMed] [Google Scholar]
- 31.Kazazic, M., J. Nissen-Meyer, and G. Fimland. 2002. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 148:2019-2027. [DOI] [PubMed] [Google Scholar]
- 32.Kieleczawa, J. 2006. Fundamentals of sequencing of difficult templates—an overview. J. Biomol. Tech. 17:207-217. [PMC free article] [PubMed] [Google Scholar]
- 33.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 34.Line, J. E., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. P. Levchuk, O. E. Svetoch, B. S. Seal, G. R. Siragusa, and N. J. Stern. 2008. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 52:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linko, M., A. Haikara, A. Ritala, and M. Penttilä. 1998. Recent advances in the malting and brewing industry. J. Biotechnol. 65:85-98. [Google Scholar]
- 36.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 37.Nes, I. F., D. B. Diep, and H. Holo. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira, C. C., J. J. van den Heuvel, and J. E. McCarthy. 1993. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the roles of the stability and position of stem-loops in the mRNA leader. Mol. Microbiol. 9:521-532. [DOI] [PubMed] [Google Scholar]
- 39.Ross, R. P., S. Morgan, and C. Hill. 2002. Preservation and fermentation: past, present and future. Int. J. Food Microbiol. 79:3-16. [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto, K., and W. N. Konings. 2003. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 89:105-124. [DOI] [PubMed] [Google Scholar]
- 41.Sánchez-Hidalgo, M., M. Maqueda, A. Gálvez, H. Abriouel, E. Valdivia, and M. Martínez-Bueno. 2003. The genes coding for enterocin EJ97 production by Enterococcus faecalis EJ97 are located on a conjugative plasmid. Appl. Environ. Microbiol. 69:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 43.Schiffer, M., C.-H. Chang, and F. J. Stevens. 1992. The function of tryptophan residues in membrane proteins. Protein Eng. 5:213-214. [DOI] [PubMed] [Google Scholar]
- 44.Schoeman, H., M. A. Vivier, M. du Toit, L. M. T. Dicks, and I. S. Pretorius. 1999. The development of bactericidal yeast strains by expressing the Pediococcus acidilactici pediocin gene (pedA) in Saccharomyces cerevisiae. Yeast 15:647-656. [DOI] [PubMed] [Google Scholar]
- 45.Schuster, M., A. Einhauer, E. Wasserbauer, F. Süßenbacher, C. Ortner, M. Paumann, G. Werner, and A. Jungbauer. 2000. Protein expression in yeast; comparison of two expression strategies regarding protein maturation. J. Biotechnol. 84:237-248. [DOI] [PubMed] [Google Scholar]
- 46.Van Reenen, C. A., M. L. Chikindas, W. H. van Zyl, and L. M. T. Dicks. 2003. Characterization and heterologous expression of a class IIa bacteriocin, plantaricin 423 from Lactobacillus plantarum 423, in Saccharomyces cerevisiae. Int. J. Food Microbiol. 81:29-40. [DOI] [PubMed] [Google Scholar]
- 47.Vaughan, A., T. O'Sullivan, and D. van Sinderen. 2005. Enhancing the microbiological stability of malt and beer—a review. J. Inst. Brew. 111:355-371. [Google Scholar]
- 48.Vega Laso, M. R., D. Zhu, F. Sagliocco, A. J. Brown, M. F. Tuite, and J. E. McCarthy. 1993. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem. 268:6453-6462. [PubMed] [Google Scholar]
- 49.Weissbach, H., F. Etienne, T. Hoshi, S. H. Heinemann, W. T. Lowther, B. Matthews, G. St John, C. Nathan, and N. Brot. 2002. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 397:172-178. [DOI] [PubMed] [Google Scholar]