Abstract
The main aim of identifying gene–environment interactions is to provide insight into mechanisms of disease development and to identify patients with an inherent vulnerability to certain conditions. This in turn may allow patients to be targeted with individualised treatment based on the knowledge of their inborn susceptibility to specific conditions. This review describes the possible effects of common genetic variation on outcome in various conditions affecting the neonate. It focuses predominantly on studies of positive association rather than non‐association to illustrate this potential influence and to highlight the potential for further study and intervention. The shortcomings of published association studies and the place of such studies in future research are also discussed.
The human genome is made up of 3.2×109 DNA base pairs. There are millions of variations of DNA sequence within the human genome, of which just under half are changes in a single base pair (or nucleotide). Much of the human genome is redundant DNA that contains no functional genetic sequences, and thus small changes found in this DNA generally have little effect. Those simple DNA changes such as single base pair changes (of which about 10 million alone have been reported so far) that do occur in or near to the genes and affect their function make us the individuals we are.1 Such changes explain why some people are more susceptible to different diseases than others and why some peoples' illnesses respond to certain drugs whereas others will get side effects from those drugs.
The potential influence of simple common genetic variation and the promise that we can develop tailor‐made treatments with fewer side effects has fuelled the excitement around the human genome project. There is mounting evidence that these factors are pertinent to the disease processes that affect the newborn infant. These findings parallel the clinical impression that some infants, within the setting of the neonatal intensive care unit, react differently from others to the insults, and often apparently identical insults, to which they are exposed. This article will attempt to provide an overview of the role that common genetic variation may have in the outcomes of the newborn infant, and its potential importance in the future. The problems with the design of the work described and the interpretation of some of the associations reported will also be discussed.
Gene expression and DNA polymorphism
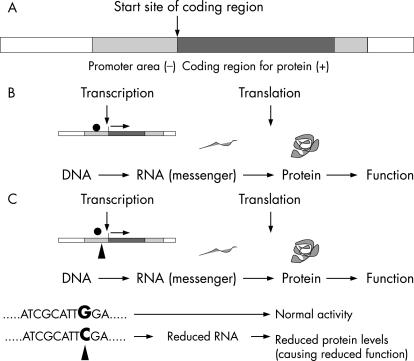
The genes of the human genome are recognised by polymerase enzymes that produce messenger molecules (made of RNA). These molecules are in turn usually decoded to produce proteins (fig 1). The protein (or peptide) produced has a functional effect—for example, an enzyme activity or a stimulus to a cell. The process of production of a messenger copy of the gene to the production of the final functional protein is known as expression. A single change in the DNA code (eg, G→C), in or near a gene can affect the amount of the message or protein produced or even the function of the protein itself. Thus a single change in the DNA code can alter the expression of the gene. A single change in the DNA sequence is known as a single nucleotide polymorphism (SNP). An SNP that affects the expression and hence the function of a gene is known, therefore, as a functional SNP (see fig 1). Functional SNPs may have only a small effect on a gene, perhaps reducing the protein produced by a small amount. However, this change may be enough to alter susceptibility to a disease or the efficacy of a drug.
Figure 1 (A) A schematic gene. A strand of DNA contains genes, areas that regulate genes, and areas that either have no function. A gene contains the coded information to make a protein. The code is written in the DNA sequence of nucleotides (bases): CTAG. The genetic information contained in a strand of DNA is determined by the sequence of the bases. (B) Gene expression. RNA polymerase (•) binds to the promoter region of the gene. Then RNA is produced (a faithful copy of the genetic code of the gene: a process known as transcription) to produce the messenger RNA, which is then converted into an amino acid chain by reading of the RNA code (translation). The final amino acid chain is then processed by the cell to produce a functional protein. (C) Functional single nucleotide polymorphism. A single nucleotide change (▴) in the DNA sequence may alter gene expression by changing levels of the protein produced or activity of the protein. Here the change in the single nucleotide or base occurs in the promoter area of the gene (the area where the RNA polymerase binds) and thus affects messenger RNA production, leading to altered protein levels and activity.
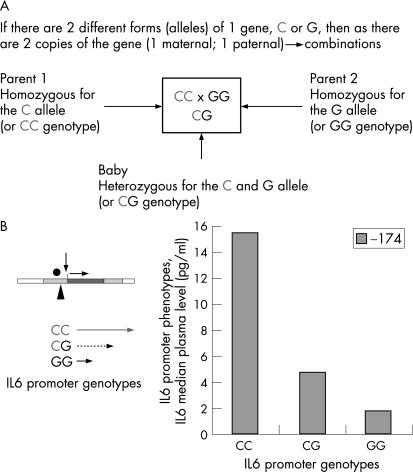
Simple functional changes in DNA sequence are inherited (fig 2). Where two different functional forms of a gene exist (alleles) the parents will pass one copy of each gene to their offspring to generate different combinations (or genotypes). The baby may then, depending on the gene combinations inherited, exhibit a different biochemical response, and perhaps physiological outcome, to a given stimulus as a result of the different genotypes they possess.
Figure 2 The effect of inherited gene combinations (using interleukin (IL) 6 as an example). (A) The different variants of a gene are known as alleles. The different combinations of the same DNA polymorphism are known as genotypes. (B) The IL6 promoter polymorphism: C or G at position −174 affects transcription (messenger RNA production) and thus IL6 production and IL6 activity. The phenotype is the effect that the polymorphic DNA change has at any level above that of the DNA in this case, an alteration in IL6 activity. Graphical data from Brull et al2 showing IL6 response after coronary artery bypass graft (CABG) in adults as influenced by genotypes.
Besides SNPs, other forms of genetic variation are also commonly found in the human genome, such as a deletion or insertion of a piece of DNA into a gene or its regulatory regions, inversions of areas of DNA sequences, and variation in the number of times a small DNA sequence is repeatedly found at one site within a gene (polymorphic copy number variants). In addition, some genes code only for an RNA molecule, and not a protein, as the RNA may have a have a structural or regulatory role—for example, ribosomal RNA. Most DNA changes do not occur within the coding region of the genes but in the non‐coding DNA sequences (the regulatory areas of the gene or the non‐coding parts of the messenger molecule—the exons). However, all these types of DNA change may alter the expression of a gene and thus its function.
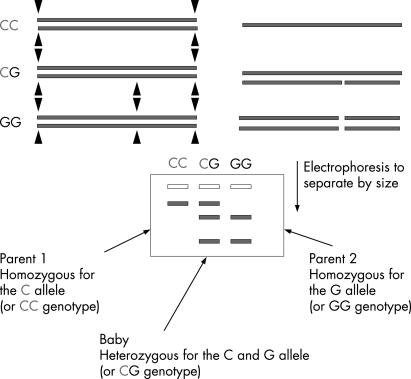
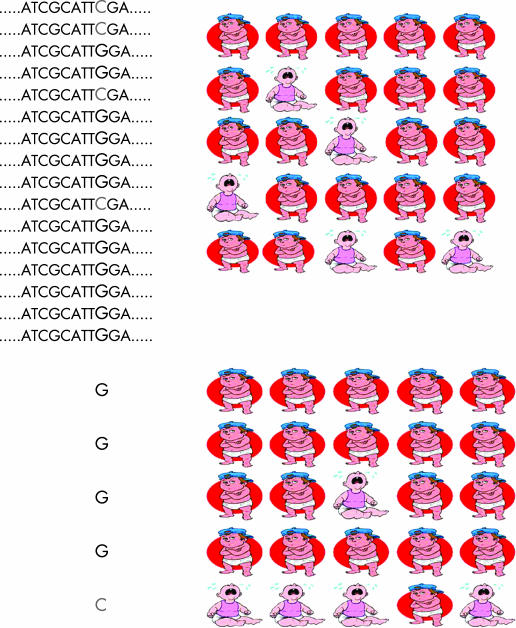
SNPs may be easily detected in the laboratory because changes to a DNA sequence may alter how the DNA is chopped up by enzymes called restriction enzymes. This is possible as restriction enzymes recognise specific DNA sequences. Abolishing a DNA cut site will increase the length of DNA produced after digestion with a restriction enzyme. This change in the gene of interest can then be seen on a gel after amplification of the DNA and electrophoresis (fig 3) or other forms of separation techniques more suited to automated, large scale analysis. DNA can be isolated from individuals in a population and then the version(s) of the DNA (the SNP for example) carried by the individuals in the population determined. Similarly, different types of polymorphism can be identified by other related techniques. The different forms of the gene of interest can then be compared with a specific disease or clinical condition in a study group. If one form of the gene is found in a higher than expected frequency in patients with the condition of interest this suggests that there is an association between that form of the gene and the condition (fig 4). Such a study is known as an association study of gene–environment interaction and is usually designed as a case–control or case‐only study.
Figure 3 Restriction fragment length polymorphism (RFLP) and detection of alleles. Restriction enzyme digestion of DNA occurs at specific DNA sequences (▴). If a polymorphism (change in DNA sequence) occurs in a restriction enzyme site near a gene of interest different sized molecules (corresponding to the alleles) will be produced that can be demonstrated on electrophoresis.
Figure 4 An association study to determine if a functional SNP (genotype) is associated with a condition (phenotype). Determine the genotypes within the population, measure the study populations characteristics and determine if the frequency of the SNP in those with the condition of interest is as expected or over‐represented.
For a much fuller review of the concepts underlying genetic epidemiological studies than is possible here see the recent series of reviews beginning with Burton et al.1 The real benefits of this type of work lie in the uncovering of functional forms of genetic variation associated with a condition. Because of the plausibility that this change and its biochemical effect is causal to the disease process, the individuals at greater risk of a condition may, in theory, be targeted with a specific treatment with greater efficacy and safety.
Common polymorphism and perinatal outcome
There is a genetic predisposition to preterm labour: approximately 20% of women with one previous preterm birth have a second preterm delivery and sisters of women who have had a preterm birth will also probably have a preterm baby. African American woman are also known to be more likely than white American women to give birth prematurely (even after accounting for differences in socio‐demographics and access to prenatal care),3 suggesting a racial predisposition and thus genetic predisposition to preterm labour. Given that preterm labour affects about 10% of all women and most neonatal mortality and morbidity is a consequence of preterm birth, it is fair to argue that the role that changes such as SNP may have in this process is an important one.
Spontaneous preterm birth is a multifactorial condition, so one individual genetic change will probably not prove to be highly predictive of the condition. Functional polymorphisms that affect key pathways will probably have a small but important role in certain clinical situations. For example, there is a body of evidence indicating that inflammation plays a major part in preterm birth, therefore, alterations to the activity of the innate immune system by functional SNPs may be important in the predisposition to preterm labour and its sequelae, particularly in the situation of evolving intrauterine infection or premature rupture of the membranes. Tumour necrosis factor (TNF) α is a proinflammatory cytokine responsible for initiating and sustaining the inflammatory response and is encoded by a polymorphic gene. The two alleles are known as TNF1 and TNF2 and result from a DNA change of a G→A located at nucleotide position –308 (ie, in the promoter region of the gene). The TNF2 allele appears to cause an increased basal TNFα gene expression and greater gene expression in response to inflammatory stimuli that may be responsible for a sevenfold increased risk of death or severe neurological outcomes from cerebral malaria in Gambian children carrying two copies of TNF2 (ie, is homozygous).4 In the context of preterm birth the same genotype (homozygous carriage of two TNF2 alleles by the fetus of women from west Kenya) was associated with a sevenfold increase in the risk of preterm birth.5 Similar but less strong effects, perhaps a doubling of the risk of preterm labour, have been observed in white Australian women, where combinations of three different TNFα polymorphisms (or haplotype) whose net effect is to enhance TNFα expression were analysed (the TNFα +488/−238/−308 AGG haplotype).6,7 A similar picture has been observed in association with (proinflammatory) interleukin (IL) 6 gene polymorphism.
Babies born small for gestational age have a greater range of medical problems across comparable gestations. Pre‐eclampsia and intrauterine growth restriction also show racial and genetic differences and several common SNPs have been implicated in the pathogenesis of these conditions. Activation of the renin–angiotensin system (by ACE and angiotensin) results in vasodilation, angiogenesis, decrease in metabolic efficiency, enhancement of the inflammatory response and prothrombosis. The pre‐proangiotensinogen gene is commonly polymorphic for a C→T change at position +704 (resulting in a methionine to threonine substitution at amino acid position 235 (M or T 235)). Women when homozygous for the T235 change (or another linked change) show a threefold increase in angiotensinogen RNA production in the uterine spiral arteries.8 This effect seems to have important consequences with 20% of white women homozygous for the T235 allele developing pre‐eclampsia compared with only 1% of those homozygous for the M235 allele.9 The fetus of a woman homozygous for T235 is three times more likely to develop intrauterine growth restriction than a woman with M235, whereas a fetus carrying two copies of T235 is twice as likely to develop intrauterine growth restriction compared with a fetus with two copies of M235. Fetal carriage of the ACE D allele (insertion/deletion polymorphism or I/D) which encodes a doubling of circulating and tissue ACE in Caucasians (and thus elevated angiotensin) has also been found in twice the expected frequency in infants with intrauterine growth restriction.10 Compatible with this finding is the raised angiotensin concentrations in the cord blood of babies born with intrauterine growth restriction.11
The purpose of identifying such associations is to provide insight into mechanism of disease development, and to identify patients groups and target them with specific treatments. Mello et al12 identified non‐thrombophilic women who had previously had pre‐eclampsia and randomised those who were genetically programmed to make more ACE (being homozygous for the ACE D allele (DD genotype)) to receive low molecular weight heparin or placebo. In the treated DD women there was a reduction, by 75–85%, of pre‐eclampsia and fetal growth restriction (number needed to treat 5 and 3, respectively). Median birth weights and gestational age at birth were also statistically and clinically significantly better in the treatment group (3.1 kg (2.6–3.5) vs 2.4 kg (1.4–3.2), p<0.001 and 37 weeks (33–39) vs 34 weeks (28–37), p<0.02, respectively). Although larger confirmatory trials are required, this one study shows the potential influence that pharmacogenomics may have on perinatal medicine in the future.
Common polymorphism and neonatal outcome
Cardiorespiratory system associations
Development of respiratory distress syndrome (RDS) is related to prematurity, use of maternal steroids and delivery mode, as well as genetic factors. The most obvious example to clinicians is that black infants have a lower incidence of RDS. This RDS is often less severe than that observed in comparable Caucasian infants. The most studied of genetic influences on newborn lung disease are the surfactant protein genes (see Clark and Clark13 for an indepth review of this subject area). Common clinically relevant DNA changes (rather than the rare defects that cause an hereditary deficiency) are found in the surfactant protein (SP) A, B, C and D genes. The SP‐A1 6A2 allele and the SP‐B 3131Thr contribute to the development of RDS (assuming causality). For instance homozygosity for the SP‐A1 6A2 allele and the SP‐B Thr131 change confers a nearly fourfold greater risk of developing RDS in preterm singletons.14 Similarly, a susceptibility to RDS is conferred by SP‐C gene polymorphisms. No such predisposition has been identified for SP‐D (although a later susceptibility to severe respiratory syncytial virus (RSV) infection by homozygosity for the Met11 allele has been revealed).15 SP‐A gene changes in the Caucasian German populations and SPA‐B gene changes in the Finnish population have also been associated with an increased risk of chronic lung disease (CLD) in the preterm infant.16,17
The activation of the inflammatory response in the preterm infant has been implicated in the development of CLD, therefore it is plausible that the many common functional proinflammatory and anti‐inflammatory cytokine gene polymorphisms may also be implicated in the development of early respiratory illness and chronic lung disease in the preterm. English Caucasian infants <33 weeks carrying the IL6 –174 CC genotype (encoding greater IL6 production in the newborn) experienced more severe early respiratory illness18 but no concomitant association was observed with the development of CLD due to lack of power. The TNFα −308 A↔G change encodes increased TNFα production in adults, but three studies have shown no association with increase CLD.18,19,20,21 Bokodi et al22 found a small increase in the duration of ventilatory support and oxygen need (<2 days in both cases) in the patients carrying the G allele, suggesting that the effect of this polymorphism in isolation may be small. Kazzi et al19 showed that the absence of an A allele (–238) within the TNFα gene, associated with decreased TNFα production, is in turn associated with protection from CLD. Taken together these observations indicate that the regulation of the TNFα gene may differ in the preterm newborn from that in adults or that the insults that predispose to CLD via a TNFα mediated pathway (if they exist) may be controlled preferentially via the effect of the change at position −238 of the TNFα promoter. Other cytokine polymorphisms (such as IL4, IL10 −1082 G/A, macrophage chemoattractant protein (MCP)‐1 −2518 A/G and transforming growth factor (TGF) β 1 +915 G/C) have not been linked with the development of long‐term lung injury in the preterm.20,23
English Caucasian infants born between 32 and 28 weeks carrying the ACE DD genotype (another proinflammatory genotype) also have a greater risk of developing RDS and are more likely to have more severe RDS.24 Compatible with this was the finding in another study that the incidence and severity of CLD of infants born weighing ⩽1250 g was threefold higher in those of ACE DD genotype compared with ID+II infants (74% vs 26%, respectively, p = 0.012).25 This finding was not replicated in another study of a similar population group.26 Unfortunately this dichotomy is not unusual and illustrates why it has been suggested that association studies should be issued with a “read at your own risk disclaimer”.27 Both of the studies of ACE I/D polymorphism in relation to CLD were racially heterogeneous, containing predominantly African American infants. They were thus underpowered to detect any difference in the Caucasian babies. It can also be argued that the bulk of available data suggests that the ACE I/D polymorphism does not have a biochemical, physiological or clinical effect in many studies of black adults, and thus these association studies may not be soundly based on biological plausibility within the population group studied. However, given the array of treatments that modify the renin‐angiotensin system, such as ACE inhibitors and angiotensin receptor antagonists, the potential for therapeutic intervention is promising should future work confirm a role for components of the renin‐angiotensin system in the development of CLD.
Common polymorphism of other genes affecting the cardiovascular system may also impact on newborn outcome. Angiotensin exerts its vasoconstrictor effects through the AT11 type 1 receptor and carriers of the AT1rCC genotype (position +1666) show a potent vasoconstrictor response (compared with AC or AA carrying counterparts). Treszl et al28 showed that infants of <32 weeks' gestation carrying the AT1rCC genotype would not probably develop a clinically important patent ductus arteriosus, and had no need for indometacin treatment at any stage in their care. This study suggests that a genotype directed prospective trial of indometacin treatment in the preterm may yield constructive results.
Abdominal associations
The risk of renal failure in the preterm is also partly genetically coded; common mutations encoding high TNFα, a low IL629,30 and also low heat shock protein 7031 (which provides a degree of ischaemic tolerance to the kidney) are all associated with acute renal failure in the preterm infant with hypotension, patent ductus arteriosus or septicaemia. These data suggest that a different threshold for intervention for hypotension in sepsis, for instance, is important for infants with the susceptible genotypes.
The development of septicaemia itself may also have a genetic predisposition. Preterm infants genetically determined to produce low basal producers of IL6 (−174GG carriers) had twice the rate of culture‐prove sepsis32,33 (IL6 acts at the granulocyte level to enhance cellular bacterial defence.) This increased rate of septicaemia, particularly of Gram‐positive organisms, was not observed in the genetically vulnerable infants who were given prophylaxis: the Gram‐positive sepsis rate in IL6 −174 GG very low birthweight infants given teicoplanin prophylaxis was 2.4% compared with 16.5% in those not given prophylaxis (non‐carriers of the GG genotype had a sepsis rate of 6.5%). However, a third, and larger, study of this polymorphism did not confirm this association.34 Hedberg et al35 showed a threefold increase in mortality from sepsis in those carrying the TNFα −308A allele, but not a greater predisposition to sepsis. Similar findings have been observed in adults.
Necrotising enterocolitis, a potentially devastating condition that seems strikingly individual in its appearance and severity, is another condition with inflammatory origins and systemic inflammatory effects. The individuality of this condition may be partly explained by genetic vulnerability of the gut to ischaemia and by modulators of the inflammatory response,36 hinting at ways in which to target treatments or even, perhaps, feeding regimens in the future.
Neurological and neurodevelopmental associations
Early studies suggest that standard neurodevelopmental outcomes too have an individual basis. Retinopathy of prematurity progressing to advanced disease was considerably higher (62% vs 38%) in Kuwaiti infants whose oxygen utilisation and control of vascular proliferation may have been rendered less competent as a result of being ACE DD genotype.37 The chances of needing treatment for severe retinopathy of prematurity were shown to be increased threefold in very low birthweight infants cared for in Budapest and homozygous for change in the vascular endothelial growth factor gene (VEGF 406+CC) and 16‐fold if carrying the VEGF haplotype −460TT/−405CC.38 Cooke et al showed a smaller effect in association with another polymorphism of the VEGF gene in infants of ⩽32 weeks' gestation of undefined race cared for in Liverpool.39
The inflammatory response has a critical role in the pathogenesis of neurological injury and subsequent developmental problems having particular, adverse, affinity for the cells of oligodendrocyte lineage. Proinflammatory pathways driven by mediators such as IL1, IL4, IL6, TNFα, have been implicated in the development of adverse neural outcomes such as intraventricular haemorrhage (IVH) and cystic periventricular leucomalacia (PVL). Polymorphisms that enhance the inflammatory effects of the environmental insults to which the preterm are often exposed such as chorioamnionitis and sepsis have, unsurprisingly therefore, also been associated with greatly increased risks of these two conditions. The IL1b −511T allele encoding increase IL1b production was shown to be strongly associated, albeit in preliminary work, with a doubling of risk of developing IVH and severe IVH.40 PVL was observed in 17% of infants with a T allele colonised with Ureaplasma urealyticum (used as a marker for chorioamnionitis in this study) compared with 4% with no T allele. Genetic predisposition to enhanced IL4 production (via the −590T rather than C allele) was also associated with an increase risk of PVL in African Americans.41 Although initial work in British Caucasians born at ⩽32 weeks indicates that endogenously higher IL6 production may also predispose to a greater risk of IVH and white matter damage,32 this was not found to be the case in a larger study of very low birthweight infants from Germany albeit with several potentially notable differences in the characteristics of study participants and outcome measures,34 or a study of predominantly American African very low birthweight infants. Furthermore, increased TNFα production encoded by the TNFα −308A allele also may double the risk of IVH 20 (but not severe IVH or cystic PVL). It is also possible that common polymorphism influencing thrombus formation and coagulation may influence the rates of IVH and PVL. See Baier41 for a more indepth review of this subject area and Hartel et al.42
Limited data are available on long‐term gene–environment neurodevelopmental follow‐up. In one case–control study 16.7% of white non‐Hispanic infants <32 weeks heterozygous for both the nitric oxide synthase −922 and factor VII 353 alleles were found to have cerebral palsy compared with 2.5% with other genotype combinations.43 Logistic regression identified a threefold increased risk of cerebral palsy for carriage of either of these genotypes individually. Disability occurred twice as often in one small preliminary study of very preterm carriers of the IL6−174CC genotype (predisposing to greater IL6 production) with no apparent effect on cognitive outcomes.24 In contrast cognitive outcome seemed to be influenced by another IL6 promoter polymorphism (−572) encoding enhanced IL6 production (Griffiths Developmental Quotient (median (interquartile range)): C allele 92.4 (89.9 to 96.6) vs GG 100.9 (96.7 to 104.8), p = 0.002; General Cognitive Ability: C allele 88.0 (80.3 to 102.8) vs GG 103.0 (92.0 to 112.0), p = 0.037.44 However, there were only 10 patients with this allele. Interestingly, infants who were presumed to be less able to induce cyclooxygenase 2 (by virtue of carrying the –765C allele) also had poorer cognitive performance at 2 and 5½ years, paralleling the lack of improvement in cognitive outcome of infants treated with indometacin despite reduction in serious cranial ultrasound abnormalities.45 The little work on cognitive outcome derives from a cohort of well‐described infants from the UK, who all survived to at least 2 years of age for follow‐up.
Mortality
It is known, also, that common polymorphisms are associated with mortality—for example, the TGFβ(1) +91520 allele has been associated with a threefold risk of death from all causes in very low birthweight infants. Associations between other SNPs and an altered risk for mortality at certain times during a baby's stay in the neonatal intensive care unit (eg, ACE I/D, TNFα) have also been revealed. Future work should, perhaps, focus on the role that common polymorphism has in the predisposition to death or adverse neurodevelopmental outcomes.
Problems with gene association studies and outcome
As things stand reading or publishing a gene–environment interaction study must be undertaken with caution. The main problems with the design of many such studies which have been hinted at above are:
lack of homogeneity of the patient group—that is, the racial and ethnic heterogeneity of the cohort of patients included (in whom different patterns of polymorphisms and different disease patterns are observed);
lack of homogeneity of the condition of interest studied (the outcome measure).
the small number of patients included in many studies leading to type I errors and inability to generalise findings;
lack of biological plausibility—any study should provide a full and thorough explanation of the biological mechanisms underlying the hypothesis driving the candidate association, in particular candidate polymorphic change should be functional (although the methods used for determining the functional effect of a polymorphism in vitro often have limitations).46
It has also been suggested27 that association studies should include a preliminary study, and a follow‐up study or contain evidence of functionality. Otherwise they should only be regarded as hypothesis generating until replicated adequately. Also the level of statistical significance that is set should be more rigorous than that usually accepted for other research. Not all studies will be able to reach all these targets, but these are important points to remember when designing or reading association studies.
It was not possible to discuss all of the—rapidly growing number of—published studies in this article. This review has focused more on positive association rather than non‐association to provide a flavour of how each body system, and thus neonatal outcomes, may be influenced by common genetic variation and also to highlight the potential for further study and intervention. Where the potential adverse effect of a SNP occurs in high frequency in the population, its clinical importance becomes more relevant.
The future
Given that many functional polymorphisms, including those described above, are common, and given the resources and effort spent on trying to improve outcomes, it is important that association studies are undertaken. There is, however, an urgent need for large, well‐designed gene association studies to further development of targeted therapeutic trials. This point has particular importance given that the potential impact of gene–environment interactions may be equal in strength to any intervention studied within the setting of such randomised controlled trials. It would therefore seem sensible when constructing clinical trials to include the potential for genetic study as few resources are required compared with the overall cost of most large multicentre studies.
Abbreviations
ACE - angiotensin‐converting enzyme
CLD - chronic lung disease
IL - interleukin
IVH - intraventricular haemorrhage
PVL - cystic periventricular leucomalacia
RDS - respiratory distress syndrome
SNP - single nucleotide polymorphism
TGF - transforming growth factor
TNF - tumour necrosis factor
Footnotes
Competing interests: None.
References
- 1.Burton P R, Tobin M D, Hopper J L. Key concepts in genetic epidemiology. Lancet 2005366941–951. [DOI] [PubMed] [Google Scholar]
- 2.Brull D J, Montgomery H E, Sanders J.et al Interleukin‐6 gene ‐174g>c and ‐572g>c promoter polymorphisms are strong predictors of plasma interleukin‐6 levels after coronary artery bypass surgery. Arterioscler Thromb Vasc Biol 2001211458–1463. [DOI] [PubMed] [Google Scholar]
- 3.Goldenberg R L, Cliver S P, Mulvihill F X.et al Medical, psychosocial, and behavioral risk factors do not explain the increased risk for low birth weight among black women. Am J Obstet Gynecol 19961751317–1324. [DOI] [PubMed] [Google Scholar]
- 4.McGuire W, Hill A V, Allsopp C E.et al Variation in the TNF‐alpha promoter region associated with susceptibility to cerebral malaria. Nature 1994371508–510. [DOI] [PubMed] [Google Scholar]
- 5.Aidoo M, McElroy P D, Kolczak M S.et al Tumor necrosis factor‐alpha promoter variant 2 (TNF2) is associated with pre‐term delivery, infant mortality, and malaria morbidity in western Kenya: Asembo Bay Cohort Project IX. Genet Epidemiol 200121201–211. [DOI] [PubMed] [Google Scholar]
- 6.Annells M F, Hart P H, Mullighan C G.et al Interleukins‐1, ‐4, ‐6, ‐10, tumor necrosis factor, transforming growth factor‐beta, FAS, and mannose‐binding protein C gene polymorphisms in Australian women: risk of preterm birth. Am J Obstet Gynecol 20041912056–2057. [DOI] [PubMed] [Google Scholar]
- 7.Engel S A, Erichsen H C, Savitz D A.et al Risk of spontaneous preterm birth is associated with common proinflammatory cytokine polymorphisms. Epidemiology 200516469–477. [DOI] [PubMed] [Google Scholar]
- 8.Morgan T, Craven C, Nelson L.et al Angiotensinogen T235 expression is elevated in decidual spiral arteries. J Clin Invest 19971001406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan T, Craven C, Lalouel J M.et al Angiotensinogen Thr235 variant is associated with abnormal physiologic change of the uterine spiral arteries in first‐trimester decidua. Am J Obstet Gynecol 199918095–102. [DOI] [PubMed] [Google Scholar]
- 10.Akisu M, Balim Z, Cetin H.et al The role of angiotensin‐converting enzyme and apolipoprotein‐E gene polymorphisms on lipid compositions in newborn infants with intrauterine growth restriction. Early Hum Dev 20047895–103. [DOI] [PubMed] [Google Scholar]
- 11.Kingdom J C, McQueen J, Connell J M.et al Fetal angiotensin II levels and vascular (type I) angiotensin receptors in pregnancies complicated by intrauterine growth retardation. BJOG 1993100476–482. [DOI] [PubMed] [Google Scholar]
- 12.Mello G, Parretti E, Fatini C.et al Low‐molecular‐weight heparin lowers the recurrence rate of pre‐eclampsia and restores the physiological vascular changes in angiotensin‐converting enzyme DD women. Hypertension 20054586–91. [DOI] [PubMed] [Google Scholar]
- 13.Clark H, Clark L S. The genetics of neonatal respiratory disease. Semin Fetal Neonatal Med 200510271–282. [DOI] [PubMed] [Google Scholar]
- 14.Marttila R, Haataja R, Guttentag S.et al Surfactant protein A and B genetic variants in respiratory distress syndrome in singletons and twins. Am J Respir Crit Care Med 20031681216–1222. [DOI] [PubMed] [Google Scholar]
- 15.Lahti M, Lofgren J, Marttila R.et al Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr Res 200251696–699. [DOI] [PubMed] [Google Scholar]
- 16.Rova M, Haataja R, Marttila R.et al Data mining and multiparameter analysis of lung surfactant protein genes in bronchopulmonary dysplasia. Hum Mol Genet 2004131095–1104. [DOI] [PubMed] [Google Scholar]
- 17.Weber B, Borkhardt A, Stoll‐Becker S.et al Polymorphisms of surfactant protein A genes and the risk of bronchopulmonary dysplasia in preterm infants. Turk J Pediatr 200042181–185. [PubMed] [Google Scholar]
- 18.Harding D R, Dhamrait S, Whitelaw A.et al Does interleukin‐6 genotype influence cerebral injury or developmental progress after preterm birth? Pediatrics 2004114941–947. [DOI] [PubMed] [Google Scholar]
- 19.Kazzi S N, Kim U O, Quasney M W.et al Polymorphism of tumor necrosis factor‐alpha and risk and severity of bronchopulmonary dysplasia among very low birth weight infants. Pediatrics 2004114e243–e248. [DOI] [PubMed] [Google Scholar]
- 20.Adcock K, Hedberg C, Loggins J.et al The TNF‐alpha ‐308, MCP‐1 ‐2518 and TGF‐beta1 +915 polymorphisms are not associated with the development of chronic lung disease in very low birth weight infants. Genes Immun 20034420–426. [DOI] [PubMed] [Google Scholar]
- 21.Lin H C, Tsai F J, Tsai C H.et al Cytokine polymorphisms and chronic lung disease in small preterm infants. Arch Dis Child Fetal Neonatal Ed 200590F93–F94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bokodi G, Treszl A, Derzbach L.et al The association of the carrier state of the tumor necrosis factor‐alpha (TNFalpha) ‐308A allele with the duration of oxygen supplementation in preterm neonates. Eur Cytokine Netw 20051678–80. [PubMed] [Google Scholar]
- 23.Lin H C, Su B H, Chang J S.et al Nonassociation of interleukin 4 intron 3 and 590 promoter polymorphisms with bronchopulmonary dysplasia for ventilated preterm infants. Biol Neonate 200587181–186. [DOI] [PubMed] [Google Scholar]
- 24.Harding D, Dhamrait S, Marlow N.et al Angiotensin‐converting enzyme DD genotype is associated with worse perinatal cardiorespiratory adaptation in preterm infants. J Pediatr 2003143746–749. [DOI] [PubMed] [Google Scholar]
- 25.Kazzi S N, Quasney M W. Deletion allele of angiotensin‐converting enzyme is associated with increased risk and severity of bronchopulmonary dysplasia. J Pediatr 2005147818–822. [DOI] [PubMed] [Google Scholar]
- 26.Yanamandra K, Loggins J, Baier R J. The angiotensin converting enzyme insertion/deletion polymorphism is not associated with an increased risk of death or bronchopulmonary dysplasia in ventilated very low birth weight infants. BMC Pediatr 2004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anon Freely associating [editorial]. Nat Genet 1999221–2. [DOI] [PubMed] [Google Scholar]
- 28.Treszl A, Szabo M, Dunai G.et al Angiotensin II type 1 receptor A1166C polymorphism and prophylactic indomethacin treatment induced ductus arteriosus closure in very low birth weight neonates. Pediatr Res 200354753–755. [DOI] [PubMed] [Google Scholar]
- 29.Treszl A, Toth‐Heyn P, Kocsis I.et al Interleukin genetic variants and the risk of renal failure in infants with infection. Pediatr Nephrol 200217713–717. [DOI] [PubMed] [Google Scholar]
- 30.Vasarhelyi B, Toth‐Heyn P, Treszl A.et al Genetic polymorphisms and risk for acute renal failure in preterm neonates. Pediatr Nephrol 200520132–135. [DOI] [PubMed] [Google Scholar]
- 31.Fekete A, Treszl A, Toth‐Heyn P.et al Association between heat shock protein 72 gene polymorphism and acute renal failure in premature neonates. Pediatr Res 200354452–455. [DOI] [PubMed] [Google Scholar]
- 32.Harding D, Dhamrait S, Millar A.et al Is interleukin‐6–174 genotype associated with the development of septicemia in preterm infants? Pediatrics 2003112800–803. [DOI] [PubMed] [Google Scholar]
- 33.Ahrens P, Kattner E, Kohler B.et al Mutations of genes involved in the innate immune system as predictors of sepsis in very low birth weight infants. Pediatr Res 200455652–656. [DOI] [PubMed] [Google Scholar]
- 34.Gopel W, Hartel C, Ahrens P.et al Interleukin‐6–174‐genotype, sepsis and cerebral injury in very low birth weight infants. Genes Immun 2006765–68. [DOI] [PubMed] [Google Scholar]
- 35.Hedberg C L, Adcock K, Martin J.et al Tumor necrosis factor alpha –308 polymorphism associated with increased sepsis mortality in ventilated very low birth weight infants. Pediatr Infect Dis J 200423424–428. [DOI] [PubMed] [Google Scholar]
- 36.Treszl A, Heninger E, Kalman A.et al Lower prevalence of IL‐4 receptor alpha‐chain gene G variant in very‐low‐birth‐weight infants with necrotizing enterocolitis. J Pediatr Surg 2003381374–1378. [DOI] [PubMed] [Google Scholar]
- 37.Haider M Z, Devarajan L V, Al‐Essa M.et al Angiotensin‐converting enzyme gene insertion/deletion polymorphism in Kuwaiti children with retinopathy of prematurity. Biol Neonate 20028284–88. [DOI] [PubMed] [Google Scholar]
- 38.Vannay A, Dunai G, Banyasz I.et al Association of genetic polymorphisms of vascular endothelial growth factor and risk for proliferative retinopathy of prematurity. Pediatr Res 200557396–398. [DOI] [PubMed] [Google Scholar]
- 39.Cooke R W, Drury J A, Mountford R.et al Genetic polymorphisms and retinopathy of prematurity. Invest Ophthalmol Vis Sci 2004451712–1715. [DOI] [PubMed] [Google Scholar]
- 40.Baeir R J, Loggins J, Yanamanadra K. Association of the interleukin‐1beta ‐511 C/T polymorphism with intraventricular haemorrhage and periventricular leukomalacia in ventilated very low birth weight infants. J PaediatrChild Health20051025B [Google Scholar]
- 41.Baeir R J. Genetics of perinatal brain injury in the preterm infant. Front Biosci 2006111371–1387. [DOI] [PubMed] [Google Scholar]
- 42.Hartel C, Konig I, Koster S.et al Genetic polymorphisms of hemostasis genes and primary outcome of very low birth weight infants. Pediatrics 2006118683–689. [DOI] [PubMed] [Google Scholar]
- 43.Nelson K B, Dambrosia J M, Iovannisci D M.et al Genetic polymorphisms and cerebral palsy in very preterm infants. Pediatr Res 200557494–499. [DOI] [PubMed] [Google Scholar]
- 44.Harding D, Brull D, Humphries S E.et al Variation in the interleukin‐6 gene is associated with impaired cognitive development in children born prematurely: a preliminary study. Pediatr Res 200558117–120. [DOI] [PubMed] [Google Scholar]
- 45.Harding D, Humphries S E, Whitelaw A.et al Cognitive outcome and cyclooxygenase‐2 Gene (‐765 G/C) variation in the preterm. Arch Dis Child Fetal Neonatal Ed 200792F108–F112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattersley A T, McCarthy M I. What makes a good genetic association study? Lancet 2005661315–1323. [DOI] [PubMed] [Google Scholar]