Abstract
Molecular analysis of black band disease of corals revealed that samples frozen immediately after collection yielded more proteobacterial 16S rRNA sequences, while unfrozen samples produced more cyanobacterial and sulfur-oxidizing bacterial sequences. These results suggest the need to use multiple approaches for preparation of samples to characterize this complex polymicrobial disease.
Black band disease (BBD) is a polymicrobial disease that affects corals on reefs worldwide. It consists of a migrating microbial mat dominated by cyanobacteria that lyses coral tissue, leading to coral colony death, and is one of the most destructive of coral diseases. Microscopic examination of BBD samples consistently reveals an abundance of nonheterocystous, filamentous cyanobacteria and colorless gliding bacteria with internal elemental sulfur granules characteristic of the genus Beggiatoa (6, 17, 18). It is thought that these are key players in the etiology of BBD. However, with one exception (2), previous molecular studies of BBD consistently detected very low proportions of cyanobacteria (4, 8, 9, 19, 20) and only one study has detected Beggiatoa (19). Instead, all molecular BBD studies indicate a highly variable and diverse composition of heterotrophic bacteria, mostly members of the Alphaproteobacteria.
It is unknown why the dominant cyanobacteria and filamentous sulfur-oxidizing bacteria observable microscopically in BBD samples are poorly or not at all detected by molecular methods. It is possible that freezing of the samples in these studies is the cause for low detection of BBD cyanobacteria and sulfur oxidizers. Freezing is the common method of sample processing to extract DNA for microbial community analysis of BBD and has been used in all previous molecular studies. However, this approach may impart a bias to detection of specific BBD bacteria. Suomalainen et al. (22) reported that freezing of samples targeting the fish pathogen Flavobacterium columnare destroyed DNA, suggested to be due to the release of DNase and other enzymes from the cell, leaving most of the F. columnare DNA undetectable by PCR. They noted that DNA from bacteria such as Escherichia coli was not affected (22). Bissett et al. (3) speculated that freezing sediments prior to DNA extraction lysed Beggiatoa filaments and caused their DNA to be lost (3). A recent report showed that algae and cyanobacteria with large cell sizes, including filamentous strains, could not be sufficiently cryopreserved (5). While the above-described studies showed or speculated that freezing of samples affects the detection of some microorganisms in environmental samples, none of these studies included detailed investigation of the mechanism responsible for the effect of freezing or of the effect of freezing on the assessment of microbial community composition.
In the present study, we investigated the effect of freezing on molecular analysis of the BBD microbial community by using DNA extracts of frozen and unfrozen BBD samples from two coral hosts (Siderastrea siderea and Diploria clivosa), using both universal and cyanobacterium-specific primers targeting the 16S rRNA gene. BBD samples (i.e., the BBD microbial mat) were collected by suctioning the mat off the coral surface using individual sterile syringes while scuba diving. Samples were transferred to 2-ml cryovials (after decanting seawater) upon return to shore and either immediately frozen and stored at −20°C until DNA extraction or maintained at ambient temperature with DNA extracted within 1 h of collection. Eleven samples were collected from reefs of the Florida Keys (United States), Lee Stocking Island (Bahamas), and St. Croix (United States Virgin Islands).
Genomic DNA was extracted by the bead-beating method as previously described (12, 19, 20). Frozen samples were first thawed at room temperature, and 500-μl aliquots were directly transferred into multimix lysing matrix tubes by using trimmed pipette tips, excluding any water. Unfrozen samples were transferred to multimix lysing matrix tubes in the same way. The extracted DNA was verified by gel electrophoresis, and DNA extracts from frozen samples were stored at −20°C, whereas DNA extracts from unfrozen samples were kept at 4°C until used for PCR amplification.
DNA extracted from both frozen and unfrozen samples was amplified by PCR using universal bacterial primers 27F and 1492R (14) and cyanobacterium-specific primers CYA359F and CYA781R(B) (15) targeting 16S rRNA genes. The purification of PCR products, cloning, and sequencing of plasmid inserts were done as described previously (20). Primer M13F (11) or CYA359F (15) was used to obtain partial sequences, and an additional primer, 518F (13), M13R (11), or CYA781R(B) (15), was used to obtain full-length sequences. Sequence editing, BLAST (1), and phylogenetic analysis using ARB (10) were done as described previously (19, 20). Sequences that matched at similarity identity values of 97% and above were considered to be of the same operational taxonomic unit. Representative gene sequences that were closely related to cyanobacterial sequences were subjected to maximum-parsimony, neighbor-joining, and maximum-likelihood phylogenetic analyses, and a consensus tree was produced based on maximum-parsimony analysis.
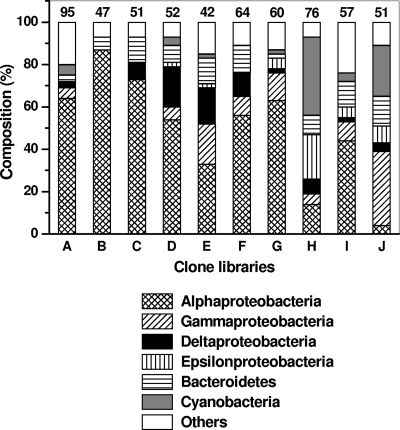
The results for universal bacterial primers indicated that all of the frozen BBD samples except one (Fig. 1, clone library E) were dominated (44 to 87%) by Alphaproteobacteria (Fig. 1; see Tables S1 and S2 in the supplemental material). We previously (19) compared the 16S rRNA gene sequences retrieved from seven of these libraries (Fig. 1, libraries A to G), all of which were obtained from frozen BBD samples from the host S. siderea, to investigate the diversity of BBD microorganisms between BBD infections. In the present study, we focus on the differences in results obtained using frozen versus unfrozen BBD samples from S. siderea (Fig. 1, libraries G and H) and a second coral host, D. clivosa (Fig. 1, libraries I and J). The S. siderea samples (libraries G and H) were taken from different host colonies on the same reef (Butler Bay Reef site), whereas the D. clivosa clone libraries were constructed from subsamples of one BBD sample.
FIG. 1.
Dominant bacterial phylogenetic groups, based on 16S rRNA gene sequence types and universal primers, present in clone libraries produced from frozen and unfrozen BBD samples from the coral hosts Siderastrea siderea and Diploria clivosa. The numbers above the bars represent the numbers of sequences in the respective clone libraries. Libraries A to H, frozen (A to G) and unfrozen (H) BBD from S. siderea. Libraries I and J, frozen (I) and unfrozen (J) BBD from D. clivosa. Sampling locations and sampling dates: libraries A and B, Horseshoe Reef, Lee Stocking Island, Bahamas, 19 July 2004; C, Rainbow Garden Reef, Lee Stocking Island, Bahamas, 16 July 2004; D, Watson's Reef, Florida Keys, 3 May 2005; E, G, and H, Butler Bay Reef site, St. Croix, U.S. Virgin Islands (USVI), 22 October 2005, 1 June 2005, and 5 June 2006, respectively; F, Frederiksted Reef site, St. Croix, USVI, 1 June 2005; I and J, Frederiksted Reef site, St. Croix, USVI, 7 August 2006. All of the sequences from clone libraries A to G have been previously published by Sekar et al. (19, 20).
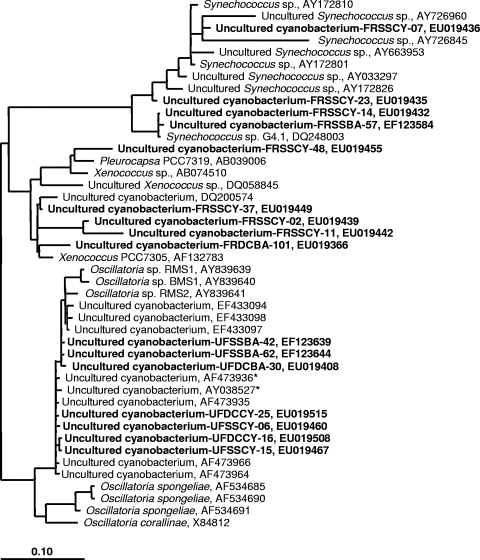
This approach yielded strikingly different results for the two methods. For example, the clone library produced from one frozen sample (Fig. 1, library G) from S. siderea contained only one (of 60) cyanobacterium-related sequence (see EF123584 [GenBank sequence accession no.] in Table S1 in the supplemental material), which was phylogenetically related to a sequence from an uncultured planktonic Synechococcus sp. (GenBank sequence accession no. AY172810; Fig. 2). In contrast, the clone library from the corresponding unfrozen sample (Fig. 1, library H) was dominated by a cyanobacterial ribotype which represented 37% of the clones. This ribotype was closely related to an Oscillatoria ribotype (GenBank sequence accession no. AY038527/AF473936) detected in almost all reported BBD molecular studies (2, 4, 7, 23). The sequence was confirmed as the BBD Oscillatoria sequence by phylogenetic analysis using two representative clone sequences (GenBank sequence accession no. EF123639 and EF123644) (Fig. 2). The unfrozen S. siderea clone library additionally produced a dominant epsilonproteobacterial ribotype (14 of 15 clones) (see Table S1 in the supplemental material) that was not detected in the corresponding frozen sample. Phylogenetic analysis of two representative sequences (GenBank sequence accession no. EF123607 and EF123613, not shown in Fig. 2) determined that the sequence was related to a sequence from the sulfur-oxidizing bacterium “Candidatus Arcobacter sulfidicus” (GenBank sequence accession no. AY035822) (24), a species known to deposit filamentous sulfur (21) and reported previously in BBD (9).
FIG. 2.
Phylogenetic tree derived from the 16S rRNA gene sequences closely related to Synechococcus spp., Xenococcus spp., and Oscillatoria spp. sequences detected in BBD and their neighbors. The tree topology is based on the maximum-parsimony analysis. The bar represents 10% estimated sequence divergence. Boldface type indicates sequences from this study, designated as follows. FRSSBA, UFSSBA, FRSSCY, and UFSSCY indicate sequences retrieved from frozen (FR) and unfrozen (UF) samples of S. siderea (SS) using universal bacterial primers (BA) and cyanobacterium-specific (CY) primers for 16S rRNA gene amplification. FRDCBA, UFDCBA, and UFDCCY indicate sequences retrieved from frozen and unfrozen samples of Diploria clivosa (DC), and the same primer designations are used as for S. siderea sequences. GenBank sequence accession numbers are listed for all sequences. Asterisks designate sequences corresponding to the sequence from the BBD Oscillatoria discussed in the text.
Again in clone library I, from the frozen subsample of D. clivosa (see Table S2 in the supplemental material), the Alphaproteobacteria were dominant (44%) and cyanobacteria represented in low percentages (4%). These cyanobacterial sequences were phylogenetically related to sequences of Leptolyngbya spp. (not shown in Fig. 2) and a planktonic cyanobacterium Xenococcus sp. (GenBank sequence accession no. AF132783) (Fig. 2; see Table S2 in the supplemental material). The library from the unfrozen BBD subsample of D. clivosa (see Table S2 in the supplemental material) was dominated by Gammaproteobacteria (35%), followed by cyanobacteria (24%) which had the same cyanobacterial sequence type (BBD Oscillatoria) observed in the unfrozen S. siderea sample (see Table S2 in the supplemental material). For corroboration of these results, we constructed an additional clone library, using universal primers, from an unfrozen BBD sample from S. siderea collected during June 2007; in this sample, 47% of the sequences were also related to the sequence from BBD Oscillatoria.
The use of cyanobacterium-specific primers produced results similar to the overall pattern we detected using universal primers. Frozen BBD from S. siderea produced 27 sequences, of which 24 were closely related to sequences from planktonic Synechococcus spp. and Xenococcus sp. (see Table S3 in the supplemental material). This was confirmed by phylogenetic analysis (Fig. 2) using representative sequences (GenBank sequence accession no. EU019432, EU019435, EU019439, EU019442, EU019449, and EU019455). In contrast, all of the sequences (n = 37) obtained from unfrozen S. siderea samples were closely related to the sequence from the BBD Oscillatoria (see Table S3 in the supplemental material). Representative sequences (GenBank sequence accession no. EU019460 and EU019467) confirmed this phylogenetic affiliation (Fig. 2). Similarly, each of 38 sequences obtained from the unfrozen subsample of D. clivosa with cyanobacterium-specific primers was closely related to the sequence from the BBD Oscillatoria (see Table S3 in the supplemental material), again confirmed by phylogenetic analysis using two representative sequences (GenBank sequence accession no. EU019508 and EU019515) (Fig. 2).
There was very little overlap (6 to 10%) between sequences obtained from frozen versus unfrozen BBD samples collected from both coral hosts when considering all of the BBD bacterial sequences detected (see Tables S1 and S2 in the supplemental material). Only four sequences were common to both frozen and unfrozen clone libraries (6% of 62 sequences detected within 136 clones) for S. siderea and seven sequences (10% of 69 sequences detected within 108 clones) for D. clivosa. Statistical analysis (ANOSIM) showed that the sequence types differed significantly between frozen and unfrozen clone libraries (R = 0.987; P = 0.022). Overall, all frozen libraries (libraries A to G and I) were 69% similar to each other, while the two unfrozen libraries (libraries H and J) were 58% similar.
The results of our study are significant for the ongoing investigations into the etiology of BBD. While it is well known that the BBD microbial community consists of photoautotrophs (cyanobacteria), sulfate-reducing bacteria, sulfur-oxidizing bacteria, and heterotrophs (16), we are just beginning to understand the roles of these functional groups in the disease process. A first step in this understanding is the valid and repeatable detection of specific members of the BBD consortium. In summary, we show here that unfrozen samples produce better results for detection of BBD cyanobacteria and sulfur-oxidizing bacteria, while frozen samples are best for detection of heterotrophic proteobacterial sequences. The latter is particularly important because of the consistent finding of Proteobacteria associated with toxic dinoflagellates (19, 20), as well as other marine invertebrate pathogens (4), in BBD. We have not studied the mechanism behind the freezing effect (e.g., release of DNase), which is outside the scope of this study. Though the current study was done with BBD samples, the effect of freezing on other microbial mats or biofilms cannot be ignored. Based on the results of this study, we suggest using multiple sample-processing approaches to characterize the microbial communities associated with BBD and other microbial mats.
Nucleotide sequence accession numbers.
The sequences reported in this study have been deposited in the GenBank database under the following accession numbers: DQ644014, DQ644015, DQ644018, EF123300 to EF123658, EU019310 to EU019417, EU019429 to EU019493, and EU019496 to EU019533.
Supplementary Material
Acknowledgments
This research was supported by a grant to L.L.R. from NIH (NIH/NIGMS SO6GM8205).
We thank T. Zimmerman, Agricultural Experiment Station, University of Virgin Islands, Project AWARE, and the ScubaWest Dive shop in St. Croix, U.S. Virgin Islands. We thank the four anonymous reviewers for their valuable comments to improve the manuscript.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barneah, O., E. Ben-Dov, E. Kramarsky-Winter, and A. Kushmaro. 2007. Characterization of black band disease in Red Sea stony corals. Environ. Microbiol. 9:1995-2006. [DOI] [PubMed] [Google Scholar]
- 3.Bissett, A., J. Bowman, and C. Burke. 2006. Bacterial diversity in organically enriched fish farm sediments. FEMS Microbiol. Ecol. 55:48-56. [DOI] [PubMed] [Google Scholar]
- 4.Cooney, R. P., O. Pantos, M. D. A. L. Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 5.Day, J. G. 2007. Cryopreservation of microalgae and cyanobacteria. Methods Mol. Biol. 368:141-151. [DOI] [PubMed] [Google Scholar]
- 6.Ducklow, H. W., and R. Mitchell. 1979. Observations on naturally and artificially diseased tropical corals: a scanning electron microscopy study. Microb. Ecol. 5:215-223. [DOI] [PubMed] [Google Scholar]
- 7.Frias-Lopez, J., G. T. Bonheyo, Q. S. Jin, and B. W. Fouke. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific Reefs. Appl. Environ. Microbiol. 69:2409-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frias-Lopez, J., J. S. Klaus, G. T. Bonheyo, and B. W. Fouke. 2004. Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 12.Mills, D. K., K. Fitzgerald, C. D. Litchfield, and P. M. Gillevet. 2003. A comparison of DNA profiling techniques for monitoring nutrient impact on microbial community composition during bioremediation of petroleum-contaminated soils. J. Microbiol. Methods 54:57-74. [DOI] [PubMed] [Google Scholar]
- 13.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 15.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson, L. L. 2004. Black band disease, p. 325-349. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
- 17.Rützler, K., and D. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of the cyanophyte pathogen. PSZNI Mar. Ecol. 4:301-319. [Google Scholar]
- 18.Rützler, K., D. L. Santavy, and A. Antonius. 1983. The black band disease of Atlantic reef corals. III. Distribution, ecology and development. PSZNI Mar. Ecol. 4:329-358. [Google Scholar]
- 19.Sekar, R., L. T. Kaczmarsky, and L. L. Richardson. 2008. Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar. Ecol. Prog. Ser. 362:85-98. [Google Scholar]
- 20.Sekar, R., D. K. Mills, E. R. Remily, J. D. Voss, and L. L. Richardson. 2006. Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl. Environ. Microbiol. 72:5963-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievert, S. M., E. B. A. Wieringa, C. O. Wirsen, and C. D. Taylor. 2007. Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ. Microbiol. 9:271-276. [DOI] [PubMed] [Google Scholar]
- 22.Suomalainen, L.-R., H. Reunanen, R. Ijas, E. T. Valtonen, and M. Tiirola. 2006. Freezing induces biased results in the molecular detection of Flavobacterium columnare. Appl. Environ. Microbiol. 72:1702-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sussman, M., D. G. Bourne, and B. L. Willis. 2006. A single cyanobacterial ribotype is associated with both red and black bands on diseased corals from Palau. Dis. Aquat. Org. 69:111-118. [DOI] [PubMed] [Google Scholar]
- 24.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.