Abstract
The Drosophila apterous (ap) gene encodes a protein of the LIM-homeodomain family. Many transcription factors of this class have been conserved during evolution; however, the functional significance of their structural conservation is generally not known. ap is best known for its fundamental role as a dorsal selector gene required for patterning and growth of the wing, but it also has other important functions required for neuronal fasciculation, fertility, and normal viability. We isolated mouse (mLhx2) and human (hLhx2) ap orthologs, and we used transgenic animals and rescue assays to investigate the conservation of the Ap protein during evolution. We found that the human protein LHX2 is able to regulate correctly ap target genes in the fly, causes the same phenotypes as Ap when ectopically produced, and most importantly rescues ap mutant phenotypes as efficiently as the fly protein. In addition, we found striking similarities in the expression patterns of the Drosophila and murine genes. Both mLhx2 and ap are expressed in the respective nerve cords, eyes, olfactory organs, brain, and limbs. These results demonstrate the conservation of Ap protein function across phyla and argue that aspects of its expression pattern have also been conserved from a common ancestor of insects and vertebrates.
As DNA sequence data generated by the genome projects fill the databases, an increasing number of genes related by sequence are being identified in the human and model systems genomes. These sequence comparisons are expected to provide invaluable insight into the function of human genes (for example see ref. 1). In some cases, genes related by sequence are known to play the same or similar functions in distantly related organisms. Some well-known examples are the Hox genes involved in antero-posterior patterning (refs. 2–4 and references therein), the dpp/bmp4, sog/chordin genes involved in embryonic dorso-ventral patterning (reviewed in refs. 5, 6), the Pax-6/eyeless genes involved in eye determination (7), the otd/otx genes (8), and genes involved in certain signaling pathways (9, 10). In most cases, however, it is not known whether structurally related genes play the same roles in different organisms. To address the question of conservation of gene function, cross-species approaches that include comparisons of expression patterns and functional assays in vivo are required.
Here we use these approaches to investigate the conservation between Drosophila apterous (ap), a member of the LIM-homeobox gene family, and its mammalian orthologs. LIM-homeobox genes encode proteins containing two N-terminal zinc-finger-like motifs, referred to as LIM domains, in addition to a homeodomain. These genes have been found in very different organisms, including vertebrates and invertebrates, and are involved in tissue patterning as well as in cell fate determination and differentiation (reviewed in refs. 11–13). Some examples are: Lhx1 required for the formation of the prechordal mesoderm (14), Lhx3 required for the specification of pituitary cell lineages (15), Isl1 required for the specification of motor neurons (16), and Lmx1 required for limb dorso-ventral patterning (17–19). In Caenorhabditis elegans, lin-11 and mec-3 are essential for the development of vulval precursors and mechanosensory neurons, respectively (20, 21). In Drosophila, islet is required for axon pathfinding and neurotransmitter identity (22) and arrowhead for the development of certain imaginal cells (23).
ap was the first LIM-homeobox gene isolated in Drosophila (24, 25) and is best known for its crucial role as a dorsal selector gene required for dorso-ventral patterning and growth of the wing (26–28). However, mutations in ap cause a variety of mutant phenotypes illustrating other functions of ap during development. For instance, ap mutant embryos lack specific muscles (28) and show neuronal fasciculation defects (29). In addition, ap mutants die within few days after eclosion from the puparium (24), and they are deficient in juvenile hormone, which leads to nonvitellogenic ovaries and low female sexual receptivity (30).
Many LIM-homeobox genes have been conserved during evolution; however, the functional significance of their structural conservation is generally not known (reviewed in ref. 12). We have isolated mouse (mLhx2) and human (hLhx2) ap orthologs. We used ectopic expression and rescue assays to investigate the extent of the functional conservation between the Drosophila and human genes. In these in vivo assays, the human and fly proteins are interchangeable. In addition, we found striking similarities in the expression patterns of the Drosophila and murine genes. These results demonstrate the conservation of Ap protein function across phyla and argue that aspects of its expression pattern have been conserved also from a common ancestor of insects and vertebrates.
MATERIALS AND METHODS
cDNA Isolation.
Two degenerate oligonucleotide pools were synthesized on the basis of two amino acid sequences conserved among Ap and other LIM-HD proteins. The sense primer [5′-GTI(G/T)TICAC(A/G)TI(A/G)AITG(T/C)TT(T/C)IIITG-3′] (I, inosine ) was directed to the amino acid sequence VxHxxCFxC of the LIM2 domain. The antisense primer [5′-CGIII(A/G)TT(T/C)TG(A/G)AACCAIAC(T/C)TG-3′] corresponds to the amino acid sequence QVWFQNxR present in the third helix of the homeodomain. Several cDNA fragments were amplified by using a mouse embryonic cDNA library. One of these fragments encoded an ORF with a high degree of similarity to the Ap homeodomain and was used to screen a λSH/lox-1 cDNA library prepared from E11 mouse embryos. The mLhx2 cDNA (1.8 kb) was recovered from clone pSH340 and used as probe to screen a human brain λZAPII cDNA library (Stratagene). The hLhx2 cDNA (2 kb) was recovered from clone p3B3A. Amino acid sequence comparisons were conducted by using the pileup program.
Generation of Upstream Activation Sequence (UAS):ap and UAS:hLhx2 Lines.
A 1.7-kb fragment containing the full ap ORF was cloned into the KpnI site of pUAST (31). The 2 kb hLhx2 cDNA was released from p3B3A and subcloned into the same vector. These constructs were introduced (32) into yw; apUGO35/CyOwglacZ (wg, wingless) flies. Several independent lines were generated in each case, all of which exhibit similar rescuing abilities (data not shown).
Drosophila Stocks.
The following stocks were used: ap-GAL4MD544 (33); 32B-GAL4 and UAS:lacZ (31); UAS:tau-GFP (34); 35UZ-1, fringe (fng)-lacZ (35), and vestigial (vg) dorsal/ventral boundary enhancer-lacZ (36). ptc-GAL4 and dpp-GAL4 were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). aprk568, an enhancer detector line that expresses lacZ in the nuclei of ap-expressing cells, as well as the ap null allele apUGO35, were described previously (24). The yw strain was used for in situ RNA hybridization and immunodetection of Ap.
Drosophila Crosses.
The experiments shown use lines UAS:apF29B and UAS:hLhx2F7A, which contain the P element insertions in the X chromosome. For rescue experiments, apUGO35/CyOwglacZ females carrying either the UAS:ap or the UAS:hLhx2 transgene were crossed to ap-GAL4MD544/CyOwglacZ males. Females lacking the UAS trangenes were used as negative control. For ectopic assays, males from these UAS lines were mated to females carrying the 32B-GAL4, ptc-GAL4, and dpp-GAL4 drivers. The P[35UZ-1] (fng-lacZ), P[vg-lacZ], and P[wg-lacZ] insertions were introduced independently into the UAS lines and then crossed to the dpp-GAL4 driver to study the regulation of ap-downstream genes. The ap VNC-GAL4 driver, which expresses GAL4 from the 2-kb ap-ventral nerve cord (VNC) enhancer (D.E. R.-L. & J. B., unpublished data) was crossed to UAS:tau-GFP to label the ap-expressing neurons in the central nervous system.
Antibody and 5-Bromo-4-chloro-3-indolyl β-d-Galactoside (X-Gal) Stainings.
Antibody staining of imaginal discs was performed as described (37). Mouse monoclonal anti-β-galactosidase antibodies (1:2,000; Promega), rat anti-Serrate serum (1:1,000; kindly provided by K. Irvine) and rat anti-Ap serum (37) were used. Ap immunodetection in adult head sections was conducted as outlined (38). For histochemical detection of β-galactosidase, wing discs and adult heads were stained with X-Gal (31) for 30 min to 2 h.
In situ RNA Hybridization.
Whole-mount in situ RNA hybridization in Drosophila embryos was carried out as outlined (39). Linearized ap cDNA was used as template in the digoxigenin-UTP RNA labeling kit (Boehringer Mannheim) to prepare sense and antisense probes. Whole-mount and sectional in situ RNA hybridization in mouse embryos was conducted as described (40). Digoxigenin- and 35S-labeled mLhx2 riboprobes were prepared from clone pSH340. Sections hybridized with sense probes did not reveal any specific signal (data not shown).
RESULTS
Cloning and Sequence of mLhx2 and hLhx2, the Murine and Human Orthologs of apterous.
The murine ortholog of ap (mLhx2) was isolated by using PCR and degenerate primers corresponding to the homeobox and LIM2 domain of the ap cDNA. The PCR product was then used to obtain a full-length cDNA from a mouse embryonic library. The human ap ortholog (hLhx2) was isolated by using the mouse cDNA to screen a human brain library (see Materials and Methods).
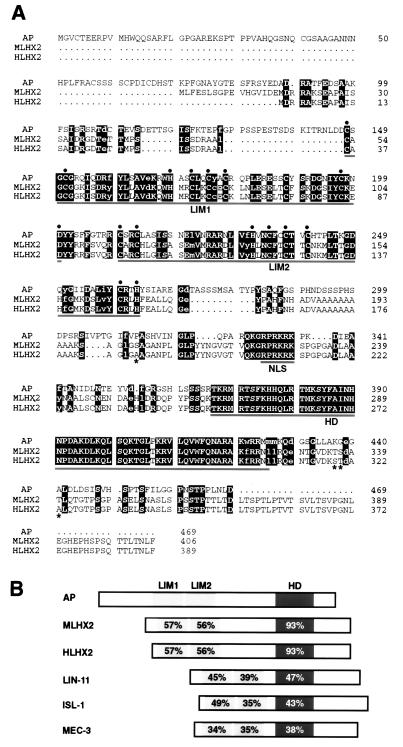
Fig. 1A shows the amino acid sequence comparison of the fly, mouse, and human proteins. MLHX2 and HLHX2 differ only in four amino acids except for an extended amino terminus of the mouse protein. The fly and mouse/human proteins show three major domains of sequence similarity that correspond to the LIM domain 1 (57% identity), LIM domain 2 (56% identity), and homeodomain (93% identity). However, conserved amino acids are found also outside these domains (Fig. 1A). Fig. 1B shows that the percent identities between Ap and its mammalian orthologs are clearly higher than between Ap and other LIM-homeodomain proteins.
Figure 1.
Amino acid sequence comparison of Drosophila Ap and its mouse (MLHX2) and human (HLHX2) orthologs. (A) Sequence alignment. Identical amino acids between the three proteins are displayed in reverse type with capital letters, whereas conservative substitions are displayed in lower case letters. Asterisks indicate the only four different residues between the mouse and human proteins in the overlapping region. The two tandem LIM domains, the putative nuclear localization signal (NLS), and the homeodomain (HD) are underlined. The consensus residues of the LIM domains are highlighted with black circles. Gaps denoted by dots have been inserted to maximize sequence alignment. (B) Domain comparisons between Drosophila Ap, its mammalian orthologs, and other LIM-homeodomain proteins. These are: ISL-1 from rat (60), LIN-11 and MEC-3 from C. elegans (20, 21). Percentage of amino acid sequence identity is indicated within the LIM domains and homeodomain.
The MLHX2 protein is 90% identical to a rat protein known as rLH2 (41), probably the rat ortholog of mLhx2. Mouse genes related to the mLhx2 gene described here have been reported elsewhere (41–43), but lack of sequence data prevents their comparison (see Discussion). The human Ap protein (HLHX2) is 92% identical to hLH2, a protein aberrantly expressed in chronic myelogenous leukemia (44). We mapped hLhx2 to chromosomal region 9q33–34.1 by fluorescent in situ hybridization (data not shown), the same chromosomal region where hLH2 maps (44).
Similarities in the Expression of apterous and mLhx2.
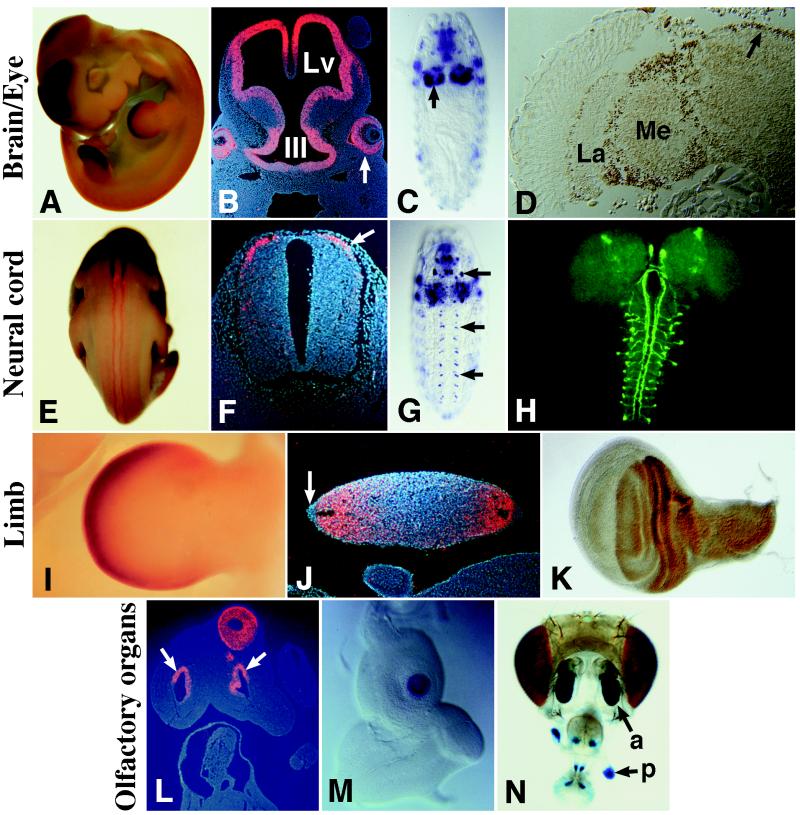
Expression of mLhx2 was investigated by in situ hybridization to whole-mounted and sectioned E9.5–12.5 embryos. mLhx2 expression was detected in the brain (Fig. 2 A and B), in the eyes (Fig. 2B), olfactory epithelium (Fig. 2L), and neural tube (Fig. 2 E and F). These patterns are reminiscent of ap expression in the embryonic and adult brain (Fig. 2 C and D), optic lobe (Fig. 2D), antenna, and maxillary palpus (the fly olfactory organs, Fig. 2 M and N), and VNC (Fig. 2G). The cells that express ap in the Drosophila VNC are interneurons, as revealed by driving expression of the tau-GFP reporter gene from the ap VNC enhancer (Fig. 2H); see also ref. 29. Thus we investigated the identity of the cells expressing mLhx2 in the mouse neural tube. Fig. 2F shows a section through the neural tube; mLhx2 label is detected in a dorso-lateral domain where dorsal commissural neurons are located. Interestingly, these are a subset of interneurons that, like Drosophila ap interneurons, send axons along longitudinal ascending tracts (45, 46). Other regions of mLhx2 expression include the liver, the infundibulum of the pituitary, and a small region of the branchial arches in E9.5 but not older embryos (data not shown).
Figure 2.
Comparison of ap and mLhx2 expression patterns. (A) mLhx2 expression in the forebrain and limbs of an E11.5 mouse embryo. (B) At this stage, mLhx2 is expressed in the walls of the lateral ventricles (Lv) and third ventricle (III) of the brain. In the eyes, mLhx2 is expressed in the future nervous layer of the retina (arrow) and in the optic stalk (not shown). (C) ap expression in the brain hemispheres (arrow) of a stage 15 fly embryo. (D) In the adult fly, Ap is immunodetected in the lamina (La) and medulla (Me) of the optic lobe and in the central brain (arrow). (E and F) At E11.5, mLhx2 is expressed along the neural tube (E) in a group of dorsal commissural interneurons (arrow in F). (G) ap expression in the VNC (arrows) of a stage 15 fly embryo. Out of focus, expression is also evident in the brain hemispheres and muscles of the body wall and pharynx. (H) Drosophila larval central nervous system showing expression of a UAS:tau-GFP responder driven by the ap-VNC enhancer. Note the axonal projections of ap-expressing interneurons along ascending longitudinal tracts. (I and J) mLhx2 expression in E11.5 mouse limbs. Label is detected in the mesenchyme, in a region roughly corresponding to the progress zone (I). In cross-sections, mLhx2 is observed in both dorsal (up) and ventral (down) regions of the limb and is excluded from the apical ectodermal ridge (arrow in J). (K) Ap immunodetection in the dorsal compartment of a Drosophila wing imaginal disc. (L) Section from an E11.5 mouse embryo showing mLhx2 expression in the olfactory epithelium surrounding the nasal pits (arrows). (M) ap expression in the center of a Drosophila antennal disc. (N) X-Gal stain of a Drosophila adult head carrying the enhancer detector aprk568, which expresses lacZ in an ap-like fashion. Note lacZ expression in the fly olfactory organs: the antenna (a) and the palpus (p).
In addition, mLhx2 expression was detected in the limb buds, specifically in the mesenchyme of the progress zone (Fig. 2 A and I) and excluded from the apical ectodermal ridge (Fig. 2J). Sections through the limb buds shows that mLhx2 is expressed both dorsally and ventrally (Fig. 2J) in contrast to the dorsal-specific expression of Drosophila ap (Fig. 2K). Also unlike ap, we did not detect mLhx2 expression in the somatic mesoderm.
hLhx2 Correctly Regulates apterous Target Genes in Drosophila and Mimics apterous in Ectopic Expression Assays.
The conservation of the Ap amino acid sequence and expression patterns from Drosophila to mammals prompted us to investigate the possible conservation of its functions using in vivo assays. The yeast GAL4/UAS system (31) was used to drive expression of a hLhx2 transgene in flies.
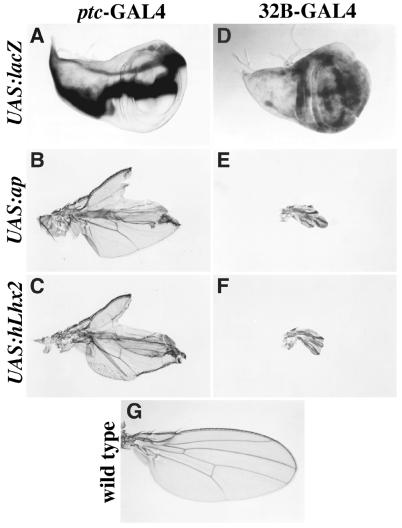
First we compared the phenotypic consequences of ectopic expression of hLhx2 and ap. Fig. 3 A and D show the lacZ expression pattern in the wing imaginal disc from two GAL4 drivers used in these experiments. Fig. 3 B and C (patched-GAL4 driver) and Fig. 3 E and F (32B-GAL4 driver) show that the severe wing mutant phenotypes produced by ectopic expression of ap or hLhx2 are virtually indistinguishable. For comparison Fig. 3G shows a wild-type wing.
Figure 3.
Ectopic expression of ap and hLhx2 in wing imaginal discs produce similar wing phenotypes. (A and D) UAS:lacZ expression from the ptc-GAL4 (A) and 32B-GAL4 (D) drivers. (B and C) Wing phenotypes caused by ectopic expression of ap (B) and hLhx2 (C) by using the ptc-GAL4 driver. (E and F) Wing phenotypes caused by ectopic expression of ap (E) and hLhx2 (F) by using the 32B-GAL4 driver. (G) Wild-type wing.
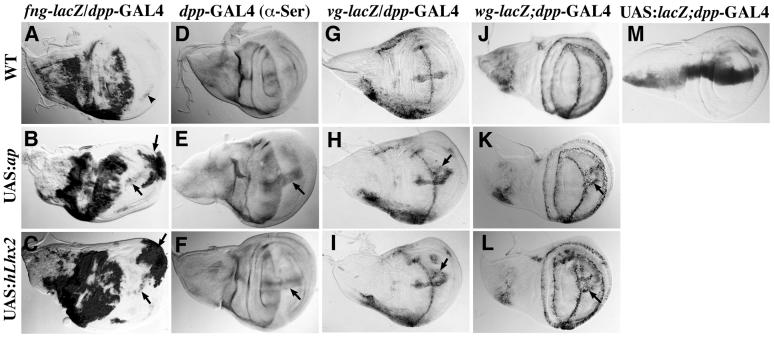
In addition, we investigated whether hLhx2 could regulate genes that are directly or indirectly under ap control, such as fng, Serrate (Ser), vg, and wg (35, 47). Using the dpp-GAL4 driver (Fig. 4M), we expressed UAS:hLhx2 and UAS:ap within the wing ventral compartment along the antero-posterior compartment boundary. Fig. 4A shows that fng expression in the wild-type wing disc is almost completely restricted to the dorsal compartment. Fig. 4 B and C show that hLhx2, like ap, activates fng expression in the ventral compartment. Fig. 4D shows the Ser wild-type expression pattern, which is also restricted to the dorsal compartment of the disc. Ectopic expression of ap and hLhx2 produced similar activation of Ser expression along the antero-posterior boundary (Fig. 4 E and F). Fig. 4 G and J show the wild-type expression patterns of vg and wg along the dorsal-ventral compartment boundary, respectively. On ap ectopic expression, vg and wg are ectopically activated as two parallel stripes within the ventral compartment (Fig. 4 H and K). These regulatory interactions are mimicked by hLhx2 ectopic expression (Fig. 4 I and L). As expected, the adult wings resulting from these crosses exhibit an ectopic wing margin along the ventral compartment (data not shown).
Figure 4.
hLhx2 correctly regulates ap target genes in Drosophila wing imaginal discs. Panels show β-galactosidase or Ser immunodetections following ectopic ap or hLhx2 expression in third instar larvae wing discs. The wild-type pattern of the fng-lacZ (A), Ser (D), vg-lacZ (G), and wg-lacZ (J) markers are depicted at the top. The dpp-GAL4 driver (M) was used to direct expression of the indicated UAS transgenes along the antero-posterior axis. Note that in all cases wing discs coexpressing dpp-GAL4 and either UAS:ap (B, E, H, K) or UAS:hLhx2 (C, F, I, L) exhibit ectopic activation of the molecular markers within the ventral compartment. Arrowhead indicates the wild-type expression of fng in the ventral compartment. Arrows point to the sites of ectopic expression.
Rescue of Drosophila apterous Mutant Phenotypes by a hLhx2 Transgene.
To test further the conservation of Ap protein functions from flies to humans, we investigated the ability of HLHX2 to substitute for Ap functions during Drosophila development.
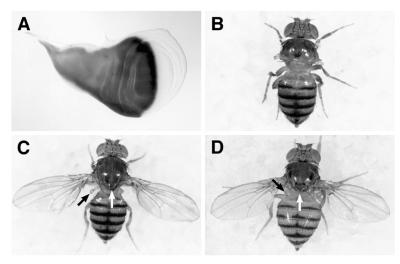
We took advantage of a GAL4 enhancer detector inserted in the ap locus (33). Insertion of the GAL4 P-element in ap causes GAL4 to be expressed like ap (Fig. 5A and data not shown). This insertion also results in a strong ap mutation leading to the lack of wings and halteres, as well as a mutant notum that lacks the scutellum and many of the bristles (Fig. 5B). In addition, these mutants also show the sterility and precocious death phenotypes associated with strong ap mutations (they have a life span of 1–3 days after eclosion from the puparium; data not shown). ap mutant flies carrying the ap-GAL4MD544 allele and the UAS:hLhx2 transgene show rescue of the ap wing, haltere, scutellum, and bristle mutant phenotypes (Fig. 5D). The sterility and precocious death phenotypes are also rescued (data not shown). We find that the fly (UAS:ap) and human (UAS:hLhx2) transgenes are equally able to rescue these phenotypes (see Fig. 5 C and D). The only difference that we detected between the two rescue transgenes is that flies that carry UAS:hLhx2 frequently develop one to two extra bristles in the scutellum, a phenotype not observed with UAS:ap.
Figure 5.
hLhx2 rescues the wing phenotype of ap mutants. (A) Wing imaginal disc expressing UAS:lacZ from the ap-GAL4MD544 driver. This GAL4 P-element insertion in ap inactivates the gene and recapitulates its expression pattern. (B) ap-GAL4MD544/apUGO35 mutant fly. Note the lack of wings, halteres, and the scutellum region of the notum. (C and D) ap-GAL4MD544/apUGO35 mutant flies carrying the UAS:ap and UAS:hLhx2 transgenes, respectively. Note that, in both cases, the wing, notum (white arrow), and haltere (black arrow) phenotypes are rescued.
DISCUSSION
Many proteins show a remarkable degree of amino acid sequence conservation between distantly related species. However, apparent structural conservation does not necessarily imply conservation of function in vivo. We have used transgenic animals and rescue assays to investigate the functional conservation of the Ap LIM-homeodomain protein during evolution. We found that the human protein HLHX2 is able to correctly regulate ap target genes in the fly, causes the same phenotypes as Ap when ectopically produced, and most importantly rescues ap mutant phenotypes as efficiently as the fly protein. These observations provide compelling evidence for the functional conservation of the Ap protein.
Other putative ap orthologs have been identified in vertebrates and invertebrates. In C. elegans, a LIM-homeobox gene closely related to ap is expressed in a specific interneuron required to mediate thermoregulation (48). In the crustacean Artemia franciscana, a putative ap ortholog is expressed in gill appendages (49). In vertebrate genomes, it appears that more than one ap-related gene is present. Paralogous genes closely related to ap have been reported in the chicken (50) and in the zebrafish (H. Okamoto, personal communication). Mouse genes related to the mLhx2 gene described here have been reported elsewhere (41–43). However, lack of reported DNA sequence and expression pattern data prevents their comparison. Thus we do not know whether these genes are the same as the mLhx2 gene described here or its paralogs.
Analysis of the expression pattern of the mouse ap ortholog described here (mLhx2) shows that it is expressed in many organs and tissues that are analogous or homologous to the organs and tissues where Drosophila ap is expressed. These include the eye, olfactory organs, limbs, brain, and neural tube. Particularly interesting are the expression patterns of the fly and mouse genes in the respective appendages (wing imaginal discs and limbs) and nerve cords. What is the significance of ap and mLhx2 similarities in their expression patterns? These similarities suggest that ap and mLhx2 may play the same or similar roles in flies and mice during differentiation of the respective limbs, brains, nerve cords, eyes, and olfactory organs.
ap expression in the Drosophila wing imaginal disc is required for specifying dorsal vs. ventral identity and for growth of the appendage (26–28, 47). These two distinct functions carried out by ap are separable. ap mutant flies in which the fringe gene is driven by the ap-GAL4 driver described here have normal-size wings that are double ventral (Jose de Celis, personal communication). As discussed below, these two functions of Drosophila ap are carried out in vertebrates by two different genes. In mice, mLhx2 does not appear to be involved in dorso-ventral specification of the limb because it is expressed on dorsal and ventral sides (Fig. 2J). In chicks, an ap/mLhx2 ortholog also shows dorsal and ventral limb expression (51). It is a different member of the LIM-homeobox family, Lmx1, the gene that specifies dorsal vs. ventral identity in vertebrates. Lack of Lmx1 function results in normal-size limbs that are double ventral (17–19).
Mice deficient for one of the ap orthologs have been generated, demonstrating a requirement of this gene in development of the eye, cerebral cortex, and efficient definitive erythropoiesis. These mutant mice do not show any limb phenotype, probably because of functional redundancy with other mouse ap paralogs (51, 52). However, a dominant negative form of a chicken ap ortholog results in arrested limb outgrowth during embryogenesis (51). These results suggest a conserved function of ap required for appendage growth in Drosophila and vertebrates.
In the Drosophila VNC, ap is expressed in a small number of cells per hemisegment. The activation of the tau-GFP reporter gene by the ap-GAL4MD544 driver allowed us to visualize the projections of these interneurons. Interestingly, in situ hybridization on mouse neural tube sections shows that the cells expressing mLhx2 precisely colocalize with a subset of interneurons that, like Drosophila ap interneurons, project along ascending longitudinal tracts to anterior segments and/or to the brain. Drosophila ap mutants show neuronal pathfinding defects (29); thus these observations suggest a conserved role for ap in interneuron identity or pathfinding.
There is still considerable controversy on the homology between the nerve cord of protostomes and deuterostomes (53–55). Although the nerve cord of deuterostomes is located dorsally instead of ventrally in protostomes, recent molecular data support the hypothesis that they are homologous (reviewed in ref. 8). The difference is explained as a consequence of an inversion of the dorso-ventral body axis between arthropods and chordates (5, 6). In addition, limbs, olfactory organs, and eyes have been classically considered to be analogous between arthropods and chordates, but see refs. 7 and 56 for novel views in the cases of eyes and limbs, respectively. Thus, according to orthodox views, it would have to be argued that ap functions were recruited independently at more than one time during evolution for the development of these analogous organs where they may carry out similar functions. Whatever the evolutionary relationship between fly and mouse organs might be, the similarities in ap expression support the idea (57) of a common set of functionally related genes involved in the development of the respective organ. This group of genes is known as a syntagma. Thus functionally related organs or tissues in flies and mice would have similar apogenomes (the combination of active genes within a given cell or tissue; ref. 57).
The similarities between the expression patterns of ap and mLhx2 leave some intriguing questions open for future investigations: What deep level of homology underlies their expression pattern similarities (58, 59)? Have ap regulatory elements been conserved between flies and mammals?
Acknowledgments
We are grateful to Mark Nalty for technical help in the initial stages of this work. We also thank Antonio Baldini for help with the fluorescence in situ hybridization (FISH) protocol, Urs Albrecht for providing reagents and advice on RNA in situ hybridization, and Ken Irvine for providing anti-Ser antibodies. We are grateful to Gerard Karsenty, Huda Zoghbi, Hugo Bellen, and Allan Bradley for providing comments to earlier versions of this manuscript. D.E.R.-L. and J.C.I.-B. are a Latin American Fellow and a Scholar of the Pew Charitable Trusts, respectively. C.-H.L. is supported by the Baylor Graduate Program in Developmental Biology. This work was supported by National Institutes of Health grant GM55681 to J.B. We also thank the Human Frontiers Science Program for supporting the collaboration between J.B. and J.C.I.-B.
ABBREVIATIONS
- ap
apterous
- VNC
ventral nerve cord
- fng, fringe
Ser, Serrate
- vg, vestigial
wg, wingless
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- UAS
upstream activation sequence
Footnotes
References
- 1.Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zollo M, et al. Nat Genet. 1996;13:167–174. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon C. Cell. 1994;78:175–180. doi: 10.1016/0092-8674(94)90288-7. [DOI] [PubMed] [Google Scholar]
- 3.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence P A, Morata G. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 5.De Robertis E M, Sasai Y. Nature (London) 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson E L. Curr Opin Genet Dev. 1996;6:424–431. doi: 10.1016/s0959-437x(96)80063-3. [DOI] [PubMed] [Google Scholar]
- 7.Halder G, Callaerts P, Gehring W J. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 8.Sharman A C, Brand M. Trends Genet. 1998;14:211–214. doi: 10.1016/s0168-9525(98)01488-7. [DOI] [PubMed] [Google Scholar]
- 9.Siegfried E, Perrimon N. BioEssays. 1994;16:395–404. doi: 10.1002/bies.950160607. [DOI] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Matsumo K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 11.Curtiss J, Heilig J S. BioEssays. 1998;20:58–69. doi: 10.1002/(SICI)1521-1878(199801)20:1<58::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–161. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 13.Jurata L W, Gill G N. Curr Top Microbiol Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 14.Shawlot W, Behringer R R. Nature (London) 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 15.Sheng H Z, Zhadanov A B, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A J, Lee E, Huang S-P, Mahon K A, Westphal H. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 16.Pfaff S L, Mendelsohn M, Stewart C L, Edlund T, Jessell T M. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 17.Riddle R D, Ensini M, Nelson C, Tsuchida T, Jessell T, Tabin C. Cell. 1995;83:631–640. doi: 10.1016/0092-8674(95)90103-5. [DOI] [PubMed] [Google Scholar]
- 18.Vogel A, Rodriguez C, Warnken W, Izpisúa-Belmonte J C. Nature (London) 1995;378:716–720. doi: 10.1038/378716a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg K C, Pepicelli C V, Gan L, Lee B, Johnson R L. Nat Genet. 1998;19:51–55. doi: 10.1038/ng0598-51. [DOI] [PubMed] [Google Scholar]
- 20.Way M C, Chalfie M. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 21.Freyd G, Kim S K, Horvitz H R. Nature (London) 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 22.Thor S, Thomas J B. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 23.Curtiss J, Heilig J S. Dev Biol. 1997;190:129–141. doi: 10.1006/dbio.1997.8659. [DOI] [PubMed] [Google Scholar]
- 24.Cohen B, McGuffin M E, Pfeifle C, Segal D, Cohen S M. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- 25.Bourgouin C, Lundgren S E, Thomas J B. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- 26.Blair S S. Development (Cambridge, UK) 1993;119:339–351. doi: 10.1242/dev.119.2.339. [DOI] [PubMed] [Google Scholar]
- 27.Diaz-Benjumea F, Cohen S M. Cell. 1993;75:741–752. doi: 10.1016/0092-8674(93)90494-b. [DOI] [PubMed] [Google Scholar]
- 28.Williams J A, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 29.Lundgren S E, Callahan C A, Thor S, Thomas J B. Development (Cambridge, UK) 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- 30.Altartz M, Applebaum S W, Richard D S, Gilbert L I, Segal D. Mol Cell Endocrinol. 1991;81:205–216. doi: 10.1016/0303-7207(91)90219-i. [DOI] [PubMed] [Google Scholar]
- 31.Brand A, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 32.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 33.Calleja M, Moreno E, Pelaz S, Morata G. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 34.Brand A. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- 35.Irvine K D, Wieschaus E. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 36.Williams J A, Paddock S W, Vorwerk K, Carroll S B. Nature (London) 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Fúnez P, Lu C-H, Rincón-Limas D E, García-Bellido A, Botas J. EMBO J. 1998;17:6846–6853. doi: 10.1093/emboj/17.23.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoulakis E M C, Davis R L. Neuron. 1996;17:931–944. doi: 10.1016/s0896-6273(00)80224-x. [DOI] [PubMed] [Google Scholar]
- 39.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 40.Albrecht U, Eichele G, Helms J A, Lu H-C. In: Molecular and Cellular Methods in Developmental Toxicology. Daston G P, editor. Boca Raton, FL: CRC; 1997. pp. 23–48. [Google Scholar]
- 41.Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz E M, Yancopoulos G D, Jessell T M, Alt F W. Proc Natl Acad Sci USA. 1993;90:227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberson M S, Schoderbek W E, Tremml G, Maurer R A. Mol Cell Biol. 1994;14:2985–2993. doi: 10.1128/mcb.14.5.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto K, Tanaka T, Furuyama T, Kashihara Y, Ishii N, Tohyama M, Kitanaka J, Takemura M, Mori T, Wanaka A. Neurosci Lett. 1996;211:147–150. doi: 10.1016/0304-3940(96)12749-x. [DOI] [PubMed] [Google Scholar]
- 44.Wu H-K, Heng H H Q, Siderovski D P, Dong W F, Okuno Y, Shi X M, Tsui L C, Minden M D. Oncogene. 1996;12:1205–1212. [PubMed] [Google Scholar]
- 45.Altman J, Bayer S A. Adv Anat Embryol Cell Biol. 1984;85:1–165. doi: 10.1007/978-3-642-69537-7. [DOI] [PubMed] [Google Scholar]
- 46.Helms A W, Johnson J E. Development (Cambridge, UK) 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, Irvine K D, Carroll S B. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- 48.Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 49.Averof M, Cohen S M. Nature (London) 1997;385:627–630. doi: 10.1038/385627a0. [DOI] [PubMed] [Google Scholar]
- 50.Nohno T, Kawakami Y, Wada N, Ishikawa T, Ohuchi H, Noji S. Biochem Biophys Res Commun. 1997;238:506–511. doi: 10.1006/bbrc.1997.7320. [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez-Esteban C, Schwabe J W R, De La Peña L, Rincón-Limas D E, Magallon J, Botas J, Izpisúa-Belmonte J C. Development (Cambridge, UK) 1998;125:3925–3934. doi: 10.1242/dev.125.20.3925. [DOI] [PubMed] [Google Scholar]
- 52.Porter F D, Drago J, Xu Y, Cheema S S, Wassif C, Huang S-P, Lee E, Grinberg A, Massalas J S, Bodine D, et al. Development (Cambridge, UK) 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 53.Nubler-Jung K, Arendt D. Wilhelm Roux’s Arch Dev Biol. 1994;203:357–366. doi: 10.1007/BF00188683. [DOI] [PubMed] [Google Scholar]
- 54.Jefferies P P, Brown N A. Nature (London) 1995;374:22. doi: 10.1038/374022a0. [DOI] [PubMed] [Google Scholar]
- 55.Lacalli T C, Peterson K J. Nature (London) 1995;373:110–112. [Google Scholar]
- 56.González-Crespo S, Abu-Shaar M, Torres M, Martínez-A C, Mann R S, Morata G. Nature (London) 1998;394:196–200. doi: 10.1038/28197. [DOI] [PubMed] [Google Scholar]
- 57.García-Bellido A. In: Genetics, Development, and Evolution. Gustafson J P, Stebbins G L, Ayala F J, editors. New York: Plenum; 1986. pp. 187–209. [Google Scholar]
- 58.Dickinson W J. Trends Genet. 1995;11:119–121. doi: 10.1016/s0168-9525(00)89015-0. [DOI] [PubMed] [Google Scholar]
- 59.Bolker J A, Raff R A. BioEssays. 1996;18:489–494. doi: 10.1002/bies.950180611. [DOI] [PubMed] [Google Scholar]
- 60.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Nature (London) 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]