Abstract
Over the past two decades, combined advances in genetics, developmental biology and biochemistry have transformed the study of human birth defects. This review describes the importance of genome architecture, parent of origin effects (imprinting), molecular pathophysiology, developmental pathways, mosaicism and cancer predisposition syndromes in the understanding of birth defects. This knowledge can be applied to improve diagnostic accuracy, prognostic information, counselling and sometimes even treatment of these conditions.
Keywords: genetics, imprinting, microarray, microdeletion, mosaicism
The modern era in the genetic study of human birth defects dates back to 1959, when Lejeune and colleagues reported that Down's syndrome was associated with an extra copy of chromosome 21. At a stroke this work identified the cause of the most common multiple congenital abnormality/mental retardation (MCA/MR) syndrome (∼1 in 700 births) and showed that syndromes could be analysed in terms of their underlying genetic origins. In the following two decades, cytogenetics was used to delineate many other chromosomal disorders. However, birth defects with a normal chromosome constitution remained difficult to study, even when a characteristic inheritance pattern in pedigrees (dominant or recessive, autosomal or X linked) indicated a genetic basis.
Two parallel developments during the 1980s gradually changed this. First, methods to isolate human genes and signpost their location on particular chromosomes (eg, using fluorescence in‐situ hybridisation to chromosomes, abbreviated “FISH”) made it possible to draw increasingly dense human gene maps. Second, geneticists and developmental biologists started to isolate and analyse the genes that specify the development of simple “model” animals, such as the fruitfly Drosophila and the nematode worm Caenorhabditis. This led to the remarkable discovery that many of these key developmental genes were so highly conserved during evolution that their direct equivalents could be identified in humans and located on the human chromosome map. This paved the way during the 1990s to the finding that mutations in many of these developmental genes cause human MCA/MR syndromes. In addition, the publication of the three billion nucleotide DNA sequence of the entire human genome (the draft version in 2001 and the “finished” sequence in 2006) has given researchers the information they need to analyse any gene, literally with the click of a mouse. Not surprisingly, several Nobel prizes have been awarded for this work (to the fly biologists in 1995 and the worm biologists in 2002), and more are bound to follow.
However, identification of genes and genetic defects is only the start. How are their functions integrated in the complex process of embryonic development, and how do mutations disrupt these processes in malformation? Although there are obvious ethical barriers to the study of these questions in humans, the basic mechanisms are conserved across a wide range of vertebrates (eg, mice, chicks and zebrafish), allowing detailed comparative analysis of development over time in these species. Sophisticated genetic techniques to manipulate the mouse genome are available and include gene addition (by inserting “transgenes”), gene interruption or “knockout” (by homologous recombination using embryonic stem cell technology), and the introduction of specific changes that directly recapitulate human disease‐causing mutations. The creation of “conditional” mutations now also allows disruption of a gene at a specified time in mouse development or in a tissue‐specific distribution, overcoming the problem of early embryonic death for certain targeted genes.1 Alternatively, sequence‐based “antisense” technologies such as morpholino oligonucleotides (using DNA) or RNA interference (RNAi) may be used to inhibit gene expression of a specific target—this often provides a more efficient and economical way of analysing loss of function than mouse genetics.2
Not surprisingly, this information has transformed the diagnosis of human birth defects and the understanding of their causes. This review aims to give a flavour of some of the major themes that have emerged in this field. Space constraints have necessitated omission of important topics such as the interactions between genes and the environment (eg, in neural tube defects) and the emerging importance of cilia in a subgroup of genetic diseases including Bardet–Biedl syndrome.3 For more details on the syndromes mentioned, we recommend the OMIM website4; the book Inborn errors of development5 provides a more comprehensive review of the molecular bases of human malformation.
Microdeletions and microduplications
Mechanisms of deletions and duplications
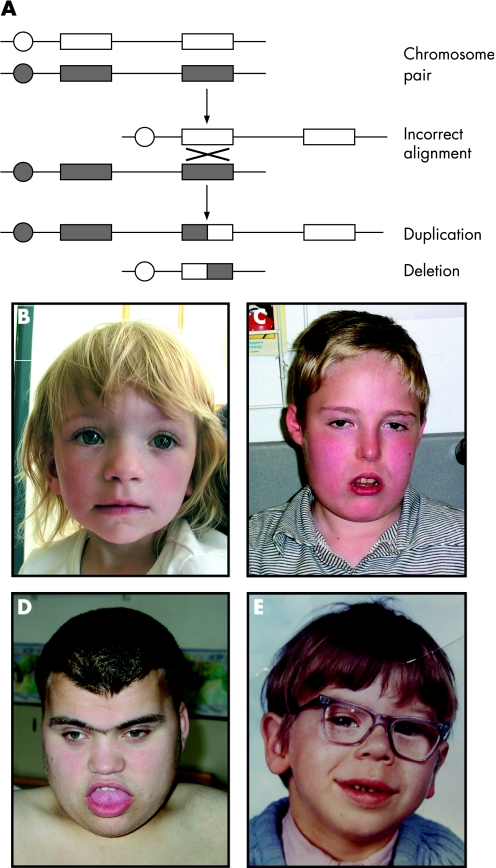
Chromosome abnormalities, including duplications and deletions, are a major cause of birth defects. Specific regions of the genome turn out to be particularly prone to these imbalances because they are flanked by blocks of low‐copy DNA repeats. Misalignment between these blocks during meiosis facilitates the occurrence of reciprocal deletion and duplication of the intervening regions (fig 1A). Well‐recognised microdeletion syndromes include 22q11 deletion syndrome (also called 22q11DelS, DiGeorge or velo‐cardio‐facial syndrome), Smith–Magenis syndrome and Williams–Beuren syndrome (fig 1C–E).
Figure 1 Duplication and deletion syndromes. (A) The mechanism of non‐allelic homologous recombination between two chromosomes resulting in unequal meiotic exchanges. Circle: centromere; rectangle: low‐copy DNA repeat. (B–E) Subtle, but recognisable dysmorphic facial features in patients with 22q11 duplication syndrome (B), 22q11 deletion syndrome (C), Smith–Magenis syndrome (17p11.2 deletion; D) and Williams–Beuren syndrome (7q11.23 deletion; E). Parental/guardian informed consent was obtained for the publication of these figures.
22q11DelS and duplication syndromes
22q11DelS is the commonest known microdeletion syndrome (∼1/4000 births). Most cases are caused by a three million base pair deletion on chromosome 22q11.2, identified by metaphase FISH. Clinically, 22q11DelS is characterised by hypocalcaemia and immunodeficiency (secondary to parathyroid and thymic hypoplasia, respectively), conotruncal heart defects, palatal abnormalities, learning difficulties and a distinct facial phenotype (fig 1C).6 Adults can develop major psychiatric illnesses including schizophrenia and bipolar affective disorder.
22q11DelS and many other recurrent microdeletion syndromes involve the deletion of multiple contiguous genes on one chromosome homologue, the other homologue being intact. For most genes, reduction from two to one functioning copy is not detrimental to the embryo, but for a minority of “haploinsufficient” genes, one copy is not enough. The question arises whether the phenotype is attributable to deletion of a single critical gene within the deleted interval or whether it reflects the combined contributions of several genes. Gene targeting in mice has identified haploinsufficiency of the transcription factor gene Tbx1 as the major cause of the 22q11DelS phenotype. Transcription factors are “master regulator” proteins that bind to DNA, affecting the expression of other genes; mutations of transcription factors are a common cause of developmental disorders. Indeed, Tbx1 knockout mice exhibit a similar phenotype to the human syndrome, and subsequently TBX1 mutations have been found in patients.7 However, these mutations only account for a few of the similarly affected cases that do not show the 22q11 deletion, leading to the theory that different mutations in other members of the TBX1 developmental pathway may be present.
More recently, interphase FISH has identified patients with duplications of the 22q11 region that, surprisingly, overlap clinically with the deletion syndrome phenotype (fig 1B).8 Whether this overlap is secondary to ascertainment bias or to a “two‐way” disturbance of gene dosage remains to be elucidated.
7q11 microdeletion and microduplication syndromes
Williams–Beuren syndrome is characterised by short stature, supravalvular aortic stenosis, characteristic facial features (fig 1E), hypercalcaemia and developmental delay with relatively preserved skills in expressive language.4 These patients have microdeletions around the elastin gene (ELN) locus at 7q11.23. Genotype–phenotype studies have suggested possible roles for the genes within the deletion.9 Mutations in ELN result in supravalvular aortic stenosis with none of the other characteristics of Williams–Beuren syndrome. GTF2IRD1 encodes a transcription factor that may contribute to some of the characteristic facial features in Williams–Beuren syndrome.10 These two examples suggest that this syndrome is truly a contiguous gene syndrome in contrast with 22q11DelS, in which the phenotype may, in large part, be due to haploinsufficiency of a single gene.
Reciprocal duplications of the Williams–Beuren syndrome interval have also been described.11 In contrast with the normal articulation and fluent expressive language observed in people with Williams–Beuren syndrome, there is a severe delay in expressive speech in people with duplication of the Williams–Beuren locus. This observation suggests that specific genes at 7q11.23 are sensitive to dosage alterations that can influence human language.
Array‐comparative genomic hybridisation
Array‐comparative genomic hybridisation (Array‐CGH) is a new, powerful technique for detection of submicroscopic chromosome duplications and deletions. The test and control sample are labelled with different fluorescent dyes and co‐hybridised to a microarray consisting of thousands of genomic clones or synthetic oligonucleotides from defined locations in the genome. The ratio of fluorescence intensities on each array element is proportional to the relative number of DNA copies between test sample and control.
This technological advance has led to the recognition that many patients with an undiagnosed MCA/MR syndrome have subtle chromosomal abnormalities that were previously undetectable by conventional karyotype analysis. If patients are selected strictly to include those with features commonly seen in chromosomal abnormalities, the detection sensitivity of causative imbalances can be around 15%.12
Array‐CGH is productive not only for diagnosis but also for discovering genes associated with diseases. CHARGE syndrome, for example, was originally defined as the association of coloboma, heart defects, atresia choanae, retardation of growth and development, genital and ear abnormalities. The identification of two patients with microdeletions on 8q12.1 using array‐CGH allowed a critical deletion interval to be defined, culminating in the screening of candidate genes in the region. Mutations in the chromodomain helicase DNA‐binding domain gene CHD7 were identified in a considerable proportion of CHARGE syndrome patients who did not have deletions, particularly those with hypoplasia of the semicircular canals.13
Imprinting (epigenetic) disorders
About a dozen birth defects do not show the usual pattern of mendelian inheritance and can be explained by the process of imprinting. Genomic imprinting occurs when the maternally and paternally inherited alleles are differentially expressed. In this situation, loss of the expressing allele by mutation or deletion is not compensated by the allele inherited from the other parent.
Prader–Willi and Angelman's syndromes
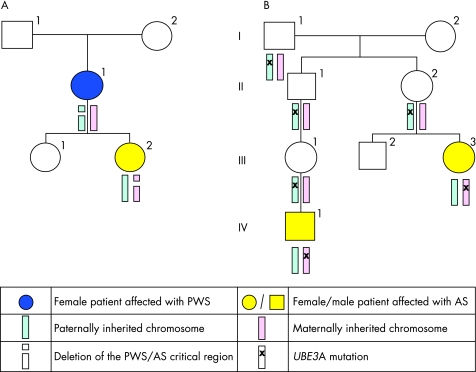
Two examples of imprinting disorders are Prader–Willi syndrome, characterised by neonatal hypotonia, subsequent hyperphagia, obesity and hypogonadism; and Angelman's syndrome, characterised by microcephaly, ataxia, severe mental retardation and an abnormal EEG pattern. Initially it was not known how two different conditions could be caused by a deletion of the same region of chromosome 15 (these are also examples of recurrent microdeletion syndromes). However, later it was realised that Prader–Willi syndrome is associated with deletion on the paternally inherited chromosome whereas Angelman's syndrome is associated with deletion on the maternally inherited chromosome.
A proportion of patients without deletions detectable by FISH, have Angelman's/Prader–Willi syndrome due to uniparental disomy (UPD), whereby two copies of chromosome 15 are inherited from one parent and none from the other parent. This seemingly improbable situation can arise when a fetus with trisomy 15, a relatively common but lethal condition, is “rescued” by the random loss of one of the three chromosome 15 s; in a third of cases, this results in UPD 15. The various molecular mechanisms of Prader–Willi and Angelman's syndromes are summarised in table 1.14 Figure 2 illustrates the inheritance patterns for two families: one with an Angelman's/Prader–Willi deletion and the other with a mutation of a specific imprinted gene within this region, UBE3A, that causes Angelman's syndrome.
Table 1 Mechanisms of selected birth defects caused by imprinting.
| Syndrome | Chromosome aneuploidy | Uniparental disomy | Imprinting centre defect | Other |
|---|---|---|---|---|
| Prader–Willi | Paternal Del 15q11–q12 (70%) | Maternal 15 (29%) | 1% | |
| Angelman's | Maternal Del 15q11–q12 (70%) | Paternal 15 (2%) | 4% | UBE3A mutations (5–10%) |
| Beckwith–Wiedemann | Paternal Dup 11p15 (2%) | Paternal 11 (15%) | 70% | CDKN1C mutations (10%) |
| Silver–Russell | Maternal Dup 11p15 (4%) | Maternal 7 (10%) | 11p15 (30%) |
Del, deletion; Dup, duplication.
Figure 2 Pedigrees illustrating the inheritance of Prader–Willi and Angelman's syndromes (PWS/AS). (A) Pedigree showing individual II:1 with a de novo deletion of the PWS/AS critical region on 15q11‐q12 on her paternal allele, resulting in Prader–Willi syndrome. One of her daughters, III:2, has inherited this deletion and has Angelman's syndrome (as the deletion is on her maternal allele). (B) Pedigree showing two individuals, III:3 and IV:1, with Angelman's syndrome due to a UBE3A mutation. The mutation only manifests if it passed through a female carrier (II:2 and III:1). Individuals I:1, II:1, II:2 and III:1 have all inherited the UBE3A mutation from their fathers. Their paternally derived chromosome is imprinted (naturally switched off) so they are phenotypically normal. III:2 has inherited the maternally derived chromosome without the UBE3A mutation.
Disorders of growth: Silver–Russell and Beckwith–Wiedemann syndromes
It is estimated that 10% of children with Silver–Russell syndrome (prenatal and postnatal growth retardation, asymmetry and minor congenital abnormalities) have maternal UPD7 (table 1).15 Despite many candidates, no specific genes on chromosome 7 have been solely implicated in the delayed growth. However, recent studies of the Beckwith–Wiedemann locus on 11p15 have found the insulin‐like growth factor 2 (IGF2) gene to be underexpressed in Silver–Russell syndrome and overexpressed in Beckwith–Wiedemann syndrome, an overgrowth disorder with predisposition to cancer. IGF2 is only expressed from the paternal allele and encodes a protein involved in prenatal and postnatal growth.16 Recent studies have suggested an increased incidence of Beckwith–Wiedemann syndrome in babies born using assisted reproduction technology, such as in vitro fertilisation and intracytoplasmic sperm injection.17 It is speculated that imprinted genes are vulnerable to epimutations when handled in an artificial environment, although the exact step in the process that is responsible is not known. Continued surveillance of growth is suggested for such children.
Single gene defects and identification of genetic pathways
Diseases caused by mutations in the fibroblast growth factor receptor genes
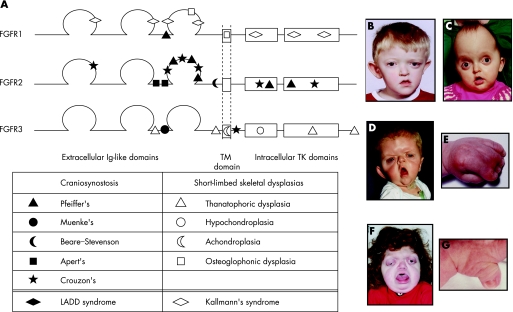
The fibroblast growth factor receptor (FGFR) gene family comprises four transmembrane tyrosine kinase receptors that bind 18 different FGF ligands. The most common form of short‐limbed dwarfism, achondroplasia, was shown in 1994 to be caused by highly specific mutations in FGFR3. Since that time, FGFR mutations have been demonstrated in more than a dozen distinct human birth defects.18 Figure 3A illustrates the molecular structure of the three FGFRs identified with mutations in human diseases (no germline mutations have been found in FGFR4). Interestingly, most of the mutations are missense (a single base pair substitution results in an amino acid exchange in the protein product).
Figure 3 Fibroblast growth factor receptor (FGFR) structure and disease. (A) Structure of FGFR1, FGFR2 and FGFR3 proteins. Positions of some of the common mutations in different diseases are shown. Ig, immunoglobulin; TM, transmembrane; TK, tyrosine kinase. (B–G) Dysmorphic facial features, in some cases associated with limb malformations, in patients with Muenke's syndrome and right unicoronal craniosynostosis (B), Crouzon's syndrome (C), Apert's syndrome (face (D) and hand (E)) and Pfeiffer's syndrome (face (F) and thumb (G)). Parental/guardian informed consent was obtained for the publication of these figures.
Generally speaking, mutations in FGFR2 cause craniosynostosis syndromes (Crouzon's (fig 3C), Apert's (fig 3D,E), Pfeiffer's (fig 3F,G) and Beare–Stevenson) and mutations in FGFR3 cause short‐limb skeletal dysplasias (achondroplasia, hypochondroplasia and thanatophoric dysplasia). However, there are some surprising exceptions. A specific mutation in FGFR3 defines Muenke's syndrome (a recently recognised disorder that is the commonest genetic cause of craniosynostosis (fig 3B)), and certain FGFR2 mutations are found in lacrimo‐auriculo‐dento‐digital (LADD) syndrome.4 The situation is also complicated for FGFR1, in which different mutations account for Kallmann's syndrome 2 (anosmia and hypogonadism), a mild form of Pfeiffer's syndrome, and osteoglophonic dysplasia (rhizomelic short stature with craniosynostosis). This extraordinary diversity of phenotypes resulting from different dominantly acting mutations in the same FGFR gene is explained by the various ways in which the mutation alters the function of the encoded protein.
Genes encoding members of the RAS/MAP kinase pathway
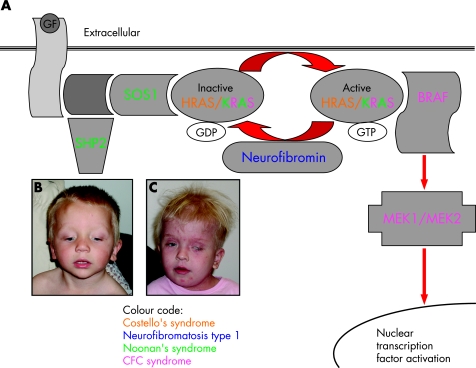
It has long been recognised that Noonan's, LEOPARD (lentigenes, ECG conduction changes, ocular hypertelorism, pulmonary stenosis, abnormal genitalia, retardation of growth and deafness), neurofibromatosis type 1, Costello's, and cardio‐facio‐cutaneous syndromes sometimes overlap phenotypically, being characterised by dysmorphic facial features, skin changes, cardiac defects and developmental delay. However, only within the past year has a genetic basis for this overlap been established. Figure 4A illustrates the RAS/MAP (mitogen‐activated protein) kinase pathway which regulates cell proliferation, differentiation and apoptosis.
Figure 4 The RAS/MAP (mitogen‐activated protein) kinase signalling pathway. (A) Binding of growth factors (GF) to the receptor stimulates binding of adaptor proteins including SHP2 and SOS1. This in turn activates RAS (HRAS and KRAS are active in different tissues) by displacing bound GDP with GTP. GTP‐bound RAS activates BRAF which activates MEK1/MEK2 via a cascade of phosphorylation, ultimately leading to activation of multiple transcription factors in the nucleus. Activity of RAS is limited by the hydrolysis of GTP back to GDP which is regulated by neurofibromin. (B,C) Characteristic facial features in children with Noonan's syndrome (B) and cardio‐facio‐cutaneous (CFC) syndrome (C). Parental/guardian informed consent was obtained for the publication of these figures.
Noonan's (fig 4B) and LEOPARD syndromes are caused by mutations in PTPN11 (encoding SHP2), and a few individuals with Noonan's have mutations in KRAS or SOS1.19 Cardio‐facio‐cutaneous syndrome, characterised by multiple congenital anomalies and a distinctive face (fig 4C), is caused by mutations in one of four genes in this pathway (MAP2K1 and MAP2K2, encoding MEK1 and MEK2, respectively, and KRAS and BRAF).20HRAS mutations have been found in patients with Costello's syndrome, associated with failure to thrive, the development of facial warts and an increased risk of malignancy. All these mutations (except for the case of LEOPARD syndrome) activate the RAS pathway. Neurofibromin is a negative regulator of the RAS pathway and loss of function mutations cause neurofibromatosis type 1, also leading to activation of the pathway. Thus the phenotypic overlap of these conditions, which often causes diagnostic dilemmas, can be explained at the molecular level.19
Mosaicism
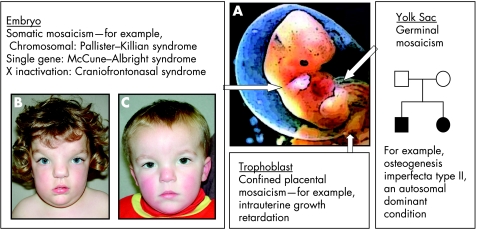
Many phenotypes can be attributed to mosaicism and are determined by the timing and tissue origin of the initial mutation (fig 5A). Chromosome aneuploidies in cells destined to become the trophoblast can result in confined placental mosaicism, which causes intrauterine growth retardation. This has to be considered when mosaicism is detected on prenatal chorionic villous sampling.
Figure 5 Manifestations of mosaicism in embryonic development. (A) The diagram illustrates three types of mosaicism resulting from mutations occurring at different times and sites in development; note that the germ cells migrate into the gonads from the yolk sac. (B,C) Photographs of a sibling pair illustrating the paradoxical effect of X inactivation on EFNB1. Note the more severe phenotype in the sister (B) with a heterozygous EFNB1 mutation and classic craniofrontonasal syndrome compared with the brother (C) who has isolated hypertelorism and a hemizygous EFNB1 mutation. Parental/guardian informed consent was obtained for the publication of these figures.
Germinal mosaicism
Germinal mosaicism describes the situation where a mutation is present in several eggs or sperm. In some instances this is combined with somatic mosaicism, but when this is not the case the mutation cannot be detected on routine blood testing. Germinal mosaicism can account for an increased recurrence risk for certain disorders such as osteogenesis imperfecta type II (fig 5A). If a mutation is identified in a fetus affected with osteogenesis imperfecta type II but is not identified in DNA extracted from the parents' blood samples, there is still a recurrence risk of approximately 7% owing to the possibility that multiple eggs or sperm nevertheless harbour the mutation.21
In contrast with this situation, in some genetic disorders caused by new mutations, such as Apert's syndrome and achondroplasia, the recurrence risk for future siblings of the first affected case is very low. Here the paternally originating gene copy virtually always carries the mutation and there is an increased birth incidence with paternal age. This does not seem to be related to imprinting; rather, a paradoxical mechanism occurs whereby the mutation, although harmful to the baby, becomes selectively enriched in the father's testes with increasing age.22 In other cellular contexts, similar alterations in growth control cause cancer, a theme to which we return later.
Somatic mosaicism
Mosaic chromosomal aneuploidies are frequently associated with birth defects. Mosaicism often results in a milder phenotype than the non‐mosaic form, allowing survival for some disorders that would otherwise result in lethality (fig 5). One example is Pallister–Killian syndrome, an MCA/MR syndrome resulting from mosaic tetrasomy 12p, often only detected by FISH analysis of buccal swabs or skin fibroblast culture.4 The same principle is illustrated by McCune–Albright syndrome (polyostotic fibrous dysplasia with “Coast of Maine” café‐au‐lait patches and endocrinopathies), which is caused by mosaic gain‐of‐function mutations in GNAS (encoding the α subunit of the stimulatory G protein), which would be lethal in the non‐mosaic state.4
X chromosome inactivation
All females are functionally mosaic because of X inactivation. Usually this permits a normal phenotype in carrier females of X linked recessive disorders (cells in which the active X chromosome bears the mutation either die during early embryonic development or are compensated by cells that express the normal gene). However an exception is craniofrontonasal syndrome (an X linked craniosynostosis syndrome resulting from ephrin‐B1 mutations), in which females have the more severe phenotype with coronal craniosynostosis and frontonasal dysplasia, and males exhibit isolated hypertelorism (fig 5B,C). EFNB1, the gene that encodes ephrin‐B1, is expressed along one side of the developing coronal suture and it is postulated that patchy loss of gene expression (due to random X inactivation in females) disturbs the accurate formation of this suture at the tissue boundary.23 In hemizygous males (who lack EFNB1 function completely) the suture boundary is maintained, perhaps by the compensatory activity of other members of the ephrin gene family.
Birth defects and cancer: overlapping pathways
Many genes associated with birth defects are also somatically mutated in certain cancers. These include genes discussed earlier in this review such as FGFR3, HRAS, KRAS, PTPN11 and BRAF. Perhaps this should not come as a surprise, because both birth defects and cancers represent disorders of cellular growth and homoeostasis. Two other genes, the tumour suppressor gene PTEN and the oncogene RET, illustrate these principles.
The many syndromes of PTEN
Somatic mutations in phosphatase and tensin homolog (PTEN) have been found in many cancers including endometrial and prostate carcinoma.4 Germline mutations in PTEN cause a spectrum of rare cancer predisposition syndromes (collectively termed PTEN hamartoma tumour syndromes) that may include congenital defects. These include Cowden syndrome (characterised by multiple hamartomas and an increased risk for breast, thyroid and endometrial cancers), Bannayan–Riley–Ruvalcaba syndrome (which features macrocephaly, learning difficulties and pigmented penile macules), autistic spectrum with extreme macrocephaly disorder and Proteus syndrome.24
PTEN indirectly regulates the important TSC‐mTOR (tuberous sclerosis complex–mammalian target of rapamycin) pathway of cell growth. If PTEN concentrations are reduced, cells are prevented from undergoing cell cycle arrest and apoptosis, leading to increased proliferation. The TSC‐mTOR signalling pathway includes genes involved in other hamartomatous diseases such as tuberous sclerosis (TSC1 and TSC2) and Peutz–Jeghers syndrome (STK11). Drugs such as mTOR inhibitors (rapamycin) are currently being evaluated in the treatment of some of these hamartomatous diseases.25
Too much or too little RET activity
Rearranged during transfection (RET) is an excellent example of a gene in which mutations cause apparently non‐overlapping phenotypes. RET, like the FGFRs, is a transmembrane receptor with an intracellular tyrosine kinase domain. Gain‐of‐function mutations in RET are responsible for the dominant human cancer syndromes multiple endocrine neoplasia (MEN) 2a and MEN2b.4 Loss‐of‐function mutations in RET cause Hirschsprung's disease.4 Curiously, patients with MEN2a have been described with Hirschsprung's disease, which requires inactivation of RET in colonic ganglion and activation in tumour tissues. This may reflect the different ways by which the mutant protein is processed in different tissues.26
Conclusions
The cytogenetic and molecular genetic analysis of birth defects is now routine and can frequently pinpoint the causative genetic lesion. However, close liaison between paediatricians and their clinical genetics colleagues remains critical, both in requesting the most appropriate and focused investigations and in interpreting the results for the benefit of their patients. Although expert systems such as the London Dysmorphology Database can assist in diagnosis, they cannot (yet) replace the clinical skill of recognising a particular facial “gestalt”.
Despite all these advances, many fundamental questions remain. To return to where this article started, why does trisomy 21 cause the particular constellation of features that we recognise as Down's syndrome? We still lack a detailed understanding, but a recent paper has suggested that the combined dysregulation of two genes, which both map to chromosome 21 and encode regulators of the nuclear factor of activated T cells (NFAT) transcription complex pathway, contribute to the phenotype.27 The work needs to be confirmed but promises to give us new insight into this common but enigmatic birth defect.
Acknowledgements
We thank all the families for their generous agreement to publication of photographs and Drs Jane Hurst, Duncan Rourke and Helen Stewart for comments on the manuscript.
Abbreviations
CGH - comparative genomic hybridisation
FISH - fluorescence in‐situ hybridisation
FGFR - fibroblast growth factor receptor
MCA/MR - multiple congenital abnormality/mental retardation
UPD - uniparental disomy
Footnotes
Funding: AW's research is supported by the Wellcome Trust.
Competing interests: None.
Parental/guardian informed consent was obtained for the publication of figures 1, 3–5.
References
- 1.Hickman‐Davis J M, Davis I C. Transgenic mice. Paediatr Respir Rev 2006749–53. [DOI] [PubMed] [Google Scholar]
- 2.Downward J. RNA interference. BMJ 20043281245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badano J L, Mitsuma N, Beales P L.et al The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 200622125–148. [DOI] [PubMed] [Google Scholar]
- 4.McKusick V and Johns Hopkins University Online Mendelian Inheritance in Man (OMIM), 2007. Available at: http://www.ncbi.nlm.nih.gov/entrez
- 5.Epstein C J, Erickson R P, Wynshaw‐Boris A. eds. Inborn errors of development. New York, NY: Oxford University Press, 2004
- 6.Ryan A K, Goodship J A, Wilson D I.et al Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 199734798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev 200515279–284. [DOI] [PubMed] [Google Scholar]
- 8.Ensenauer R E, Adeyinka A, Flynn H C.et al Microduplication 22q11. 2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients, Am J Hum Genet 2003731027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tassabehji M. Williams–Beuren syndrome: a challenge for genotype‐phenotype correlations. Hum Mol Genet 200312(Spec No 2)R229–R237. [DOI] [PubMed] [Google Scholar]
- 10.Tassabehji M, Hammond P, Karmiloff‐Smith A.et al GTF2IRD1 in craniofacial development of humans and mice. Science 20053101184–1187. [DOI] [PubMed] [Google Scholar]
- 11.Somerville M J, Mervis C B, Young E J.et al Severe expressive‐language delay related to duplication of the Williams–Beuren locus. N Engl J Med 20053531694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vissers L E, Veltman J A, van Kessel A G.et al Identification of disease genes by whole genome CGH arrays. Hum Mol Genet 200514(Spec No. 2)R215–R223. [DOI] [PubMed] [Google Scholar]
- 13.Vissers L E, van Ravenswaaij C M, Admiraal R.et al Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 200436955–957. [DOI] [PubMed] [Google Scholar]
- 14.Horsthemke B, Buiting K. Imprinting defects on human chromosome 15. Cytogenet Genome Res 2006113292–299. [DOI] [PubMed] [Google Scholar]
- 15.Eggermann T, Wollmann H A, Kuner R.et al Molecular studies in 37 Silver‐Russell syndrome patients: frequency and etiology of uniparental disomy. Hum Genet 1997100415–419. [DOI] [PubMed] [Google Scholar]
- 16.Gicquel C, Rossignol S, Cabrol S.et al Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver‐Russell syndrome. Nat Genet 2005371003–1007. [DOI] [PubMed] [Google Scholar]
- 17.Maher E R, Brueton L A, Bowdin S C.et al Beckwith–Wiedemann syndrome and assisted reproduction technology (ART). J Med Genet 20034062–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkie A O M. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev 200516187–203. [DOI] [PubMed] [Google Scholar]
- 19.Gelb B D, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS‐mitogen activated protein kinase transduction. Hum Mol Genet 200615(Spec No 2)R220–R226. [DOI] [PubMed] [Google Scholar]
- 20.Roberts A, Allanson J, Jadico S K.et al The cardio‐facio‐cutaneous (CFC) syndrome: a review. J Med Genet 200643833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole W G, Dalgleish R. Perinatal lethal osteogenesis imperfecta. J Med Genet 199532284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goriely A, McVean G A, Rojmyr M.et al Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science 2003301643–646. [DOI] [PubMed] [Google Scholar]
- 23.Twigg S R F, Kan R, Babbs C.et al Mutations of ephrin‐B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci U S A 20041018652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eng C. PTEN: one gene, many syndromes. Hum Mutat 200322183–198. [DOI] [PubMed] [Google Scholar]
- 25.Inoki K, Corradetti M N, Guan K L. Dysregulation of the TSC‐mTOR pathway in human disease. Nat Genet 20053719–24. [DOI] [PubMed] [Google Scholar]
- 26.Arighi E, Borrello M G, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev 200516441–467. [DOI] [PubMed] [Google Scholar]
- 27.Arron J R, Winslow M M, Polleri A.et al NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 2006441595–600. [DOI] [PubMed] [Google Scholar]