Abstract
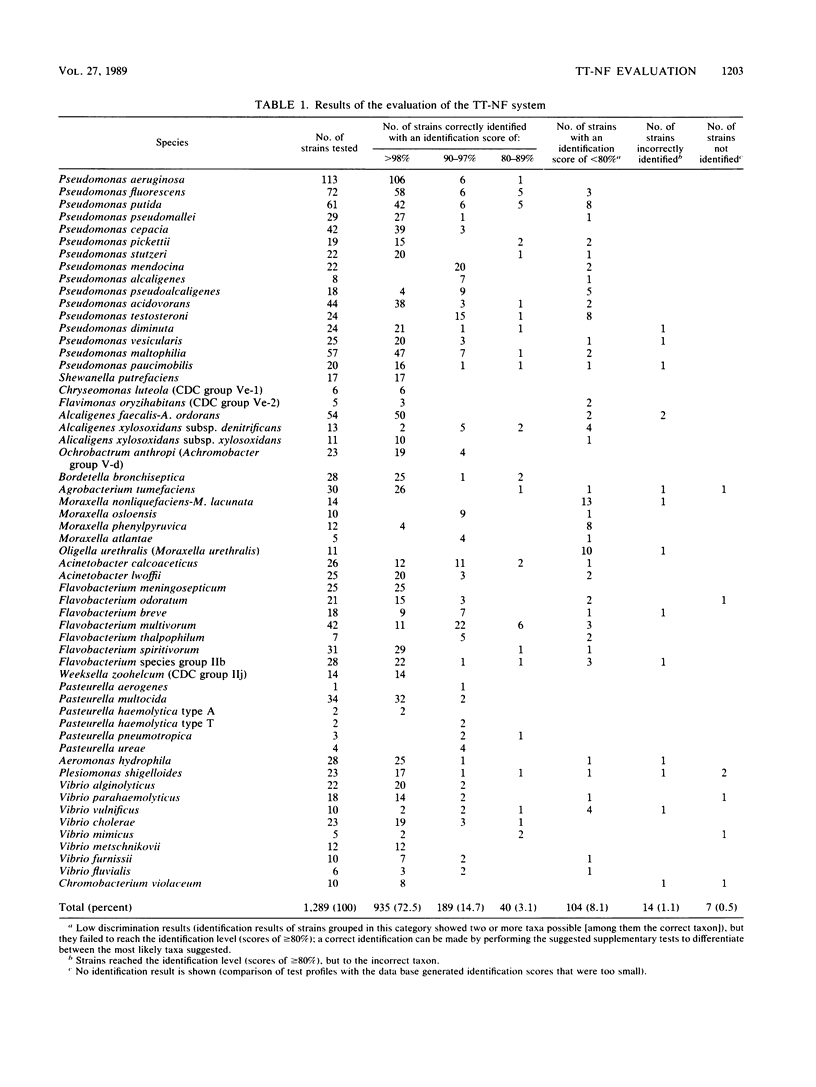
The Titertek-NF (TT-NF) system (Flow Laboratories GmbH, Meckenheim, Federal Republic of Germany) was evaluated for the identification of 1,289 strains of gram-negative, nonfermentative bacteria and some gram-negative, oxidase-positive bacteria. The oxidase test was also performed. Identifications were classified as correct, not identified (two or more taxa possible and identification score of less than 80%; supplementary tests for furthering the identification were not performed), and incorrect. Correct identification results were further subdivided by the correct level of species or biotype identification as greater than or equal to 98% (category a), 90 to 97% (category b), and 80 to 89% (category c). When compared with conventional identification results, the TT-NF system correctly identified 90.3% of strains (1,164 of 1,289 strains), with 72.5% (935 strains) belonging to category a, 14.7% of strains (189 strains) belonging to category b, and 3.1% of strains (40 strains) belonging to category c. Among the remaining strains, 104 (8.1%) were not identified and 14 (1.1%) were misidentified, and the system failed to generate identification results for 7 strains (0.5%). Reactions within the TT-NF system were reproducible, with an estimated probability of erroneous test results of 0.2%.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Algorta G., Monteil V., Popoff M. Y. Profils d'activités enzymatiques des genres Agrobacterium, Alcaligenes, Alteromonas, Flavobacterium et Pseudomonas. Ann Inst Pasteur Microbiol. 1987 May-Jun;138(3):297–302. doi: 10.1016/0769-2609(87)90117-7. [DOI] [PubMed] [Google Scholar]
- Appelbaum P. C., Leathers D. J. Evaluation of the rapid NFT system for identification of gram-negative, nonfermenting rods. J Clin Microbiol. 1984 Oct;20(4):730–734. doi: 10.1128/jcm.20.4.730-734.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard R. W., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic psuedomonads: Pseudomonas diminuta and P. vesiculare. J Gen Microbiol. 1968 Oct;53(3):349–361. doi: 10.1099/00221287-53-3-349. [DOI] [PubMed] [Google Scholar]
- Ballard R. W., Palleroni N. J., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970 Feb;60(2):199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- Bascomb S., Lapage S. P., Curtis M. A., Willcox W. R. Identification of bacteria by computer: identification of reference strains. J Gen Microbiol. 1973 Aug;77(2):291–315. doi: 10.1099/00221287-77-2-291. [DOI] [PubMed] [Google Scholar]
- Bruun B. Studies on a collection of strains of the genus Flavobacterium. 2. Nutritional studies. Acta Pathol Microbiol Immunol Scand B. 1983 Feb;91(1):35–41. doi: 10.1111/j.1699-0463.1983.tb00006.x. [DOI] [PubMed] [Google Scholar]
- Freney J., Laban P., Desmonceaux M., Gayral J. P., Fleurette J. Differentiation of gram negative rods other than Enterobacteriaceae and Vibrionaceae by a micromethod for determination of carbon substrate assimilation. Zentralbl Bakteriol Mikrobiol Hyg A. 1984 Dec;258(2-3):198–212. doi: 10.1016/s0176-6724(84)80038-3. [DOI] [PubMed] [Google Scholar]
- Holmes B., Dawson C. A., Pinning C. A. A revised probability matrix for the identification of gram-negative, aerobic, rod-shaped, fermentative bacteria. J Gen Microbiol. 1986 Nov;132(11):3113–3135. doi: 10.1099/00221287-132-11-3113. [DOI] [PubMed] [Google Scholar]
- Holmes B., Pinning C. A., Dawson C. A. A probability matrix for the identification of gram-negative, aerobic, non-fermentative bacteria that grow on nutrient agar. J Gen Microbiol. 1986 Jul;132(7):1827–1842. doi: 10.1099/00221287-132-7-1827. [DOI] [PubMed] [Google Scholar]
- Holmes B., Willcox W. R., Lapage S. P., Malnick H. Test reproducibility of the API (20E), Enterotube, and Pathotec systems. J Clin Pathol. 1977 Apr;30(4):381–387. doi: 10.1136/jcp.30.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer P., Bette W., Dott W. Automatisierte Mikromethode zur Bestimmung der Verwertbarkeit organischer Kohlenstoffquellen durch klinisch bedeutsame Pseudomonas-Arten. Zentralbl Bakteriol Mikrobiol Hyg A. 1987 Jun;265(1-2):62–73. [PubMed] [Google Scholar]
- Kämpfer P., Bette W., Dott W. Phenotypic differentiation of members of the family Vibrionaceae using miniaturized biochemical tests. Zentralbl Bakteriol Mikrobiol Hyg A. 1987 Oct;266(3-4):425–437. doi: 10.1016/s0176-6724(87)80223-7. [DOI] [PubMed] [Google Scholar]
- Lampe A. S., van der Reijden T. J. Evaluation of commercial test systems for the identification of nonfermenters. Eur J Clin Microbiol. 1984 Aug;3(4):301–305. doi: 10.1007/BF01977477. [DOI] [PubMed] [Google Scholar]
- Lapage S. P., Bascomb S., Willcox W. R., Curtis M. A. Identification of bacteria by computer: general aspects and perspectives. J Gen Microbiol. 1973 Aug;77(2):273–290. doi: 10.1099/00221287-77-2-273. [DOI] [PubMed] [Google Scholar]
- Martin R., Riley P. S., Hollis D. G., Weaver R. E., Krichevsky M. I. Characterization of some groups of gram-negative nonfermentative bacteria by the carbon source alkalinization technique. J Clin Microbiol. 1981 Jul;14(1):39–47. doi: 10.1128/jcm.14.1.39-47.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Siavoshi F., McDougal D. L. Comparison of Rapid NFT system and conventional methods for identification of nonsaccharolytic gram-negative bacteria. J Clin Microbiol. 1986 Dec;24(6):1089–1092. doi: 10.1128/jcm.24.6.1089-1092.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni N. J., Doudoroff M., Stanier R. Y., Solánes R. E., Mandel M. Taxonomy of the aerobic pseudomonads: the properties of the Pseudomonas stutzeri group. J Gen Microbiol. 1970 Feb;60(2):215–231. doi: 10.1099/00221287-60-2-215. [DOI] [PubMed] [Google Scholar]
- Pichinoty F., Véron M., Mandel M., Durand M., Job C., Garcia J. L. Etude physiologique et taxonomique du genre Alcaligenes: A. denitrificans, A. odorans et A. faecalis. Can J Microbiol. 1978 Jun;24(6):743–753. doi: 10.1139/m78-123. [DOI] [PubMed] [Google Scholar]
- Pickett M. J., Greenwood J. R. Identification of oxidase-positive, glucose-negative, motile species of nonfermentative bacilli. J Clin Microbiol. 1986 May;23(5):920–923. doi: 10.1128/jcm.23.5.920-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintér M., Kántor M. Comparison of Alcaligenes faecalis and Alcaligenes odorans var. viridans by carbon source utilization test. Acta Microbiol Acad Sci Hung. 1974;21(3-4):293–295. [PubMed] [Google Scholar]
- Rarick H. R., Riley P. S., Martin R. Carbon substrate utilization studies of some cultures of Alcaligenes denitrificans, Alcaligenes faecalis, and Alcaligenes odorans isolated from clinical specimens. J Clin Microbiol. 1978 Sep;8(3):313–319. doi: 10.1128/jcm.8.3.313-319.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoden D. L., Schable B., Smith P. B. Evaluation of PASCO MIC-ID system for identifying gram-negative bacilli. J Clin Microbiol. 1987 Dec;25(12):2363–2366. doi: 10.1128/jcm.25.12.2363-2366.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell J. J., Lapage S. P. Carbon source utilization tests as an aid to the classification of non-fermenting gram-negative bacteria. J Gen Microbiol. 1973 Jan;74(1):9–20. doi: 10.1099/00221287-74-1-9. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Willcox W. R., Lapage S. P., Bascomb S., Curtis M. A. Identification of bacteria by computer: theory and programming. J Gen Microbiol. 1973 Aug;77(2):317–330. doi: 10.1099/00221287-77-2-317. [DOI] [PubMed] [Google Scholar]
- Willcox W. R., Lapage S. P., Holmes B. A review of numerical methods in bacterial identification. Antonie Van Leeuwenhoek. 1980;46(3):233–299. doi: 10.1007/BF00453024. [DOI] [PubMed] [Google Scholar]


